Transcriptome Response of Metallicolous and a Non-Metallicolous Ecotypes of Noccaea goesingensis to Nickel Excess
Abstract
1. Introduction
2. Results
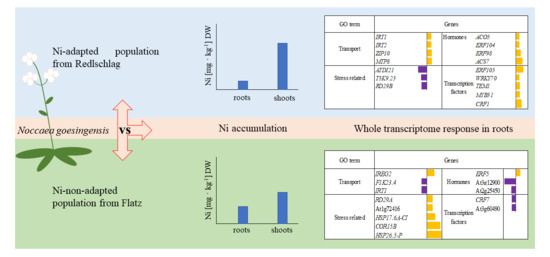
2.1. Plant Growth Response to Nickel Supplementation and Ni Accumulation
2.2. RNA-seq Analysis of N. goesingensis Root Transcriptomes
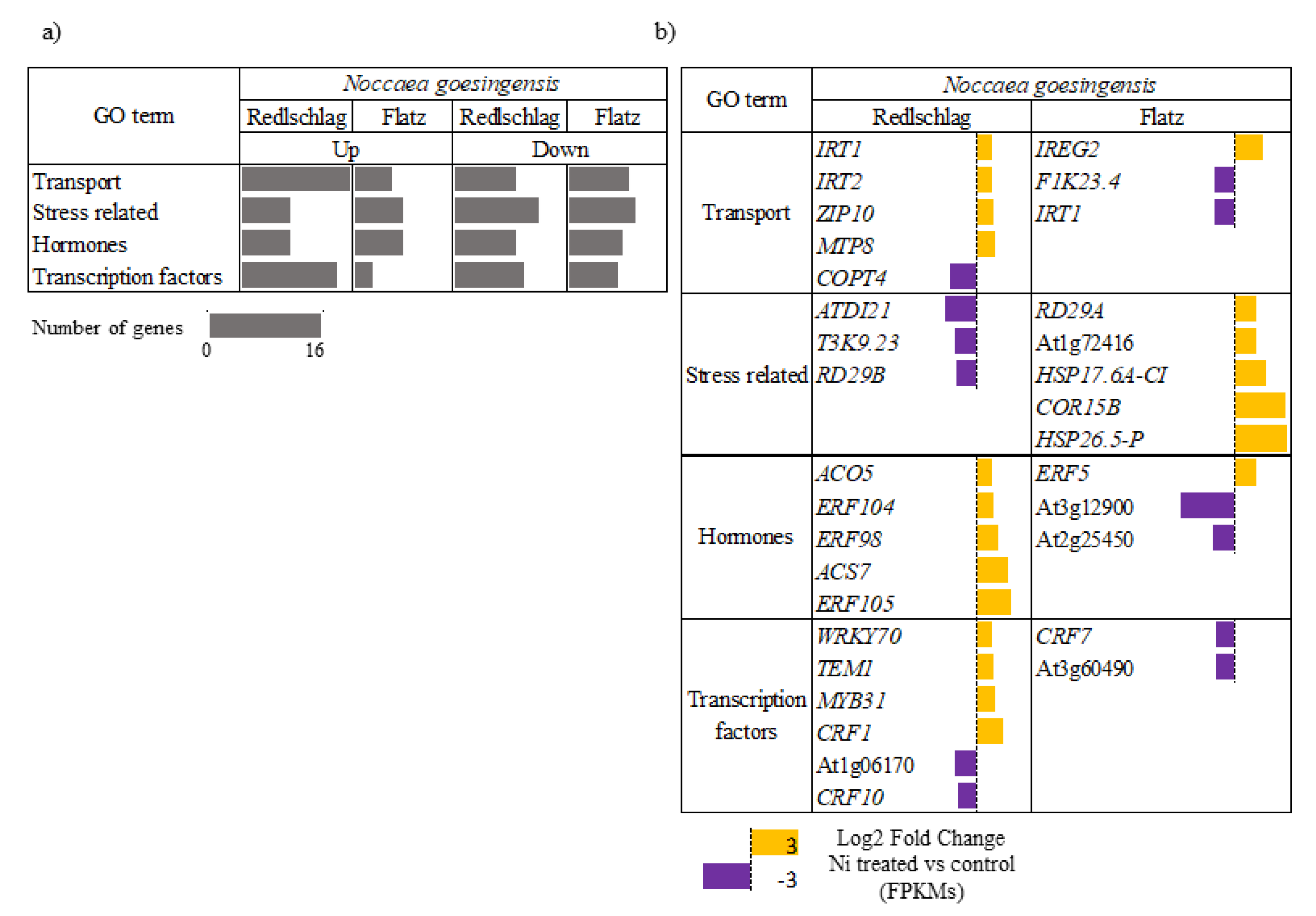
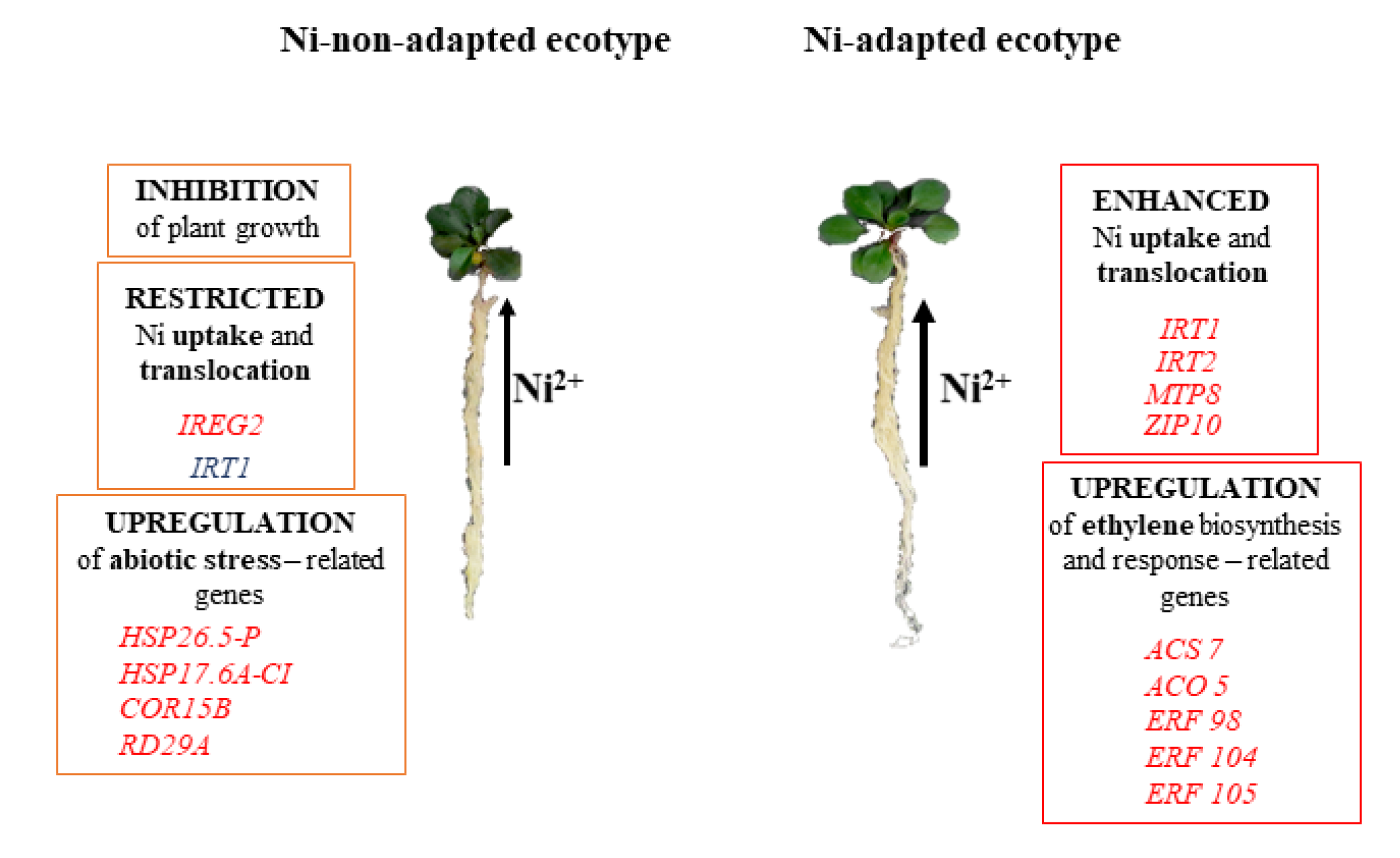
2.3. Up-Regulated Genes
2.4. Down-Regulated Genes
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation
4.2. Plant Nickel Concentration
4.3. RNA Sequencing and Functional Data Mining
4.4. Data Availability
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Baker, A.J.M.; Ernst, W.H.O.; van der Ent, A.; Malaisse, F.; Ginocchio, R. Metallophytes: The unique biological resource, its ecology and conservational status in Europe, central Africa and Latin America. Ecol. Ind. Pollut. 2012, 7–40. [Google Scholar] [CrossRef]
- Pauwels, M.; Vekemans, X.; Godé, C.; Frérot, H.; Castric, V.; Saumitou-Laprade, P. Nuclear and chloroplast DNA phylogeography reveals vicariance among European populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae). New Phytol. 2012, 193, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Ernst, W.H.O. Evolution of metal tolerance in higher plants. For. Snow Landsc. Res. 2006, 80, 251–274. [Google Scholar]
- Wójcik, M.; Gonnelli, C.; Selvi, F.; Dresler, S.; Rostański, A.; Vangronsveld, J. Metallophytes of Serpentine and Calamine Soils—Their Unique Ecophysiology and Potential for Phytoremediation. Adv. Bot. Res. 2017, 83, 1–42. [Google Scholar] [CrossRef]
- Lambinon, J.; Auquier, P. La flore et la végétation des terrains calaminaires de la Wallonie septentrionale et de la Rhénanie aixoise. Types chorologiques et groupes écologiques. Nat. Mosana 1964, 16, 13–131. [Google Scholar]
- Willems, G.; Dräger, D.B.; Courbot, M.; Godé, C.; Verbruggen, N.; Saumitou-Laprade, P. The Genetic Basis of Zinc Tolerance in the Metallophyte Arabidopsis halleri ssp. halleri (Brassicaceae): An Analysis of Quantitative Trait Loci. Genetics 2007, 176, 659–674. [Google Scholar] [CrossRef]
- Bothe, H. Plants in Heavy Metal Soils. Soil biology. In Detoxification of Heavy Metals; Sherameti, I., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 35–57. [Google Scholar]
- Bothe, H.; Słomka, A. Divergent biology of facultative heavy metal plants. J. Plant Physiol. 2017, 219, 45–61. [Google Scholar] [CrossRef]
- Krämer, U. Metal Hyperaccumulation in Plants. Annu. Rev. Plant Biol. 2010, 61, 517–534. [Google Scholar] [CrossRef]
- Reeves, R.D.; Baker, A.J.M.; Jaffré, T.; Erskine, P.D.; Echevarria, G.; van der Ent, A. A global database for plants that hyperaccumulate metal and metalloid trace elements. New Phytol. 2018, 218, 407–411. [Google Scholar] [CrossRef]
- Reeves, R.D.; Brooks, R.R. European species of Thlaspi L. (Cruciferae) as indicators of nickel and zinc. J. Geochem. Explor. 1983, 18, 275–283. [Google Scholar] [CrossRef]
- Persans, M.W.; Yan, X.; Patnoe, J.M.M.L.; Krämer, U.; Salt, D.E. Molecular dissection of the role of histidine in nickel hyperaccumulation in Thlaspi goesingense (Halacsy). Plant Physiol. 1999, 121, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Rosenkranz, T.; Hipfinger, C.; Ridard, C.; Puschenreiter, M. A nickel phytomining field trial using Odontarrhena chalcidica and Noccaea goesingensis on an Austrian serpentine soil. J. Environ. Manag. 2019, 242, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Krämer, U.; Smith, R.D.; Wenzel, W.W.; Raskin, I.; Salt, D.E. The role of metal transport and tolerance in nickel hyperaccumulation by Thlaspi goesingense Halacsy. Plant Physiol. 1997, 115, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.D.; Baker, A.J.M. Studies on metal uptake by plants from serpentine and non-serpentine populations of Thlaspi goesingense halácsy (Crycuferae). New Phytol. 1984, 98, 191–204. [Google Scholar] [CrossRef]
- Kidd, P.S.; Bani, A.; Benizri, E.; Gonnelli, C.; Hazotte, C.; Kisser, J.; Konstantinou, M.; Kuppens, T.; Kyrkas, D.; Laubie, B.; et al. Developing Sustainable Agromining Systems in Agricultural Ultramafic Soils for Nickel Recovery. Front. Environ. Sci. 2018, 6. [Google Scholar] [CrossRef]
- Krämer, U.; Pickering, I.J.; Prince, R.C.; Raskin, I.; Salt, D.E. Subcellular Localization and Speciation of Nickel in Hyperaccumulator and Non-Accumulator Thlaspi Species. Plant Physiol. 2000, 122, 1343–1354. [Google Scholar] [CrossRef]
- Küpper, H.; Lombi, E.; Zhao, F.; Wieshammer, G.; McGrath, S.P. Cellular compartmentation of nickel in the hyperaccumulators Alyssum lesbiacum, Alyssum bertolonii and Thlaspi goesingense. J. Exp. Bot. 2001, 52, 2291–2300. [Google Scholar] [CrossRef]
- Persans, M.W.; Nieman, K.; Salt, D.E. Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc. Natl. Acad. Sci. USA 2001, 98, 9995–10000. [Google Scholar] [CrossRef]
- Freeman, J.L.; Persans, M.W.; Nieman, K.; Albrecht, C.; Peer, W.; Pickering, I.J.; Salt, D.E. Increased Glutathione Biosynthesis Plays a Role in Nickel Tolerance in Thlaspi Nickel Hyperaccumulators. Plant Cell 2004, 16, 2176–2191. [Google Scholar] [CrossRef]
- Freeman, J.L.; Salt, D.E. The metal tolerance profile of Thlaspi goesingense is mimicked in Arabidopsis thaliana heterologously expressing serine acetyl-transferase. BMC Plant Biol. 2007, 7, 63. [Google Scholar] [CrossRef]
- Himmelbauer, M.L.; Puschenreiter, M.; Schnepf, A.; Loiskandl, W.; Wenzel, W.W. Root morphology of Thlaspi goesingense Hálácsy grown on a serpentine soil. J. Plant Nutr. Soil Sci. 2005, 168, 138–144. [Google Scholar] [CrossRef]
- Plessl, M.; Rigola, D.; Hassinen, V.H.; Tervahauta, A.; Kärenlampi, S.; Schat, H.; Aarts, M.G.M.; Ernst, D. Comparison of two ecotypes of the metal hyperaccumulator Thlaspi caerulescens (J. & C. PRESL) at the transcriptional level. Protoplasma 2010, 239, 81–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Halimaa, P.; Lin, Y.F.; Ahonen, V.H.; Blande, D.; Clemens, S.; Gyenesei, A.; Häikiö, E.; Kärenlampi, S.O.; Laiho, A.; Aarts, M.G.M.; et al. Gene expression differences between noccaea caerulescens ecotypes help to identify candidate genes for metal phytoremediation. Environ. Sci. Technol. 2014, 48, 3344–3353. [Google Scholar] [CrossRef] [PubMed]
- Schvartzman, M.S.; Corso, M.; Fataftah, N.; Scheepers, M.; Nouet, C.; Bosman, B.; Carnol, M.; Motte, P.; Verbruggen, N.; Hanikenne, M. Adaptation to high zinc depends on distinct mechanisms in metallicolous populations of Arabidopsis halleri. New Phytol. 2018, 218, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Chmielowska-Bąk, J.; Lefèvre, I.; Lutts, S.; Deckert, J. Short term signaling responses in roots of young soybean seedlings exposed to cadmium stress. J. Plant Physiol. 2013, 170, 1585–1594. [Google Scholar] [CrossRef]
- Maksymiec, W. Effects of jasmonate and some other signalling factors on bean and onion growth during the initial phase of cadmium action. Biol. Plant. 2011, 55, 112–118. [Google Scholar] [CrossRef]
- Masood, A.; Iqbal, N.; Khan, N.A. Role of ethylene in alleviation of cadmium-induced photosynthetic capacity inhibition by sulphur in mustard. Plant. Cell Environ. 2012, 35, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Khan, N.A. Ethylene reverses photosynthetic inhibition by nickel and zinc in mustard through changes in PS II activity, photosynthetic nitrogen use efficiency, and antioxidant metabolism. Protoplasma 2014, 251, 1007–1019. [Google Scholar] [CrossRef]
- Khan, N.A.; Khan, M.I.R.; Ferrante, A.; Poor, P. Editorial: Ethylene: A Key Regulatory Molecule in Plants. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Cao, S.; Chen, Z.; Liu, G.; Jiang, L.; Yuan, H.; Ren, G.; Bian, X.; Jian, H.; Ma, X. The Arabidopsis Ethylene-Insensitive 2 gene is required for lead resistance. Plant Physiol. Biochem. 2009, 47, 308–312. [Google Scholar] [CrossRef]
- Monteiro, C.C.; Carvalho, R.F.; Gratão, P.L.; Carvalho, G.; Tezotto, T.; Medici, L.O.; Peres, L.E.P.; Azevedo, R.A. Biochemical responses of the ethylene-insensitive Never ripe tomato mutant subjected to cadmium and sodium stresses. Environ. Exp. Bot. 2011, 71, 306–320. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54 and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Response. Plant Cell 2017. [Google Scholar] [CrossRef] [PubMed]
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 2011, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Halimaa, P.; Blande, D.; Baltzi, E.; Aarts, M.G.M.; Granlund, L.; Keinänen, M.; Kärenlampi, S.O.; Kozhevnikova, A.D.; Peräniemi, S.; Schat, H.; et al. Transcriptional effects of cadmium on iron homeostasis differ in calamine accessions of Noccaea caerulescens. Plant J. 2019, 97, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wu, K.; Tang, Z.; Guo, Q.; Guo, X.; Wang, H. Exogenous ethylene enhanced the cadmium resistance and changed the alkaloid biosynthesis in Catharanthus roseus seedlings. Acta Physiol. Plant. 2017, 39, 267. [Google Scholar] [CrossRef]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef]
- Rogers, E.E.; Eide, D.J.; Guerinot, M.L. Altered selectivity in an Arabidopsis metal transporter. Proc. Natl. Acad. Sci. USA 2000, 97, 12356–12360. [Google Scholar] [CrossRef]
- Nishida, S.; Tsuzuki, C.; Kato, A.; Aisu, A.; Yoshida, J.; Mizuno, T. AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1433–1442. [Google Scholar] [CrossRef]
- Korshunova, Y.O.; Eide, D.; Gregg Clark, W. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 1999, 40, 37–44. [Google Scholar] [CrossRef]
- Vert, G.; Briat, J.-F.; Curie, C. Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J. 2001, 26, 181–189. [Google Scholar] [CrossRef]
- Talke, I.N.; Hanikenne, M.; Krämer, U. Zinc-Dependent Global Transcriptional Control, Transcriptional Deregulation, and Higher Gene Copy Number for Genes in Metal Homeostasis of the Hyperaccumulator Arabidopsis halleri. Plant Physiol. 2006, 142, 148–167. [Google Scholar] [CrossRef] [PubMed]
- Assuncao, A.G.L.; Martins, P.D.C.; De Folter, S.; Vooijs, R.; Schat, H.; Aarts, M.G.M. Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2001, 24, 217–226. [Google Scholar] [CrossRef]
- Ghasemi, R.; Ghaderian, S.M.; Krämer, U. Interference of nickel with copper and iron homeostasis contributes to metal toxicity symptoms in the nickel hyperaccumulator plant Alyssum inflatum. New Phytol. 2009, 184, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Persson, D.P.; Duan, F.; Jørgensen, K.; Yuan, L.; Schjoerring, J.K.; Pedas, P.R. The iron-regulated transporter 1 plays an essential role in uptake, translocation and grain-loading of manganese, but not iron, in barley. New Phytol. 2018, 217, 1640–1653. [Google Scholar] [CrossRef] [PubMed]
- Dubeaux, G.; Neveu, J.; Zelazny, E.; Vert, G. Metal Sensing by the IRT1 Transporter-Receptor Orchestrates Its Own Degradation and Plant Metal Nutrition. Mol. Cell 2018, 69, 953–964.e5. [Google Scholar] [CrossRef]
- Arteca, R.N.; Arteca, J.M. Heavy-metal-induced ethylene production in Arabidopsis thaliana. J. Plant Physiol. 2007, 164, 1480–1488. [Google Scholar] [CrossRef]
- Thao, N.P.; Khan, M.I.R.; Anh Thu, N.B.; Thi Hoang, X.L.; Asgher, M.; Khan, N.A.; Tran, L.S.P. Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress. Plant Physiol. 2015, 169, 73–84. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Landa, P.; Vankova, R.; Andrlova, J.; Hodek, J.; Marsik, P.; Storchova, H.; White, J.C.; Vanek, T. Nanoparticle-specific changes in Arabidopsis thaliana gene expression after exposure to ZnO, TiO2, and fullerene soot. J. Hazard. Mater. 2012, 241–242, 55–62. [Google Scholar] [CrossRef]
- Rajewska, I.; Talarek, M.; Bajguz, A. Brassinosteroids and Response of Plants to Heavy Metals Action. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Fornalé, S.; Shi, X.; Chai, C.; Encina, A.; Irar, S.; Capellades, M.; Fuguet, E.; Torres, J.-L.; Rovira, P.; Puigdomènech, P.; et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 2010, 64, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Goda, H.; Sawa, S.; Asami, T.; Fujioka, S.; Shimada, Y.; Yoshida, S. Comprehensive Comparison of Auxin-Regulated and Brassinosteroid-Regulated Genes in Arabidopsis. Plant Physiol. 2004, 134, 1555–1573. [Google Scholar] [CrossRef]
- García, S.; Prado, M.; Dégano, R.; Domínguez, A. A Copper-responsive Transcription Factor, CRF1, Mediates Copper and Cadmium Resistance in Yarrowia lipolytica. J. Biol. Chem. 2002, 277, 37359–37368. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Peer, W.A.; Mamoudian, M.; Lahner, B.; Reeves, R.D.; Murphy, A.S.; Salt, D.E. Identifying model metal hyperaccumulating plants: Germplasm analysis of 20 Brassicaceae accessions from a wide geographical area. New Phytol. 2003, 159, 421–430. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domka, A.; Rozpądek, P.; Ważny, R.; Jędrzejczyk, R.J.; Hubalewska-Mazgaj, M.; Gonnelli, C.; Benny, J.; Martinelli, F.; Puschenreiter, M.; Turnau, K. Transcriptome Response of Metallicolous and a Non-Metallicolous Ecotypes of Noccaea goesingensis to Nickel Excess. Plants 2020, 9, 951. https://doi.org/10.3390/plants9080951
Domka A, Rozpądek P, Ważny R, Jędrzejczyk RJ, Hubalewska-Mazgaj M, Gonnelli C, Benny J, Martinelli F, Puschenreiter M, Turnau K. Transcriptome Response of Metallicolous and a Non-Metallicolous Ecotypes of Noccaea goesingensis to Nickel Excess. Plants. 2020; 9(8):951. https://doi.org/10.3390/plants9080951
Chicago/Turabian StyleDomka, Agnieszka, Piotr Rozpądek, Rafał Ważny, Roman Jan Jędrzejczyk, Magdalena Hubalewska-Mazgaj, Cristina Gonnelli, Jubina Benny, Federico Martinelli, Markus Puschenreiter, and Katarzyna Turnau. 2020. "Transcriptome Response of Metallicolous and a Non-Metallicolous Ecotypes of Noccaea goesingensis to Nickel Excess" Plants 9, no. 8: 951. https://doi.org/10.3390/plants9080951
APA StyleDomka, A., Rozpądek, P., Ważny, R., Jędrzejczyk, R. J., Hubalewska-Mazgaj, M., Gonnelli, C., Benny, J., Martinelli, F., Puschenreiter, M., & Turnau, K. (2020). Transcriptome Response of Metallicolous and a Non-Metallicolous Ecotypes of Noccaea goesingensis to Nickel Excess. Plants, 9(8), 951. https://doi.org/10.3390/plants9080951