Autophagy Dances with Phytohormones upon Multiple Stresses
Abstract
:1. Introduction
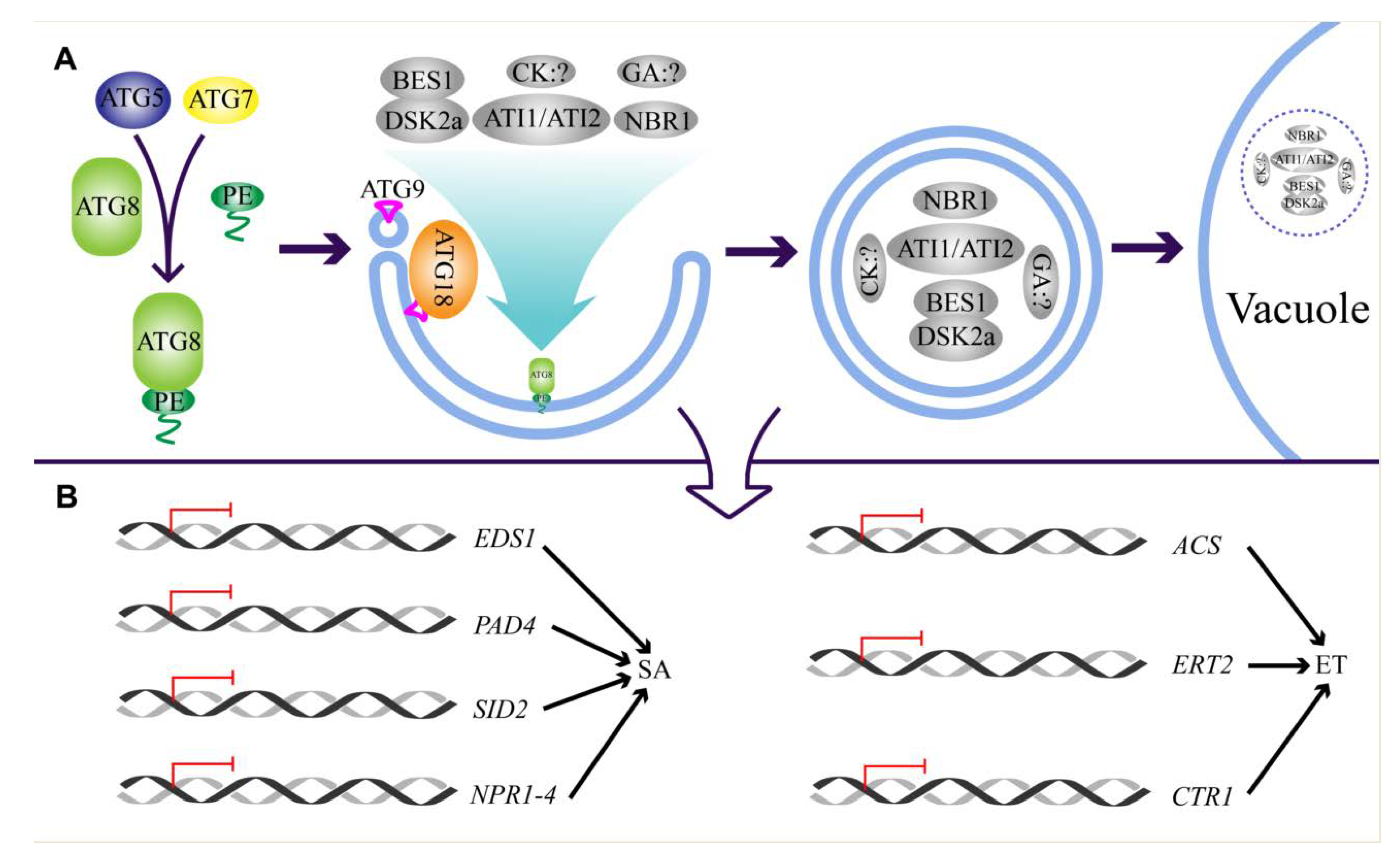
2. Regulation of Autophagy
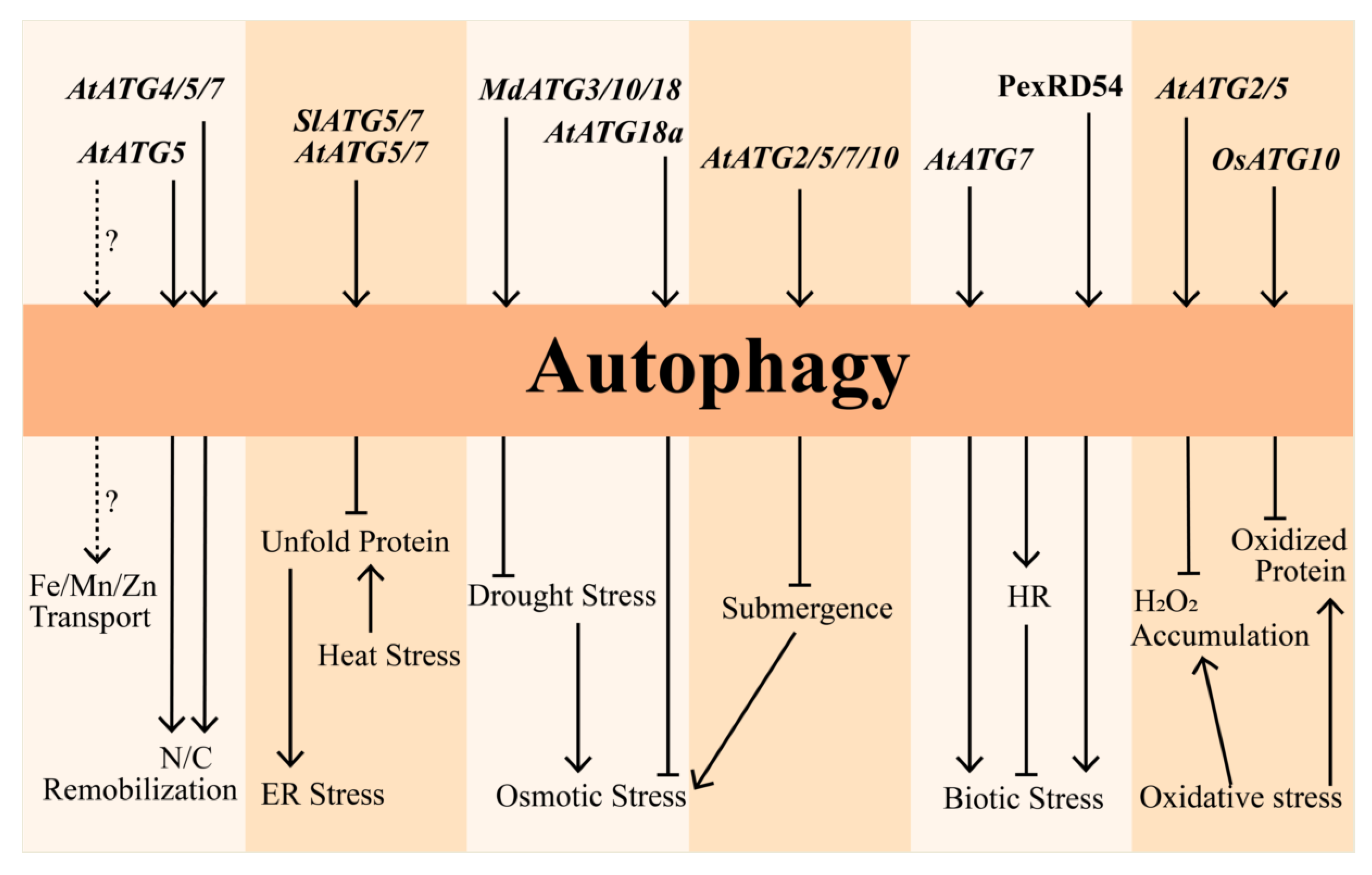
3. Role of Autophagy under Abiotic Stress
3.1. Oxidative Stress
3.2. Nutrition Starvation
3.3. Osmotic Stress
3.4. Heat and ER Stresses
4. Regulation of Autophagy under Biotic Stress
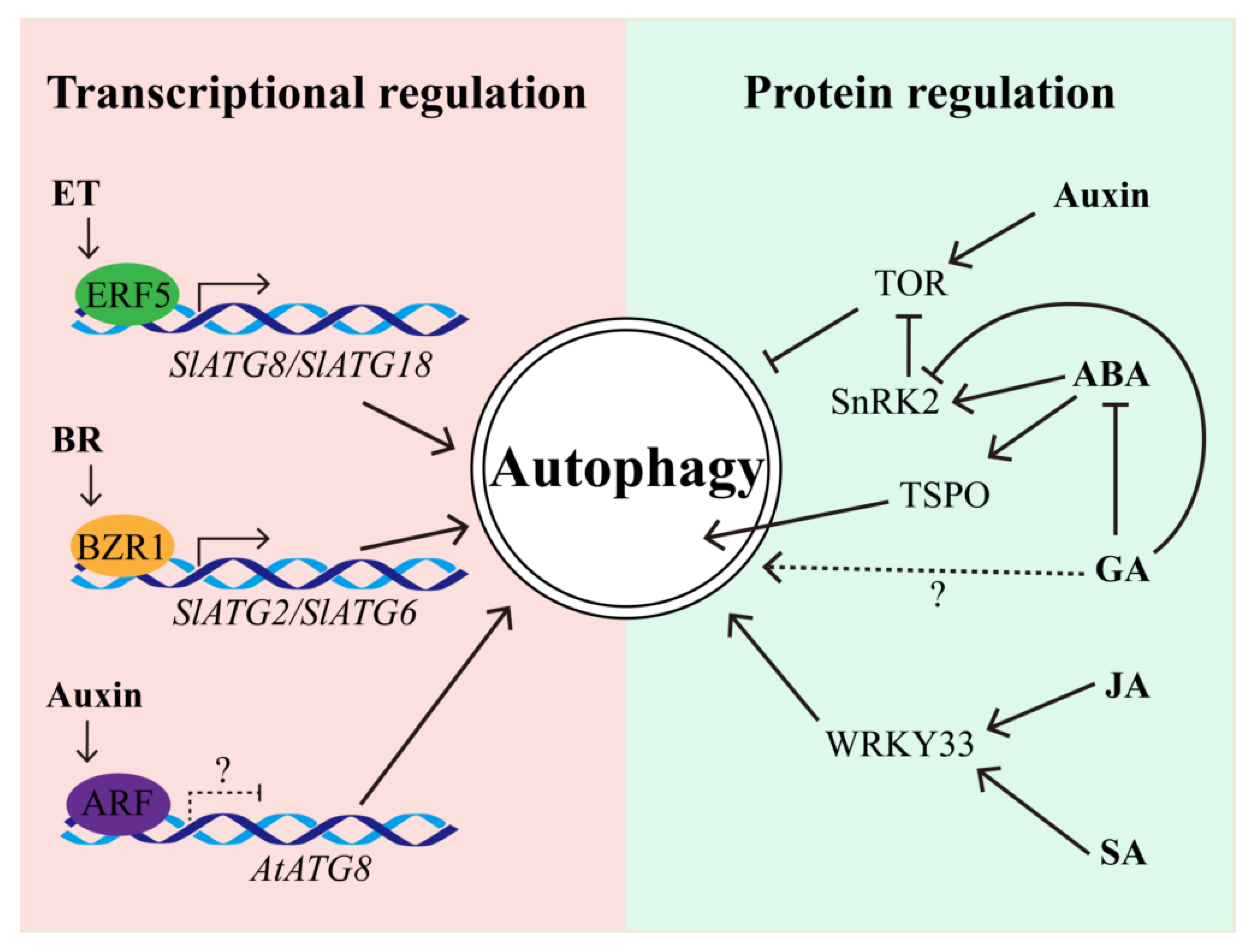
5. Autophagy Interplays with Plant Hormones upon Multiple Stresses
5.1. The Regulation of Autophagy by Hormones
5.2. Autophagy Regulates Hormone Biosynthesis and Signaling
6. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leng, G.Y.; Hall, J. Crop yield sensitivity of global major agricultural countries to droughts and the projected changes in the future. Sci. Total Environ. 2019, 654, 811–821. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Van Doorn, W.G.; Papini, A. Ultrastructure of autophagy in plant cells: A review. Autophagy 2013, 9, 1922–1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Pleskot, R.; Zang, J.; Winkler, J.; Wang, J.; Yperman, K.; Zhang, T.; Wang, K.; Gong, J.; Guan, Y.; et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 2019, 10, 5132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, X.; Wang, H.; Lam, S.K.; Gao, C.; Wang, X.; Cai, Y.; Jiang, L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 2013, 25, 4596–4615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Bars, R.; Marion, J.; Le Borgne, R.; Satiat-Jeunemaitre, B.; Bianchi, M.W. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 2014, 5, 4121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, W.; Li, X.; Guo, S.; Liu, Y.; Li, F.; Xie, Q. Autophagy in plant: A new orchestrator in the regulation of the phytohormones homeostasis. Int. J. Mol. Sci. 2019, 20, 2900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sienko, K.; Poormassalehgoo, A.; Yamada, K.; Goto-Yamada, S. Microautophagy in plants: Consideration of its molecular mechanism. Cells 2020, 9, 887. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. Eur. Mol. Biol. Organ. J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Chanoca, A.; Kovinich, N.; Burkel, B.; Stecha, S.; Bohorquez-Restrepo, A.; Ueda, T.; Eliceiri, K.W.; Grotewold, E.; Otegui, M.S. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell 2015, 27, 2545–2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.; Yu, J.Q.; Chen, Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 2013, 9, e1003196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.H.; Yoshimoto, K.; Ohsumi, Y.; Jeon, J.S.; An, G. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 2009, 27, 67–74. [Google Scholar] [CrossRef]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Nolan, T.M.; Yin, Y.; Bassham, D.C. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 2020, 16, 123–139. [Google Scholar] [CrossRef]
- Liao, C.Y.; Bassham, D.C. Combating stress: The interplay between hormone signaling and autophagy in plants. J. Exp. Bot. 2020, 71, 1723–1733. [Google Scholar] [CrossRef]
- Yue, W.; Nie, X.; Cui, L.; Zhi, Y.; Zhang, T.; Du, X.; Song, W. Genome-wide sequence and expressional analysis of autophagy Gene family in bread wheat (Triticum aestivum L.). Plant Physiol. 2018, 229, 7–21. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.M.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.H.; Peng, X.B.; Sun, M.X. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015, 22, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Chung, T.; Suttangkakul, A.; Vierstra, R.D. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009, 149, 220–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, D.; Zhang, W.; Sun, H.; Wei, X.; Yue, J.; Wang, H. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 2014, 33, 1697–1710. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Ohsumi, Y. Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol. 2018, 59, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, H.; Noda, T.; Shirano, Y.; Kato, T.; Hayashi, H.; Shibata, D.; Tabata, S.; Ohsumi, Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002, 129, 1181–1193. [Google Scholar] [CrossRef] [Green Version]
- Kurusu, T.; Koyano, T.; Hanamata, S.; Kubo, T.; Noguchi, Y.; Yagi, C.; Nagata, N.; Yamamoto, T.; Ohnishi, T.; Okazaki, Y.; et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 2014, 10, 878–888. [Google Scholar] [CrossRef]
- Yu, J.; Zhen, X.; Li, X.; Li, N.; Xu, F. Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 2019, 10, 584. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [Green Version]
- Guiboileau, A.; Yoshimoto, K.; Soulay, F.; Bataille, M.P.; Avice, J.C.; Masclaux-Daubresse, C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012, 194, 732–740. [Google Scholar] [CrossRef]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR Signaling and Nutrient Sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef]
- Zhu, T.; Li, L.; Feng, L.; Mo, H.; Ren, M. Target of rapamycin regulates genome methylation reprogramming to control plant growth in Arabidopsis. Front. Genet. 2020, 11, 186. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Wang, P.; Xiong, Y. Target of rapamycin signaling in plant stress responses. Plant Physiol. 2020, 182, 1613–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montane, M.H.; Menand, B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 2013, 64, 4361–4374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, Y.; Luo, X.; Bassham, D.C. TOR-dependent and-independent pathways regulate autophagy in Arabidopsis thaliana. Front. Plant. Sci. 2017, 8, 1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Bassham, D.C. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 2010, 5, e11883. [Google Scholar] [CrossRef] [Green Version]
- Dong, P.; Xiong, F.; Que, Y.; Wang, K.; Yu, L.; Li, Z.; Ren, M. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 2015, 6, 677. [Google Scholar] [CrossRef] [Green Version]
- Avin-Wittenberg, T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019, 42, 1045–1053. [Google Scholar] [CrossRef]
- Li, X.; Cai, W.; Liu, Y.; Li, H.; Fu, L.; Liu, Z.; Xu, L.; Liu, H.; Xu, T.; Xiong, Y. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc. Natl. Acad. Sci. USA 2017, 114, 2765–2770. [Google Scholar] [CrossRef] [Green Version]
- Schepetilnikov, M.; Dimitrova, M.; Mancera-Martinez, E.; Geldreich, A.; Keller, M.; Ryabova, L.A. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. Eur. Mol. Biol. Organ. J. 2013, 32, 1087–1102. [Google Scholar] [CrossRef] [Green Version]
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. Eur. Mol. Biol. Organ. J. 2017, 36, 886–903. [Google Scholar] [CrossRef]
- Kravchenko, A.; Citerne, S.; Jehanno, I.; Bersimbaev, R.I.; Veit, B.; Meyer, C.; Leprince, A.S. Mutations in the Arabidopsis Lst8 and Raptor genes encoding partners of the TOR complex, or inhibition of TOR activity decrease abscisic acid (ABA) synthesis. Biochem. Biophys. Res. Commun. 2015, 467, 992–997. [Google Scholar] [CrossRef]
- Crozet, P.; Margalha, L.; Confraria, A.; Rodrigues, A.; Martinho, C.; Adamo, M.; Elias, C.A.; Baena-Gonzalez, E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant. Sci. 2014, 5, 190. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Tarkowski, L.P.; Van den Ende, W.; Bassham, D.C. Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lastdrager, J.; Hanson, J.; Smeekens, S. Sugar signals and the control of plant growth and development. J. Exp. Bot. 2014, 65, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.; Gazzarrini, S. Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: The emerging picture. Front. Plant Sci. 2014, 5, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Su, Z.Z.; Huang, L.; Xia, F.N.; Qi, H.; Xie, L.J.; Xiao, S.; Chen, Q.F. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 2017, 8, 1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 2016, 85, 83–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Sun, X.; Yue, Z.; Liang, D.; Wang, N.; Ma, F. Isolation and characterization of MdATG18a, a WD40-repeat AuTophaGy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci. Hortic. 2014, 165, 51–61. [Google Scholar] [CrossRef]
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. Role of plant autophagy in stress response. Protein Cell 2011, 2, 784–791. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, K.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Consonni, C.; Panstruga, R.; Ohsumi, Y.; Shirasu, K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009, 21, 2914–2927. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Contento, A.L.; Nguyen, P.Q.; Bassham, D.C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007, 143, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Masclaux-Daubresse, C.; Chen, Q.; Have, M. Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 2017, 39, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Shinozaki, D.; Luo, J.; Pottier, M.; Have, M.; Marmagne, A.; Reisdorf-Cren, M.; Chardon, F.; Thomine, S.; Yoshimoto, K.; et al. Autophagy and nutrients management in plants. Cells 2019, 8, 1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.R.; Doelling, J.H.; Suttangkakul, A.; Vierstra, R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005, 138, 2097–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, T.; Phillips, A.R.; Vierstra, R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010, 62, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Avin-Wittenberg, T.; Bajdzienko, K.; Wittenberg, G.; Alseekh, S.; Tohge, T.; Bock, R.; Giavalisco, P.; Fernie, A.R. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 2015, 27, 306–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumann, C.; Muller, J.; Sakhonwasee, S.; Wieghaus, A.; Hause, G.; Heisters, M.; Burstenbinder, K.; Abel, S. The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant Physiol. 2019, 179, 460–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Sun, X.; Jia, X.; Ma, F. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017, 256, 53–64. [Google Scholar] [CrossRef]
- Huo, L.; Guo, Z.; Jia, X.; Sun, X.; Wang, P.; Gong, X.; Ma, F. Increased autophagic activity in roots caused by overexpression of the autophagy-related gene MdATG10 in apple enhances salt tolerance. Plant Sci. 2020, 294, 110444. [Google Scholar] [CrossRef]
- Sun, X.; Wang, P.; Jia, X.; Huo, L.; Che, R.; Ma, F. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J 2018, 16, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Song, W.M.; Wang, P.; Yu, X.; Li, B.; Jiang, C.; Shiu, S.H.; Zhang, H.; Bassham, D.C. COST1 regulates autophagy to control plant drought tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 7482–7493. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liao, B.; Qi, H.; Xie, L.J.; Huang, L.; Tan, W.J.; Zhai, N.; Yuan, L.B.; Zhou, Y.; Yu, L.J.; et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 2015, 11, 2233–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wang, J.; Yu, J.Q.; Chen, Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014, 5, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Burgos, J.S.; Deng, Y.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012, 24, 4635–4651. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofius, D.; Schultz-Larsen, T.; Joensen, J.; Tsitsigiannis, D.I.; Petersen, N.H.; Mattsson, O.; Jorgensen, L.B.; Jones, J.D.; Mundy, J.; Petersen, M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 2009, 137, 773–783. [Google Scholar] [CrossRef] [Green Version]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife 2017, 6, e23897. [Google Scholar] [CrossRef]
- Leary, A.Y.; Sanguankiattichai, N.; Duggan, C.; Tumtas, Y.; Pandey, P.; Segretin, M.E.; Salguero Linares, J.; Savage, Z.D.; Yow, R.J.; Bozkurt, T.O. Modulation of plant autophagy during pathogen attack. J. Exp. Bot. 2018, 69, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Dinesh-Kumar, S.P. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 2008, 4, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Lenz, H.D.; Vierstra, R.D.; Nurnberger, T.; Gust, A.A. ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal. Behav. 2011, 6, 1040–1042. [Google Scholar] [CrossRef] [Green Version]
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarnowski, L.; Rodriguez, M.C.; Brzywczy, J.; Piecho-Kabacik, M.; Krckova, Z.; Martinec, J.; Wawrzynska, A.; Sirko, A. A selective autophagy cargo receptor NBR1 modulates abscisic acid signalling in Arabidopsis thaliana. Sci. Rep. 2020, 10, 7778. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Smidler, A.; Puigvert, M.; Popa, C.; Valls, M.; Dangl, J.L. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: Functional linkage with autophagy. Cell Death Differ. 2014, 21, 1399–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Wang, Y.; Zheng, X.; Jia, Q.; Zhao, J.; Bai, F.; Hong, Y.; Liu, Y. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 2015, 27, 1316–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wu, Y.; Tang, D. The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal. Behav. 2011, 6, 1408–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagdas, Y.F.; Belhaj, K.; Maqbool, A.; Chaparro-Garcia, A.; Pandey, P.; Petre, B.; Tabassum, N.; Cruz-Mireles, N.; Hughes, R.K.; Sklenar, J.; et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 2016, 5, e10856. [Google Scholar] [CrossRef]
- Quijia Pillajo, J.O.; Chapin, L.J.; Jones, M.L. Senescence and abiotic stress induce expression of autophagy-related genes in Petunia. J. Am. Soc. Hortic. Sci. 2018, 143, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Shibuya, K.; Niki, T.; Ichimura, K. Pollination induces autophagy in petunia petals via ethylene. J. Exp. Bot. 2013, 64, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Masclaux-Daubresse, C.; Clement, G.; Anne, P.; Routaboul, J.M.; Guiboileau, A.; Soulay, F.; Shirasu, K.; Yoshimoto, K. Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development and plant responses to the environment in Arabidopsis. Plant Cell 2014, 26, 1857–1877. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Liu, W.; Hu, W.; Liu, G.; Wu, C.; Liu, W.; Zeng, H.; He, C.; Shi, H. Genome-wide analysis of autophagy-related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep. 2017, 36, 1237–1250. [Google Scholar] [CrossRef]
- Okuda, M.; Nang, M.P.; Oshima, K.; Ishibashi, Y.; Zheng, S.H.; Yuasa, T.; Iwaya-Inoue, M. The ethylene signal mediates induction of GmATG8i in soybean plants under starvation stress. Biosci. Biotechnol. Biochem. 2011, 75, 1408–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, T.; Zou, L.; Li, Y.; Yao, X.; Xu, F.; Deng, X.; Zhang, D.; Lin, H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 2063–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuroh, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, M.A.; Li, Y.; Bajdzienko, K.; Fisahn, J.; Watanabe, M.; Hoefgen, R.; Schottler, M.A.; Giavalisco, P. RAPTOR controls developmental growth transitions by altering the hormonal and metabolic balance. Plant Physiol. 2018, 177, 565–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Cai, S.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachez, C.; Veljanovski, V.; Reinhardt, H.; Guillaumot, D.; Vanhee, C.; Chaumont, F.; Batoko, H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 2014, 26, 4974–4990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhee, C.; Zapotoczny, G.; Masquelier, D.; Ghislain, M.; Batoko, H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 2011, 23, 785–805. [Google Scholar] [CrossRef] [Green Version]
- Honig, A.; Avin-Wittenberg, T.; Ufaz, S.; Galili, G. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 2012, 24, 288–303. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H.; et al. The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef] [Green Version]
- Maruri-Lopez, I.; Aviles-Baltazar, N.Y.; Buchala, A.; Serrano, M. Intra and extracellular journey of the phytohormone salicylic acid. Front. Plant. Sci. 2019, 10, 423. [Google Scholar] [CrossRef]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.J.; Wang, K.X.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Zhou, J. BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 2019, 179, 671–685. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Huo, L.; Jia, X.; Che, R.; Gong, X.; Wang, P.; Ma, F. Overexpression of MdATG18a in apple improves resistance to Diplocarpon mali infection by enhancing antioxidant activity and salicylic acid levels. Hortic. Res. 2018, 5, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurusu, T.; Koyano, T.; Kitahata, N.; Kojima, M.; Hanamata, S.; Sakakibara, H.; Kuchitsu, K. Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal. Behav. 2017, 12, e1365211. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 2017, 41, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalvari, I.; Tsompanis, S.; Mulakkal, N.C.; Osgood, R.; Johansen, T.; Nezis, I.P.; Promponas, V.J. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 2014, 10, 913–925. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Tzfadia, O.; Levy, M.; Weithorn, E.; Peled-Zehavi, H.; Van Parys, T.; Van de Peer, Y.; Galili, G. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy 2016, 12, 876–887. [Google Scholar] [CrossRef]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 2019, 177, 766–781. [Google Scholar] [CrossRef] [Green Version]
- Zhen, X.; Xu, F.; Zhang, W.; Li, N.; Li, X. Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS ONE 2019, 14, e0223011. [Google Scholar] [CrossRef] [Green Version]
- Zhen, X.; Li, X.; Yu, J.; Xu, F. OsATG8c-mediated increased autophagy regulates the yield and nitrogen use efficiency in rice. Int. J. Mol. Sci. 2019, 20, 4596. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Lin, Y.; Li, X.; Guo, S.; Huang, Y.; Xie, Q. Autophagy Dances with Phytohormones upon Multiple Stresses. Plants 2020, 9, 1038. https://doi.org/10.3390/plants9081038
Li Y, Lin Y, Li X, Guo S, Huang Y, Xie Q. Autophagy Dances with Phytohormones upon Multiple Stresses. Plants. 2020; 9(8):1038. https://doi.org/10.3390/plants9081038
Chicago/Turabian StyleLi, Yifan, Yanni Lin, Xi Li, Shaoying Guo, Yifeng Huang, and Qingjun Xie. 2020. "Autophagy Dances with Phytohormones upon Multiple Stresses" Plants 9, no. 8: 1038. https://doi.org/10.3390/plants9081038
APA StyleLi, Y., Lin, Y., Li, X., Guo, S., Huang, Y., & Xie, Q. (2020). Autophagy Dances with Phytohormones upon Multiple Stresses. Plants, 9(8), 1038. https://doi.org/10.3390/plants9081038




