Complete Chloroplast Genome Characterization of Oxalis Corniculata and Its Comparison with Related Species from Family Oxalidaceae
Abstract
1. Introduction
2. Results
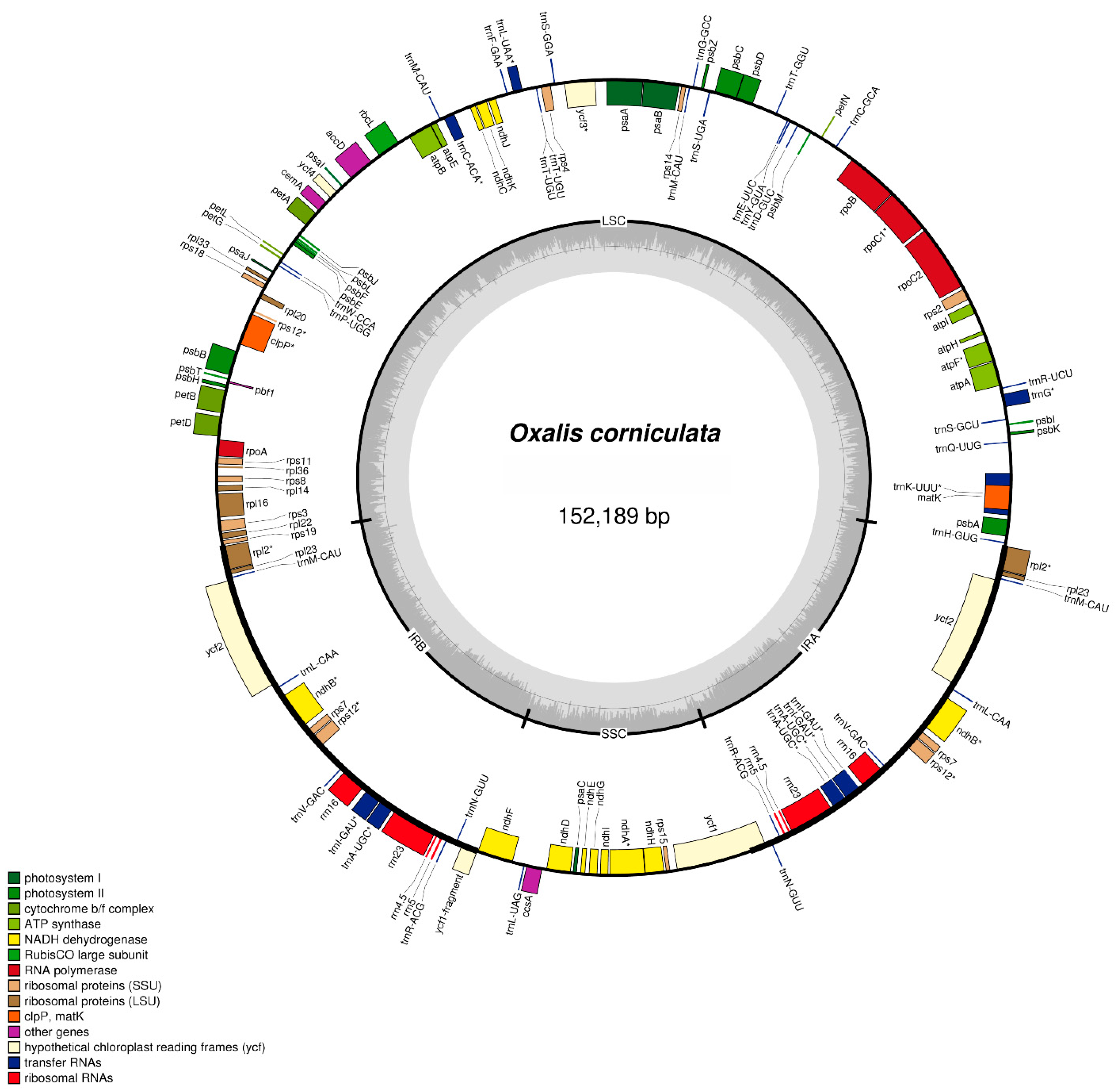
2.1. Chloroplast Genome Structure of O. corniculata
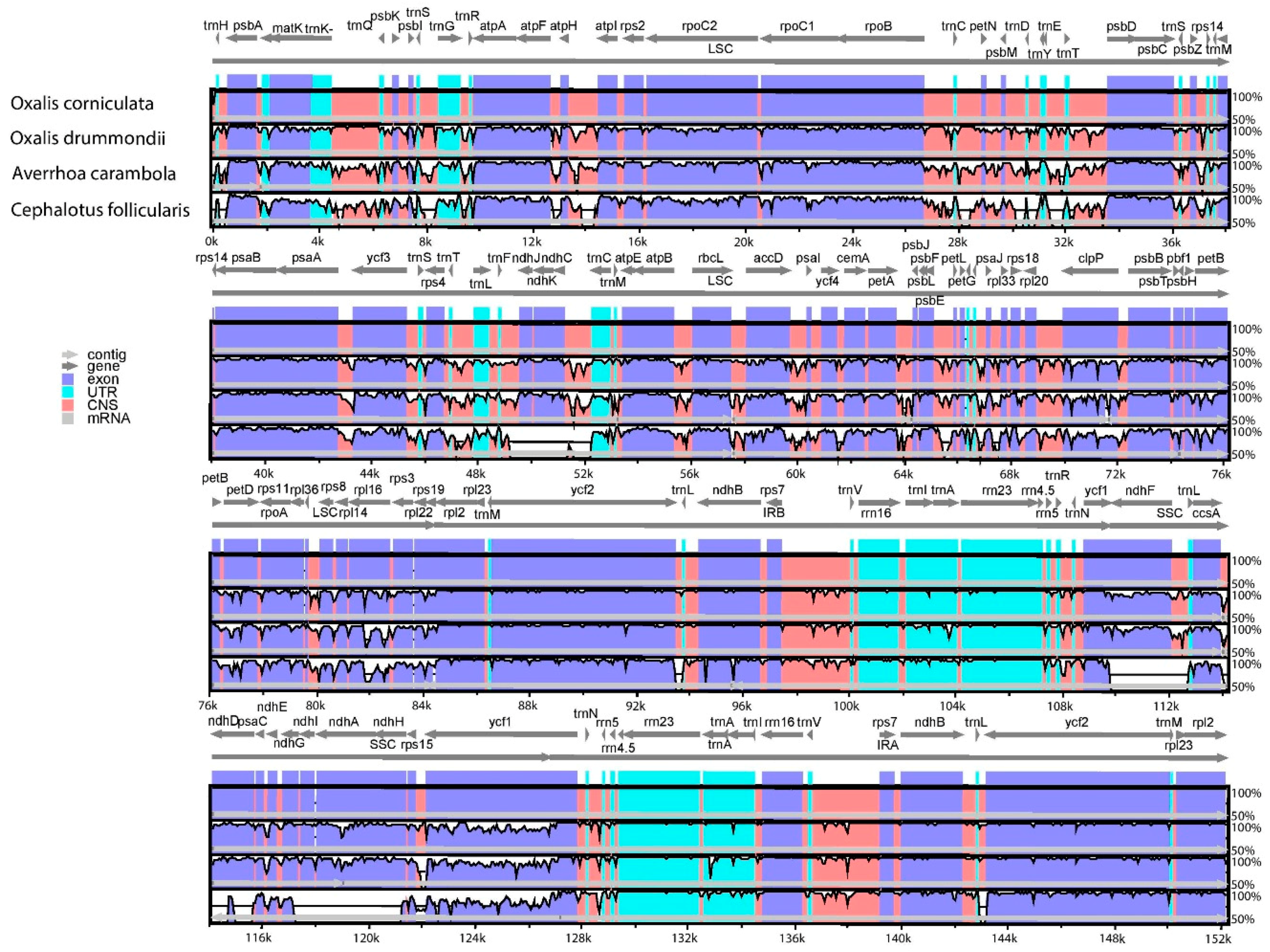
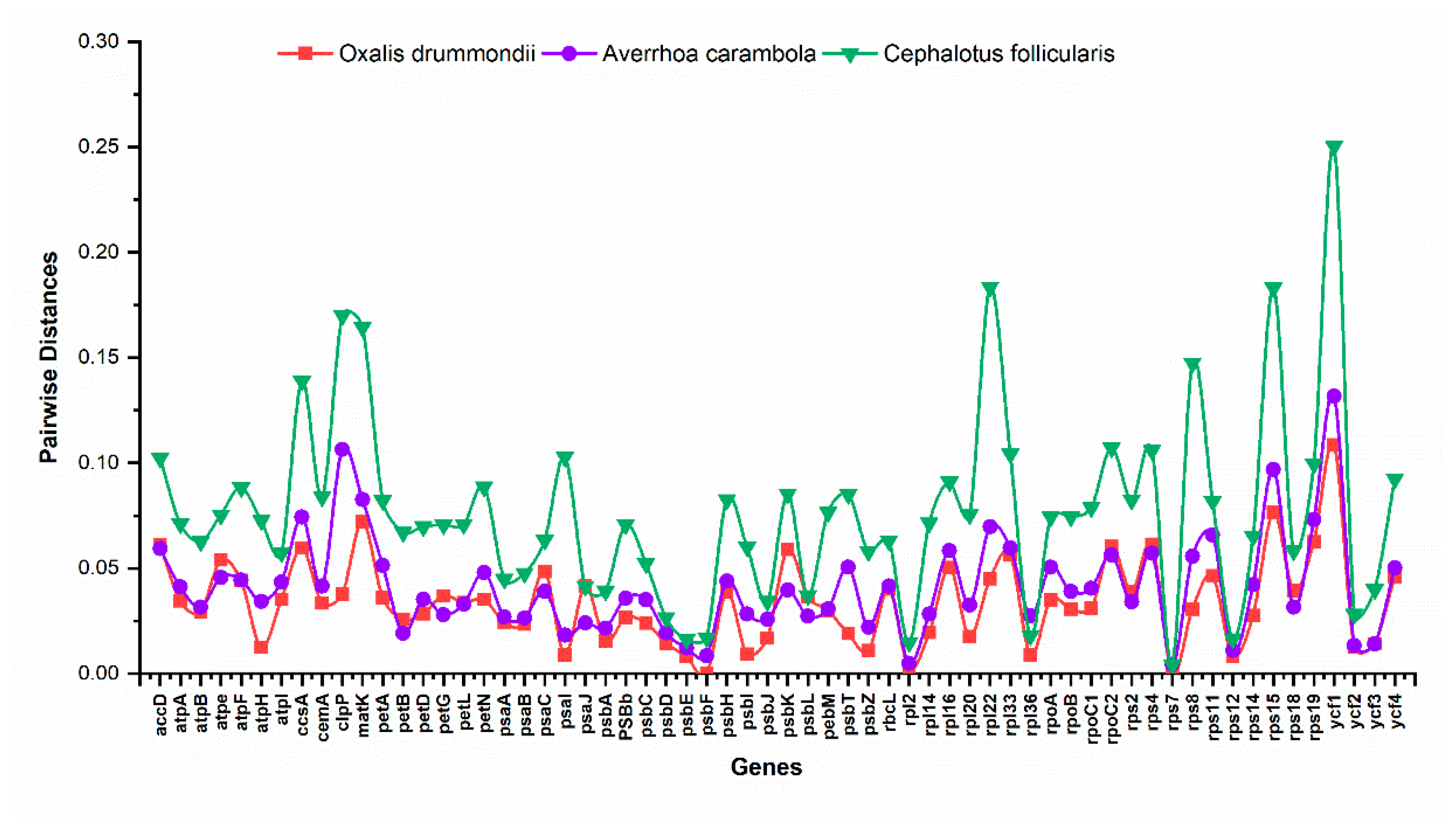
2.2. Comparative Analysis of O. corniculata Chloroplast Genome with Related Species
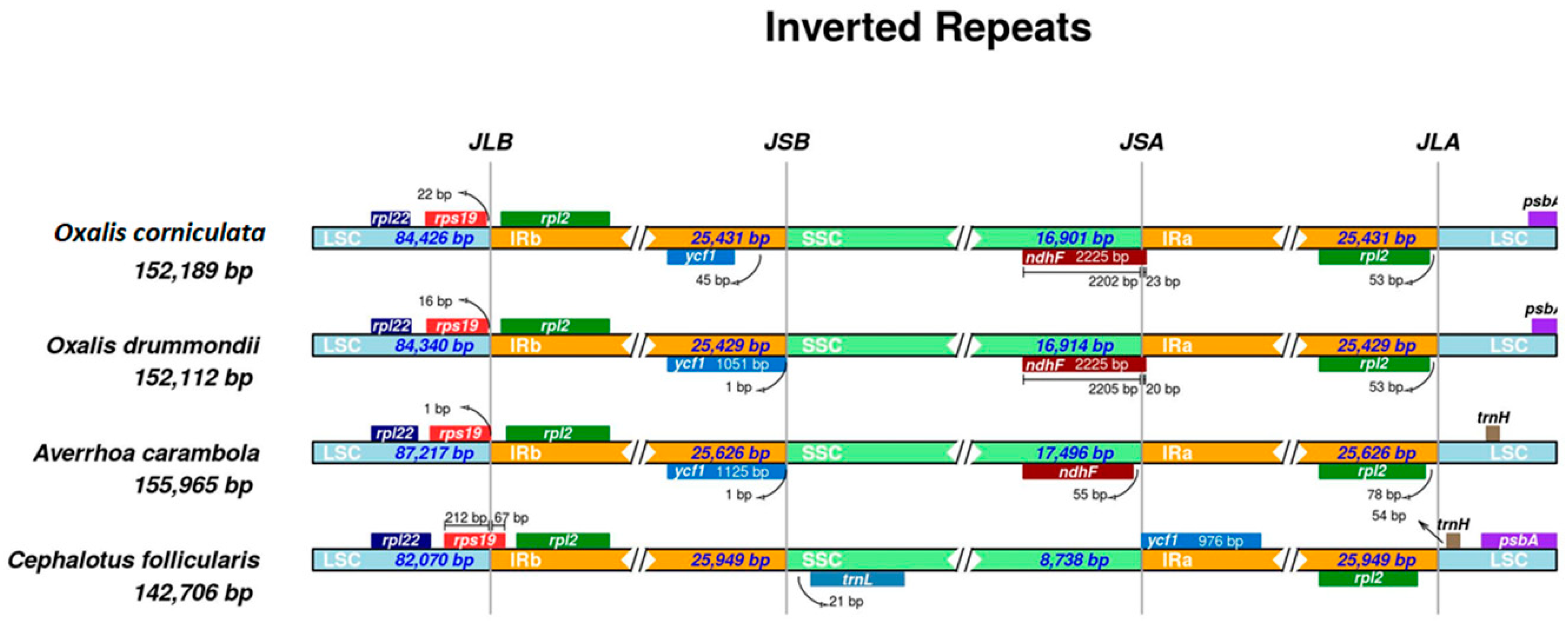
2.3. Expansion and Contraction of IR Regions
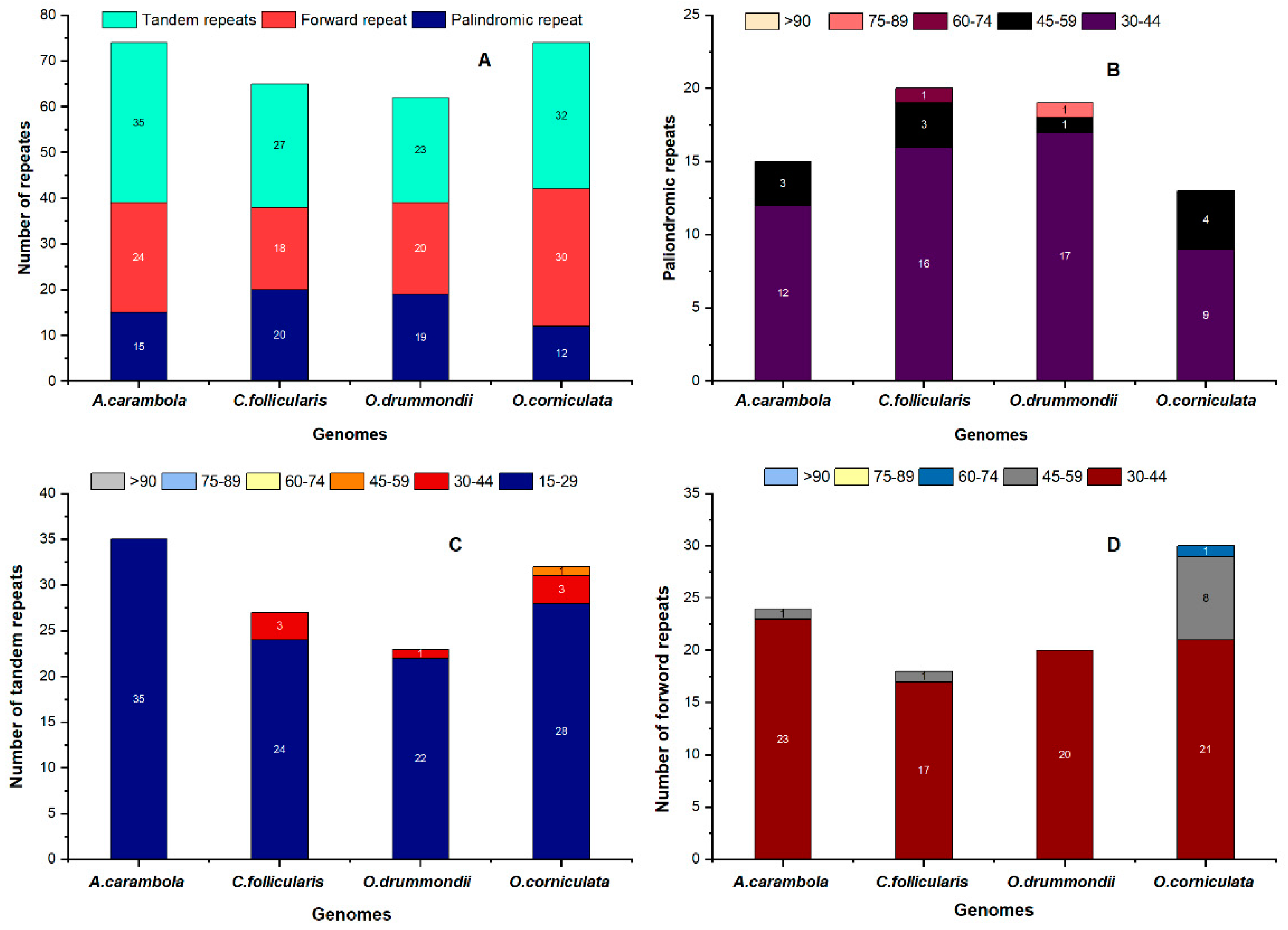
2.4. Repeat Sequence Analysis
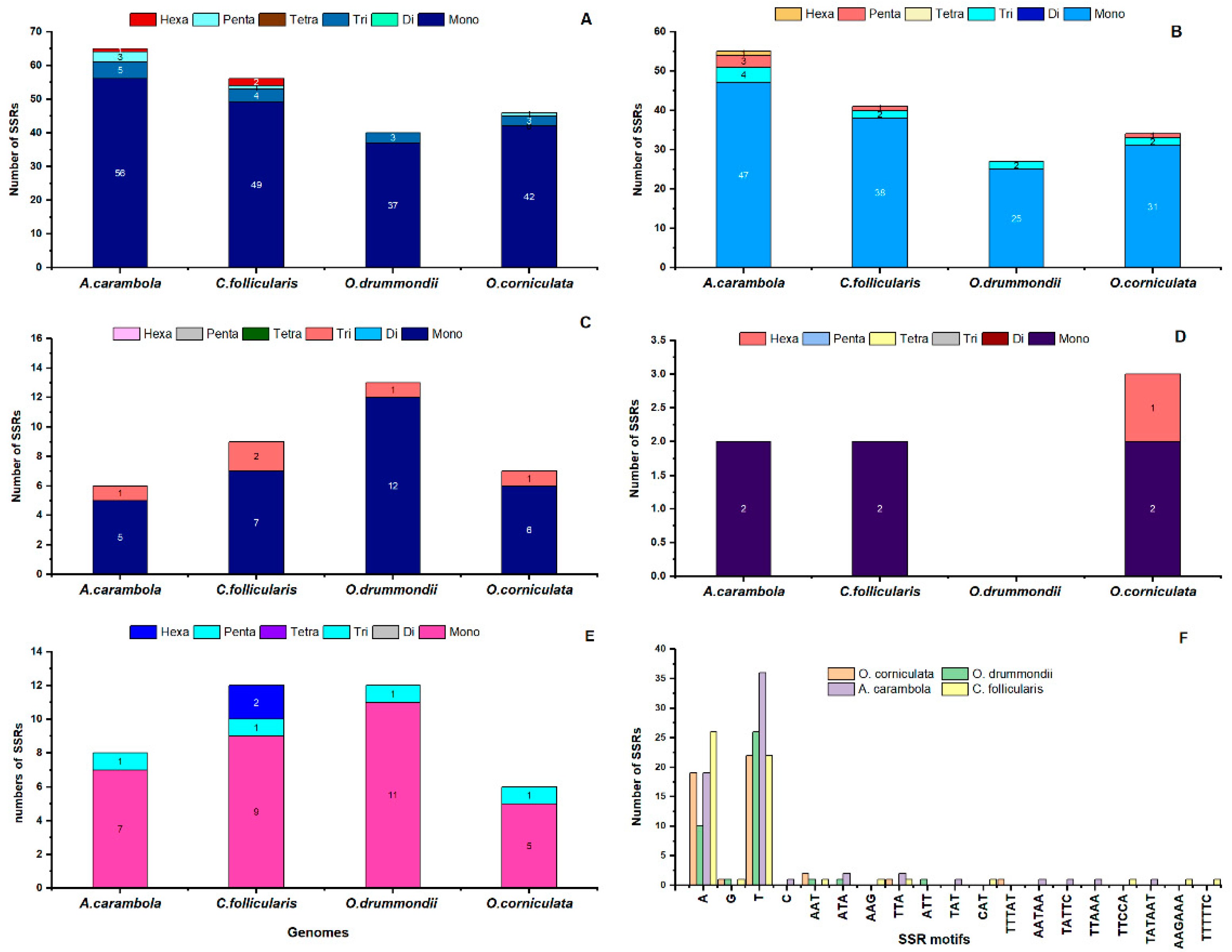
2.5. Simple Sequence Repeat (SSR) Analysis
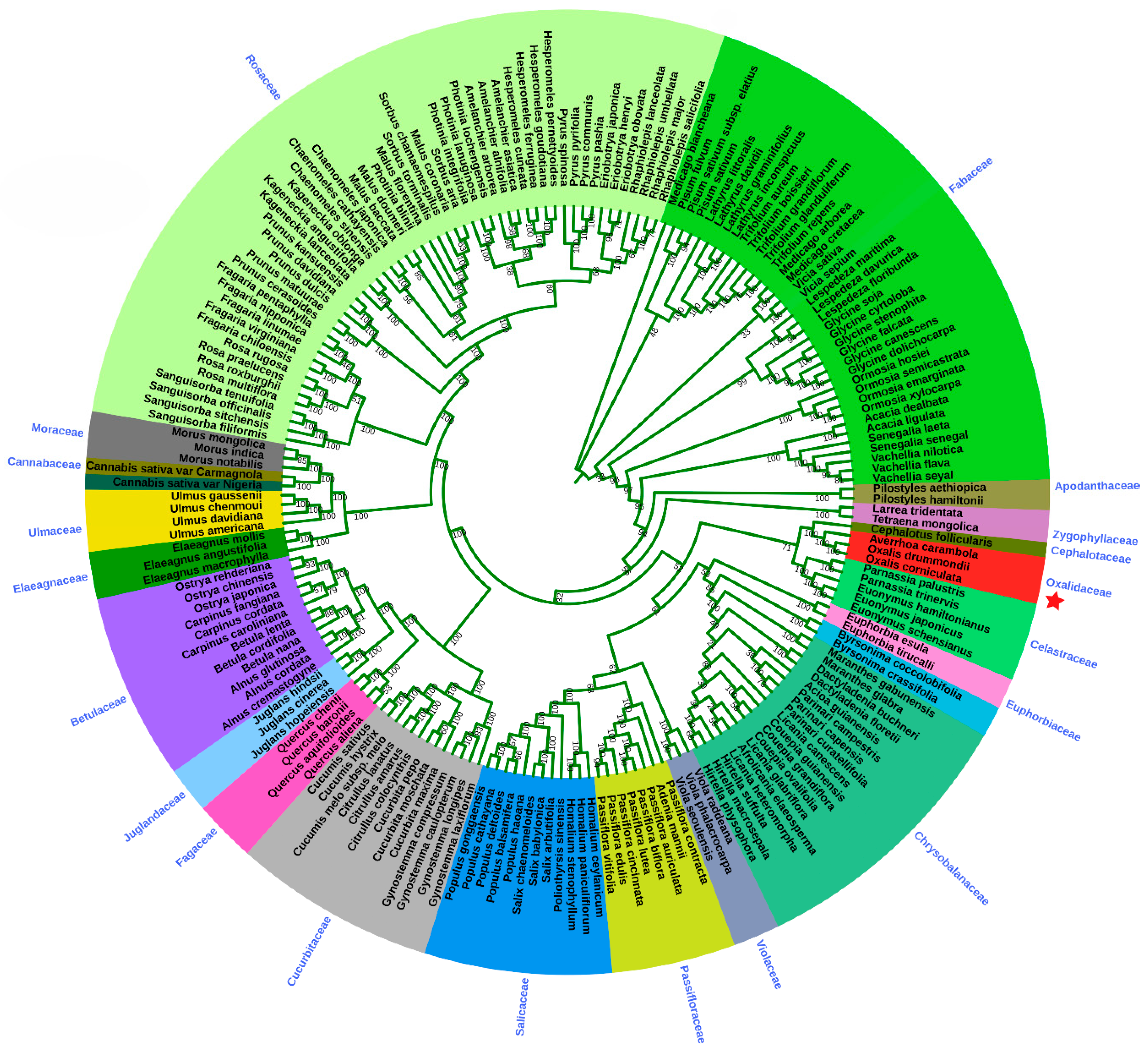
2.6. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Chloroplast DNA Extraction, Sequencing, and Assembly
4.2. Genome Annotation
4.3. Characterization of Repetitive Sequences and SSR
4.4. Sequence Divergence and Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Availability of Data and Materials
References
- Denton, M.F. Monograph of Oxalis, Section Ionoxalis (Oxalidaceae) in North America; Michigan State University: East Lansing, MI, USA, 1973. [Google Scholar]
- Lourteig, A. Oxalis, L. subg é nero Monoxalis (Small) Lourteig, Oxalis y Trifidus Lourteig; Herbarium Bradeanum: Rio de Janeiro, Brazil, 2000. [Google Scholar]
- De Azkue, D. Chromosome diversity of South American Oxalis (Oxalidaceae). Bot. J. Linn. Soc. 2000, 132, 143–152. [Google Scholar] [CrossRef]
- Groom, Q.J.; Van der Straeten, J.; Hoste, I. The origin of Oxalis corniculata L. PeerJ 2019, 7, e6384. [Google Scholar] [CrossRef]
- Lourteig, A. Oxalidaceae extra-austroamericanae. II. Oxalis, L. Sectio Corniculatae DC. Phytologia 1979, 42, 57–198. [Google Scholar]
- Marks, G. Chromosome numbers in the genus Oxalis. New Phytol. 1956, 55, 120–129. [Google Scholar] [CrossRef]
- Naranjo, C. Estudios citotaxonomicos y evolutivos en especies herbaceas sudamericanas de Oxalis (Oxalidaceae) I. Bol. Soc. Argent. Bot 1982, 20, 183–200. [Google Scholar]
- Groom, Q.; Hoste, I.; Janssens, S. Observación confirmada de Oxalis dillenii en España. Collect. Bot. 2017, 36, 4. [Google Scholar] [CrossRef]
- Pyšek, P.; Pergl, J.; Essl, F.; Lenzner, B.; Dawson, W.; Kreft, H.; Weigelt, P.; Winter, M.; Kartesz, J.; Nishino, M.; et al. Naturalized alien flora of the world. Preslia 2017, 89, 203–274. [Google Scholar] [CrossRef]
- Marble, S.C.; Andrew, K.K.; Gitta, H.; Drew, M.; Annette, C. Efficacy and estimated annual cost of common weed control methods in landscape planting beds. HortTechnology 2017, 27, 199–211. [Google Scholar] [CrossRef]
- Rehman, A.; Rehman, A.; Ahmad, I. Antibacterial, antifungal, and insecticidal potentials of Oxalis corniculata and its isolated compounds. Int. J. Anal. Chem. 2015, 2015, 1–5. [Google Scholar] [CrossRef]
- Duke, J.A.; Ayensu, E.S. Medicinal Plants of China; Reference Publications: Algonac, MI, USA, 1985; Volume 2. [Google Scholar]
- Neuhaus, H.; Emes, M. Nonphotosynthetic metabolism in plastids. Ann. Rev. Plant Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef]
- Rodríguez-Ezpeleta, N.; Brinkmann, H.; Burey, S.C.; Roure, B.; Burger, G.; Löffelhardt, W.; Bohnert, H.J.; Philippe, H.; Lang, B.F. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr. Biol. 2005, 15, 1325–1330. [Google Scholar] [CrossRef]
- McCauley, D.E.; Sundby, A.K.; Bailey, M.F.; Welch, M.E. Inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae): Evidence from experimental crosses and natural populations. Am. J. Bot. 2007, 94, 1333–1337. [Google Scholar] [CrossRef]
- Reboud, X.; Zeyl, C. Organelle inheritance in plants. Heredity 1994, 72, 132–140. [Google Scholar] [CrossRef]
- Allen, J.F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 10231–10238. [Google Scholar] [CrossRef]
- Olmstead, R.G.; Palmer, J.D. Chloroplast DNA systematics: A review of methods and data analysis. Am. J. Bot. 1994, 81, 1205–1224. [Google Scholar] [CrossRef]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium × hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; de Pamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant. Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Lee, H.L.; Jansen, R.K.; Chumley, T.W.; Kim, K.J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 2007, 24, 1161–1180. [Google Scholar] [CrossRef]
- Greiner, S.; Wang, X.; Herrmann, R.G.; Rauwolf, U.; Mayer, K.; Haberer, G.; Meurer, J. The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol. Biol. Evol. 2008, 25, 2019–2030. [Google Scholar] [CrossRef]
- Frailey, D.C.; Chaluvadi, S.R.; Vaughn, J.N.; Coatney, C.G.; Bennetzen, J.L. Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant. Biol. 2018, 18, 30. [Google Scholar] [CrossRef]
- Barrett, C.F.; Freudenstein, J.V.; Li, J.; Mayfield-Jones, D.R.; Perez, L.; Pires, J.C.; Santos, C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 2014, 31, 3095–3112. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.-B.; Wu, Y.; Yang, J.; Shahzad, K.; Li, Z.-H. Comparative Chloroplast Genomics of Dipsacales Species: Insights Into Sequence Variation, Adaptive Evolution, and Phylogenetic Relationships. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef]
- Koehler, S.; Cabral, J.S.; Whitten, W.M.; Williams, N.H.; Singer, R.B.; Neubig, K.M.; Guerra, M.; Souza, A.P.; Amaral, M.D.C.E. Molecular phylogeny of the neotropical genus Christensonella (Orchidaceae, Maxillariinae): Species delimitation and insights into chromosome evolution. Ann. Bot. 2008, 102, 491–507. [Google Scholar] [CrossRef]
- Stuessy, T.F.; Blöch, C.; Villaseñor, J.L.; Rebernig, C.A.; Weiss-Schneeweiss, H. Phylogenetic analyses of DNA sequences with chromosomal and morphological data confirm and refine sectional and series classification within Melampodium (Asteraceae, Millerieae). Taxon 2011, 60, 436–449. [Google Scholar] [CrossRef]
- Ruhfel, B.R.; Gitzendanner, M.A.; Soltis, P.S.; Soltis, D.E.; Burleigh, J.G. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 2014, 14, 23. [Google Scholar] [CrossRef]
- Soltis, D.E.; Smith, S.A.; Cellinese, N.; Wurdack, K.J.; Tank, D.C.; Brockington, S.F.; Refulio-Rodriguez, N.F.; Walker, J.B.; Moore, M.J.; Carlsward, B.S.; et al. Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 2011, 98, 704–730. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 84. [Google Scholar] [CrossRef]
- Carbonell-Caballero, J.; Alonso, R.; Ibañez, V.; Terol, J.; Talon, M.; Dopazo, J. A Phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus citrus. Mol. Biol. Evol. 2015, 32, 2015–2035. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Bell, C.D.; Soltis, P.S.; Soltis, D.E. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA 2007, 104, 19363–19368. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Khan, A.L.; Khan, A.; Al-Harrasi, A. Unraveling the chloroplast genomes of two prosopis species to identify its genomic information, comparative analyses and phylogenetic relationship. Int. J. Mol. Sci. 2020, 21, 3280. [Google Scholar] [CrossRef] [PubMed]
- Oberlander, K.C.; Dreyer, L.L.; Bellstedt, D.U. Molecular phylogenetics and origins of southern African Oxalis. TAXON 2011, 60, 1667–1677. [Google Scholar] [CrossRef]
- PM, M. Cytology of Oxalidaceae. Cytologia 1958, 23, 200–210. [Google Scholar]
- Vaio, M.; Gardner, A.; Emshwiller, E.; Guerra, M. Molecular phylogeny and chromosome evolution among the creeping herbaceous Oxalis species of sections Corniculatae and Ripariae (Oxalidaceae). Mol. Phyl. Evol. 2013, 68, 199–211. [Google Scholar] [CrossRef]
- Ohnishi, O.; Matsuoka, Y. Search for the wild ancestor of buckwheat II. Taxonomy of Fagopyrum (Polygonaceae) species based on morphology, isozymes and cpDNA variability. Genes Genet. Syst. 1996, 71, 383–390. [Google Scholar] [CrossRef]
- Cuénoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Yamane, K.; Yasui, Y.; Ohnishi, O. Intraspecific cpDNA variations of diploid and tetraploid perennial buckwheat, Fagopyrum cymosum (Polygonaceae). Am. J. Bot. 2003, 90, 339–346. [Google Scholar] [CrossRef]
- Cui, Y.; Qin, S.; Jiang, P. Chloroplast transformation of Platymonas (Tetraselmis) subcordiformis with the bar gene as selectable marker. PLoS ONE 2014, 9, e98607. [Google Scholar] [CrossRef]
- McPherson, H.; Van der Merwe, M.; Delaney, S.K.; Edwards, M.A.; Henry, R.J.; McIntosh, E.; Rymer, P.D.; Milner, M.L.; Siow, J.; Rossetto, M. Capturing chloroplast variation for molecular ecology studies: A simple next generation sequencing approach applied to a rainforest tree. BMC Ecol. 2013, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- McNeal, J.R.; Kuehl, J.V.; Boore, J.L.; de Pamphilis, C.W. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, D.; Senalik, D.; Ames, M.; Zhu, H.; Steffan, S.A.; Harbut, R.; Polashock, J.; Vorsa, N.; Gillespie, E.; Kron, K. Complete plastid genome sequence of Vaccinium macrocarpon: Structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet. Genomes 2013, 9, 489–498. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Cheng, T.; Zhou, S. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE 2013, 8, e77965. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; De Paola, D.; Danzi, D.; Vendramin, G.G.; Sonnante, G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS ONE 2015, 10, e0120589. [Google Scholar] [CrossRef]
- Yang, J.-B.; Yang, S.-X.; Li, H.-T.; Yang, J.; Li, D.-Z. Comparative chloroplast genomes of Camellia species. PLoS ONE 2013, 8, e73053. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, A.; Khan, G.; Lee, I.-J.; Al-Harrasi, A. Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef]
- Jansen, R.K.; Saski, C.; Lee, S.-B.; Hansen, A.K.; Daniell, H. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2011, 28, 835–847. [Google Scholar] [CrossRef]
- Nie, X.; Lv, S.; Zhang, Y.; Du, X.; Wang, L.; Biradar, S.S.; Tan, X.; Wan, F.; Weining, S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar] [CrossRef]
- Kim, K.-J.; Lee, H.-L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef]
- Jansen, R.K.; Wojciechowski, M.F.; Sanniyasi, E.; Lee, S.-B.; Daniell, H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenetics Evol. 2008, 48, 1204–1217. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; He, P.; Li, P.; Lee, J.; Soltis, D.E.; Fu, C. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom. 2018, 19, 235. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-S.; Chaw, S.-M. Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol. Evol. 2015, 7, 2000–2009. [Google Scholar] [CrossRef] [PubMed]
- Goulding, S.E.; Wolfe, K.; Olmstead, R.; Morden, C. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. MGG 1996, 252, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Park, C.-W.; Kim, K.-J. Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia, and its evolutionary implications. Mol. Cells 2009, 27, 365. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Waqas, M.; Kang, S.-M.; Yun, B.-W.; Lee, I.-J. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Khan, A.; Asaf, S.; Khan, A.L.; Al-Harrasi, A.; Al-Sudairy, O.; AbdulKareem, N.M.; Khan, A.; Shehzad, T.; Alsaady, N.; Al-Lawati, A.; et al. First complete chloroplast genomics and comparative phylogenetic analysis of Commiphora gileadensis and C. foliacea: Myrrh producing trees. PLoS ONE 2019, 14, e0208511. [Google Scholar] [CrossRef]
- Khan, A.; Asaf, S.; Khan, A.L.; Khan, A.; Al-Harrasi, A.; Al-Sudairy, O.; AbdulKareem, N.M.; Al-Saady, N.; Al-Rawahi, A. Complete chloroplast genomes of medicinally important Teucrium species and comparative analyses with related species from Lamiaceae. PeerJ 2019, 7, e7260. [Google Scholar] [CrossRef]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H.; Sun, C.; Li, X.E.; Li, C.; et al. The Complete Chloroplast Genome Sequence of the Medicinal Plant Salvia miltiorrhiza. PLoS ONE 2013, 8, e57607. [Google Scholar] [CrossRef]
- Leclercq, S.; Rivals, E.; Jarne, P. Detecting microsatellites within genomes: Significant variation among algorithms. BMC Bioinf. 2007, 8, 125. [Google Scholar] [CrossRef]
- Echt, C.S.; Deverno, L.L.; Anzidei, M.; Vendramin, G.G. Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Mol. Ecol. 1998, 7, 307–316. [Google Scholar] [CrossRef]
- Kuang, D.-Y.; Wu, H.; Wang, Y.-L.; Gao, L.-M.; Zhang, S.-Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Burke, S.V.; Lin, C.-S.; Wysocki, W.P.; Clark, L.G.; Duvall, M.R. Phylogenomics and Plastome Evolution of Tropical Forest Grasses (Leptaspis, Streptochaeta: Poaceae). Front. Plant Sci. 2016, 7, 1993. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, W.; Xie, X.; Lu, Y.; Liu, Y.; Jin, X.; Suo, Z. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Du, Y.-P.; Bi, Y.; Yang, F.-P.; Zhang, M.-F.; Chen, X.-Q.; Xue, J.; Zhang, X.-H. Complete chloroplast genome sequences of Lilium: Insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Sun, L.; Fang, L.; Zhang, Z.; Chang, X.; Penny, D.; Zhong, B. Chloroplast phylogenomic inference of green algae relationships. Sci. Rep. 2016, 6, 20528. [Google Scholar] [CrossRef]
- Majure, L.C.; Puente, R.; Griffith, M.P.; Judd, W.S.; Soltis, P.S.; Soltis, D.E. Phylogeny of Opuntia s.s. (Cactaceae): Clade delineation, geographic origins, and reticulate evolution. Am. J. Bot. 2012, 99, 847–864. [Google Scholar] [CrossRef]
- Hilu, K.W.; Alice, L.A. A phylogeny of Chloridoideae (Poaceae) based on matK sequences. Syst. Bot. 2001, 26, 386–405. [Google Scholar]
- Goremykin, V.V.; Nikiforova, S.V.; Biggs, P.J.; Zhong, B.; Delange, P.; Martin, W.; Woetzel, S.; Atherton, R.A.; McLenachan, P.A.; Lockhart, P.J. The evolutionary root of flowering plants. Syst. Biol. 2013, 62, 50–61. [Google Scholar] [CrossRef]
- Luo, Y.; Ma, P.-F.; Li, H.-T.; Yang, J.-B.; Wang, H.; Li, D.-Z. Plastid Phylogenomic Analyses Resolve Tofieldiaceae as the Root of the Early Diverging Monocot Order Alismatales. Genome Biol. Evol. 2016, 8, 932–945. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Gen. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinf. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucl. Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucl. Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucl. Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
| A. carambola | C. follicularis | O. drummondii | O. corniculata | |
|---|---|---|---|---|
| Size (bp) | 155,965 | 142,706 | 152,112 | 152,189 |
| Overall GC contents | 36.5 | 37.1 | 36.5 | 36.7 |
| LSC size in bp | 86,218 | 81,071 | 83,341 | 83,427 |
| SSC size in bp | 17,497 | 8739 | 16915 | 16,990 |
| IR size in bp | 25,626 | 25,949 | 25,429 | 25,387 |
| Protein coding regions size in bp | 78,528 | 66,672 | 77,826 | 79,239 |
| tRNA size in bp | 2790 | 2790 | 2790 | 3042 |
| rRNA size in bp | 9046 | 9050 | 9046 | 9048 |
| Number of genes | 131 | 122 | 127 | 131 |
| Number of protein coding genes | 83 | 71 | 82 | 83 |
| Number of rRNA | 8 | 8 | 8 | 8 |
| Number of tRNA | 37 | 37 | 37 | 40 |
| Category | Group of Genes | Name of Genes |
|---|---|---|
| Self-replication | Large subunit of ribosomal proteins | rpl2 *, rpl14, rpl16, rpl20, rpl22, rpl23 *, rpl33, rpl36 |
| Small subunit of ribosomal proteins | rps2, rps3, rps4, rps7 *, rps8, rps11, rps12 *, rps14, rps15, rps18, rps19 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| rRNA genes | rrn4.5, 5, 16, 23 | |
| tRNA genes | trnA-UGC *, trnC-GCA, trnD-GUC, trnE-UUC , trnF-GAA, trnfM-CAU, trnG-GCC *, trnH-GUG, trnI-GAU *, trnK-UUU, trnL-CAA *, trnL-UAA, trnL-UAG, trnM-CAU *, trnN-GUU *, trnP-UGG, trnQ-UUG, trnR-ACG *, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU *, trnV-GAC *, trnW-CCA, trnY-GUA | |
| Photosynthesis | Photosystem I | psaA, B, C, I, J |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbT, psbZ | |
| NadH oxidoreductase | ndhA, ndhB *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Cytochrome b6/f complex | petA, petB, petD, pet G, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Rubisco | rbcL | |
| Other genes | Maturase | matK |
| Protease | clpP | |
| Envelop membrane protein | cemA | |
| Subunit Acetyl- CoA-Carboxylate | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Unknown | Conserved Open reading frames | ycf1 *, 2 *, 3, 4, |
| Gene | Location | Exon I (bp) | Intron 1 (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O. c | O. d | A. c | C. f | O. c | O. d | A. c | C. f | O. c | O. d | A. c | C. f | O. c | O. d | A. c | C. f | O. c | O. d | A. c | C. f | ||
| atpF | LSC | 144 | 145 | 145 | 145 | 717 | 714 | 714 | 717 | 411 | 410 | 410 | 410 | ||||||||
| petB | LSC | 6 | 6 | 6 | 6 | 746 | 746 | 785 | 793 | 645 | 642 | 642 | 642 | ||||||||
| PetD | LSC | 8 | 8 | 7 | 7 | 746 | 778 | 707 | 721 | 645 | 475 | 473 | 473 | ||||||||
| rpl2 * | IR | 391 | 391 | 391 | 391 | 658 | 661 | 659 | 671 | 434 | 434 | 434 | 525 | ||||||||
| rps16 | LSC | -- | -- | 40 | 40 | -- | -- | 935 | 929 | -- | -- | 224 | 227 | ||||||||
| rpoC1 | LSC | 430 | 432 | 453 | 459 | 727 | 729 | 751 | 759 | 1634 | 1629 | 1608 | 1617 | ||||||||
| rps12 * | IR/LSC | 391 | 527 | 232 | 434 | 25 | |||||||||||||||
| clpP | LSC | 71 | 71 | 69 | 69 | 808 | 812 | 833 | 833 | 289 | 289 | 291 | 291 | 603 | 612 | 568 | 228 | 228 | 228 | ||
| ndhA | SSC | 557 | 557 | 559 | -- | 1077 | 1067 | 1037 | -- | 541 | 541 | 557 | -- | ||||||||
| ndhB* | IR | 777 | 777 | 777 | -- | 685 | 685 | 685 | -- | 756 | 756 | 756 | -- | ||||||||
| ycf3 | LSC | 124 | 124 | 126 | 126 | 718 | 712 | 718 | 714 | 228 | 230 | 228 | 228 | 684 | 678 | 685 | 678 | 155 | 153 | 153 | 150 |
| trnA-UGC * | IR | 38 | 38 | 38 | 38 | 799 | 798 | 763 | 811 | 35 | 35 | 35 | 35 | ||||||||
| trnI –GAU * | IR | 37 | 37 | 37 | 37 | 925 | 924 | 932 | 946 | 35 | 35 | 35 | 35 | ||||||||
| trnL-UAA | LSC | 35 | 35 | 35 | 35 | 497 | 497 | 507 | 492 | 50 | 50 | 50 | 50 | ||||||||
| trnK -UUU | LSC | 37 | 34 | 37 | 35 | 2519 | 2515 | 2545 | 2558 | 35 | 37 | 35 | 37 | ||||||||
| trnG-GCC | LSC | 71 | 71 | 71 | 71 | -- | -- | -- | -- | -- | -- | -- | -- | ||||||||
| trnV-UAC | LSC | -- | -- | 35 | 35 | -- | -- | 617 | 618 | -- | -- | 39 | 39 | ||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lubna; Asaf, S.; Jan, R.; Khan, A.L.; Lee, I.-J. Complete Chloroplast Genome Characterization of Oxalis Corniculata and Its Comparison with Related Species from Family Oxalidaceae. Plants 2020, 9, 928. https://doi.org/10.3390/plants9080928
Lubna, Asaf S, Jan R, Khan AL, Lee I-J. Complete Chloroplast Genome Characterization of Oxalis Corniculata and Its Comparison with Related Species from Family Oxalidaceae. Plants. 2020; 9(8):928. https://doi.org/10.3390/plants9080928
Chicago/Turabian StyleLubna, Sajjad Asaf, Rahmatullah Jan, Abdul Latif Khan, and In-Jung Lee. 2020. "Complete Chloroplast Genome Characterization of Oxalis Corniculata and Its Comparison with Related Species from Family Oxalidaceae" Plants 9, no. 8: 928. https://doi.org/10.3390/plants9080928
APA StyleLubna, Asaf, S., Jan, R., Khan, A. L., & Lee, I.-J. (2020). Complete Chloroplast Genome Characterization of Oxalis Corniculata and Its Comparison with Related Species from Family Oxalidaceae. Plants, 9(8), 928. https://doi.org/10.3390/plants9080928







