Identification of Three Monofunctional Diterpene Synthases with Specific Enzyme Activities Expressed during Heartwood Formation in Western Redcedar (Thuja plicata) Trees
Abstract
1. Introduction
2. Results
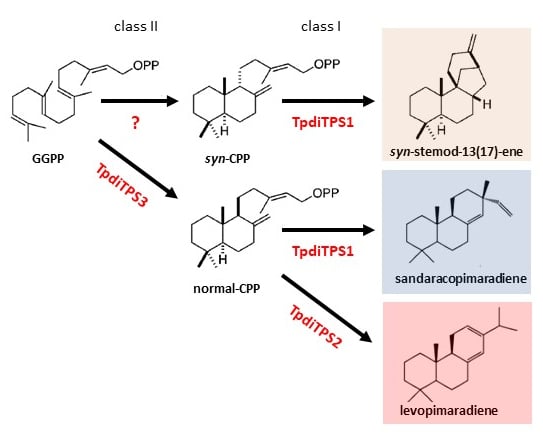
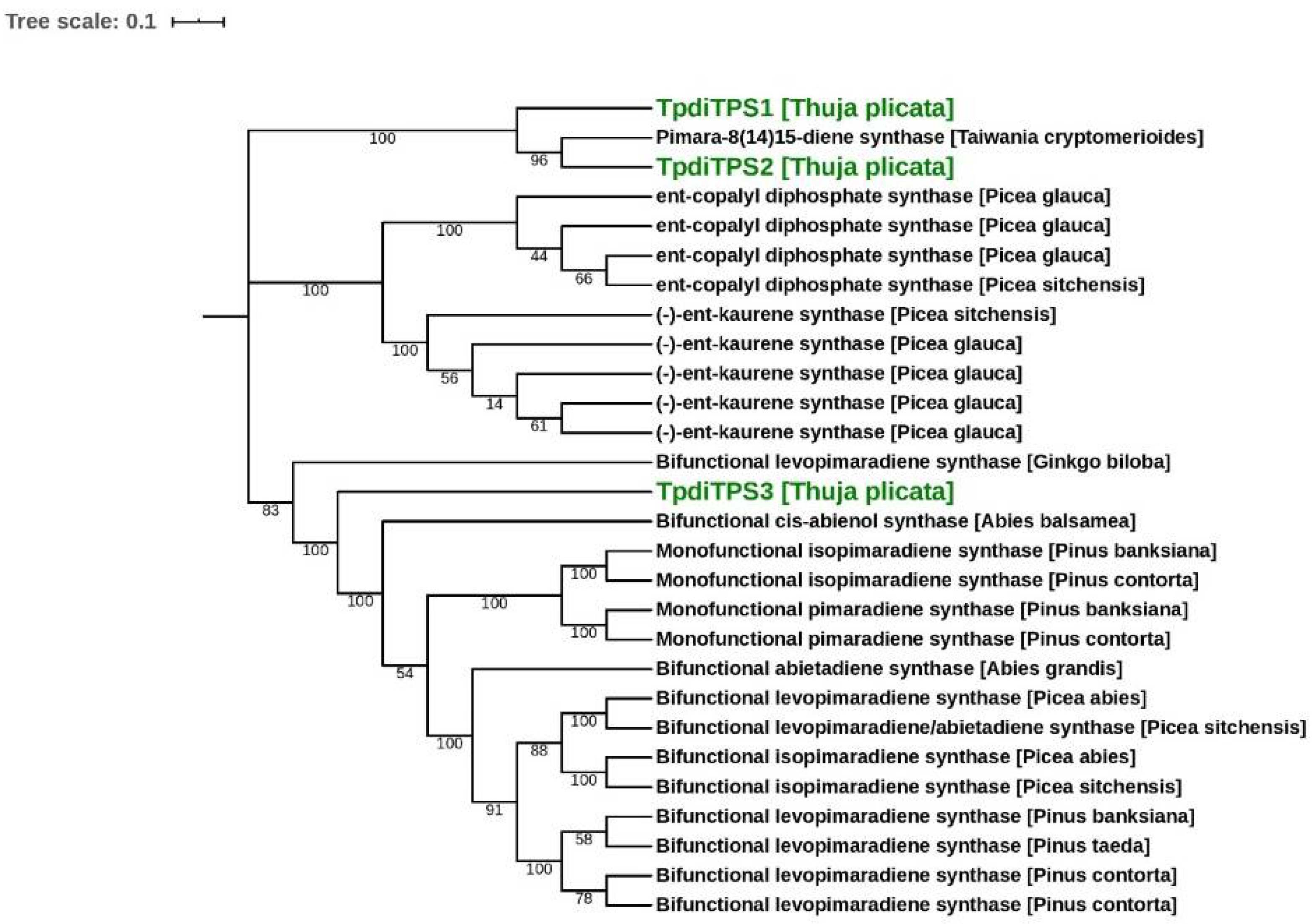
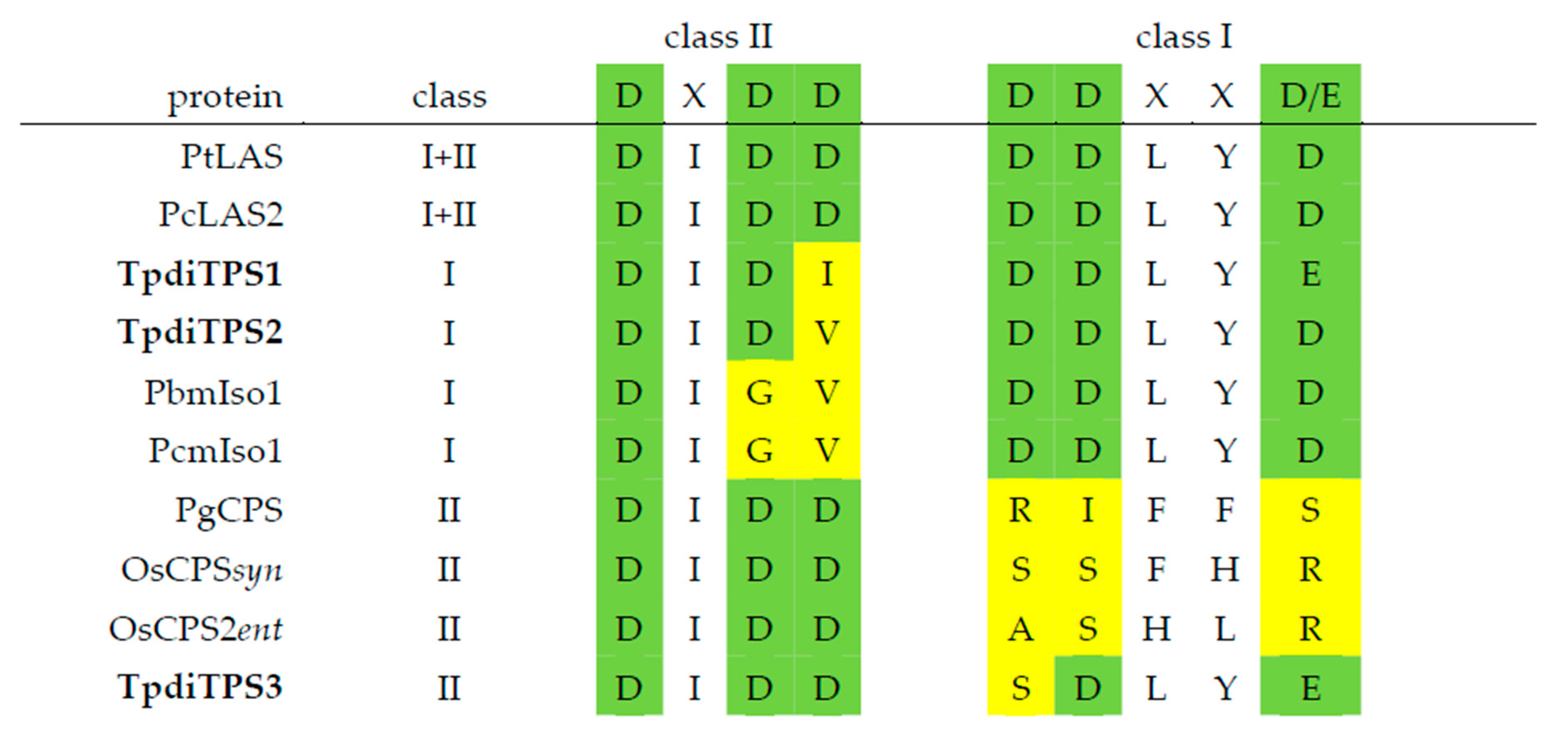
2.1. Identification of Three Putative Diterpene Synthases Expressed in WRC Sap to Heartwood Transition Zone
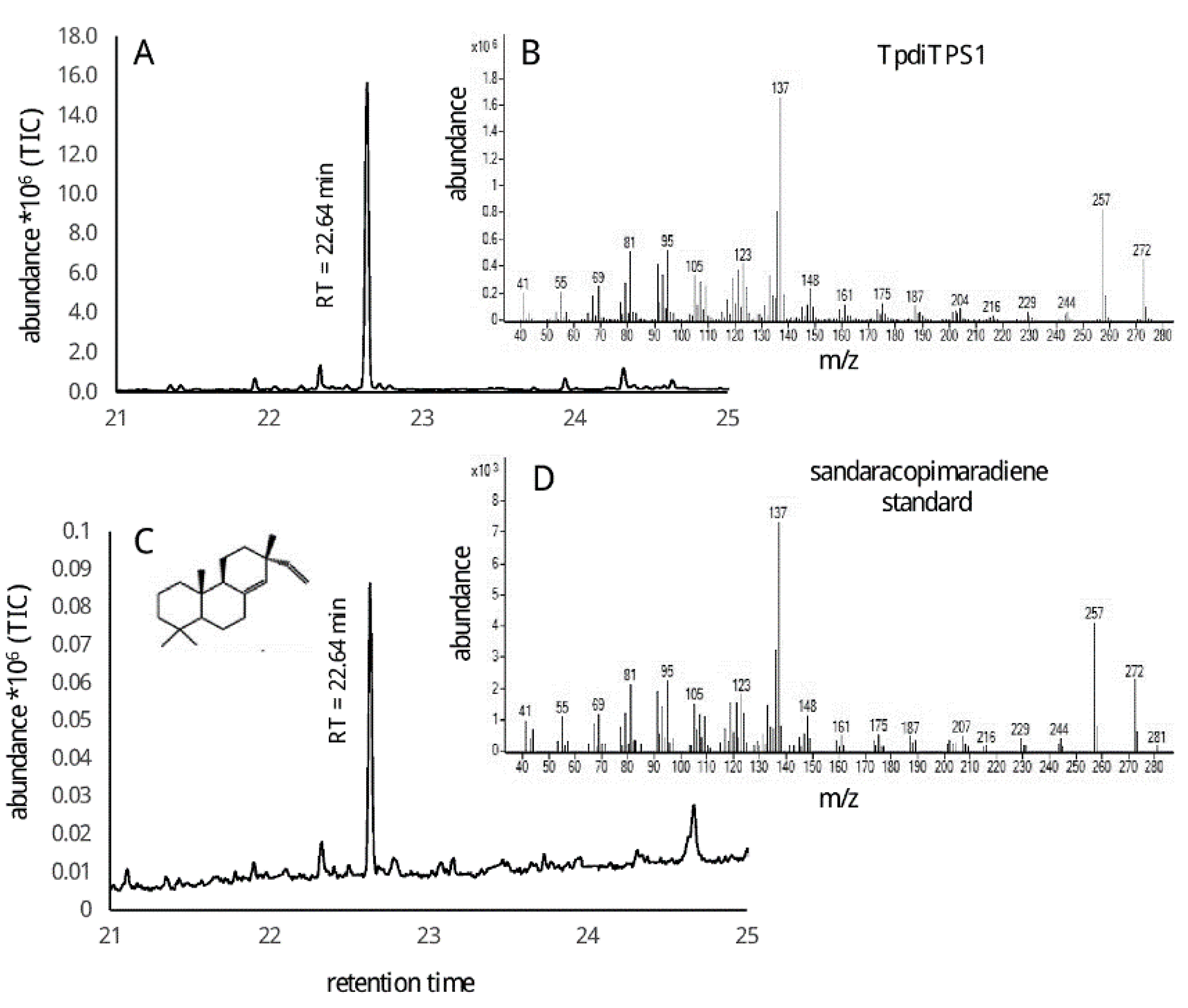
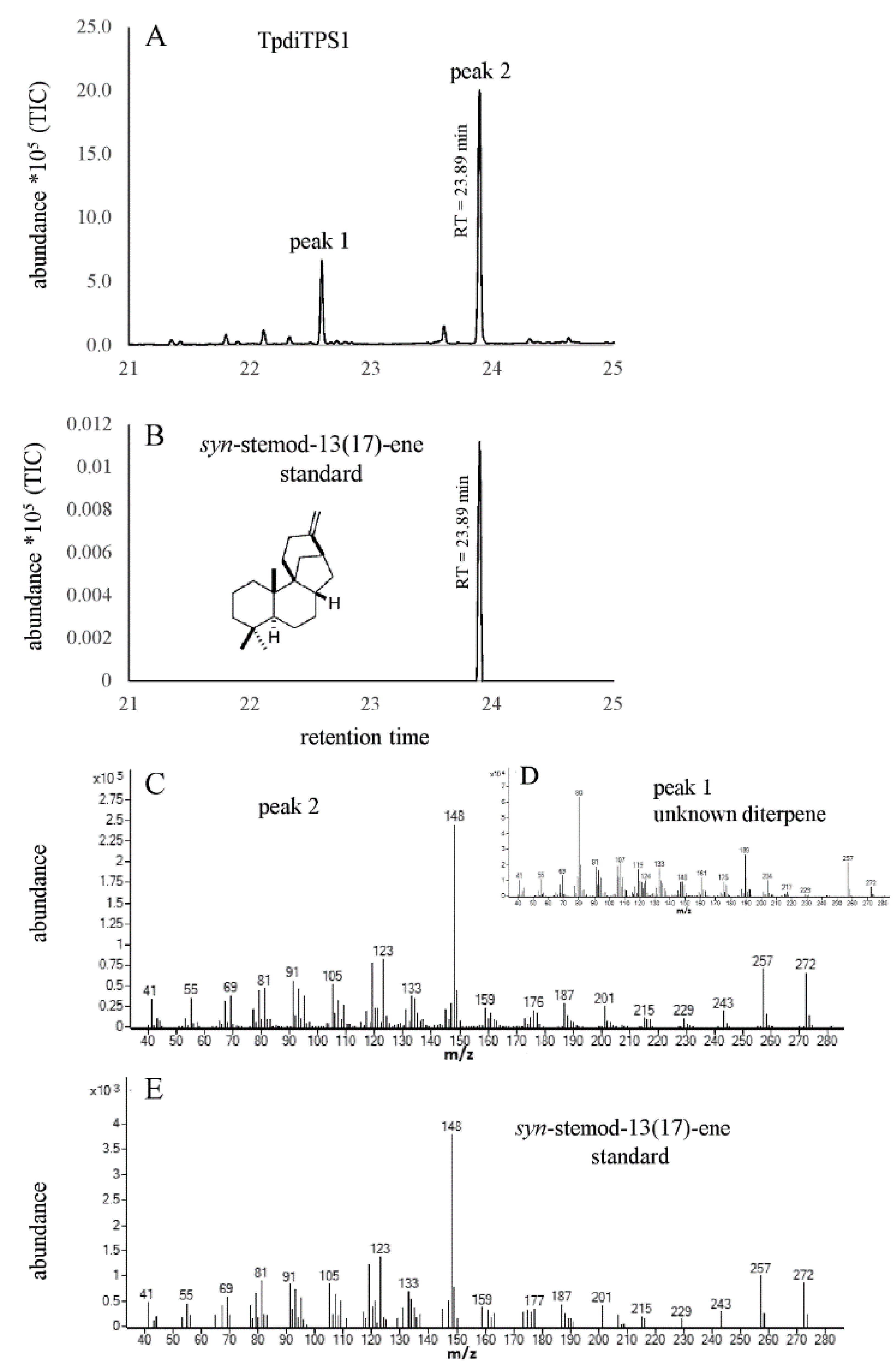
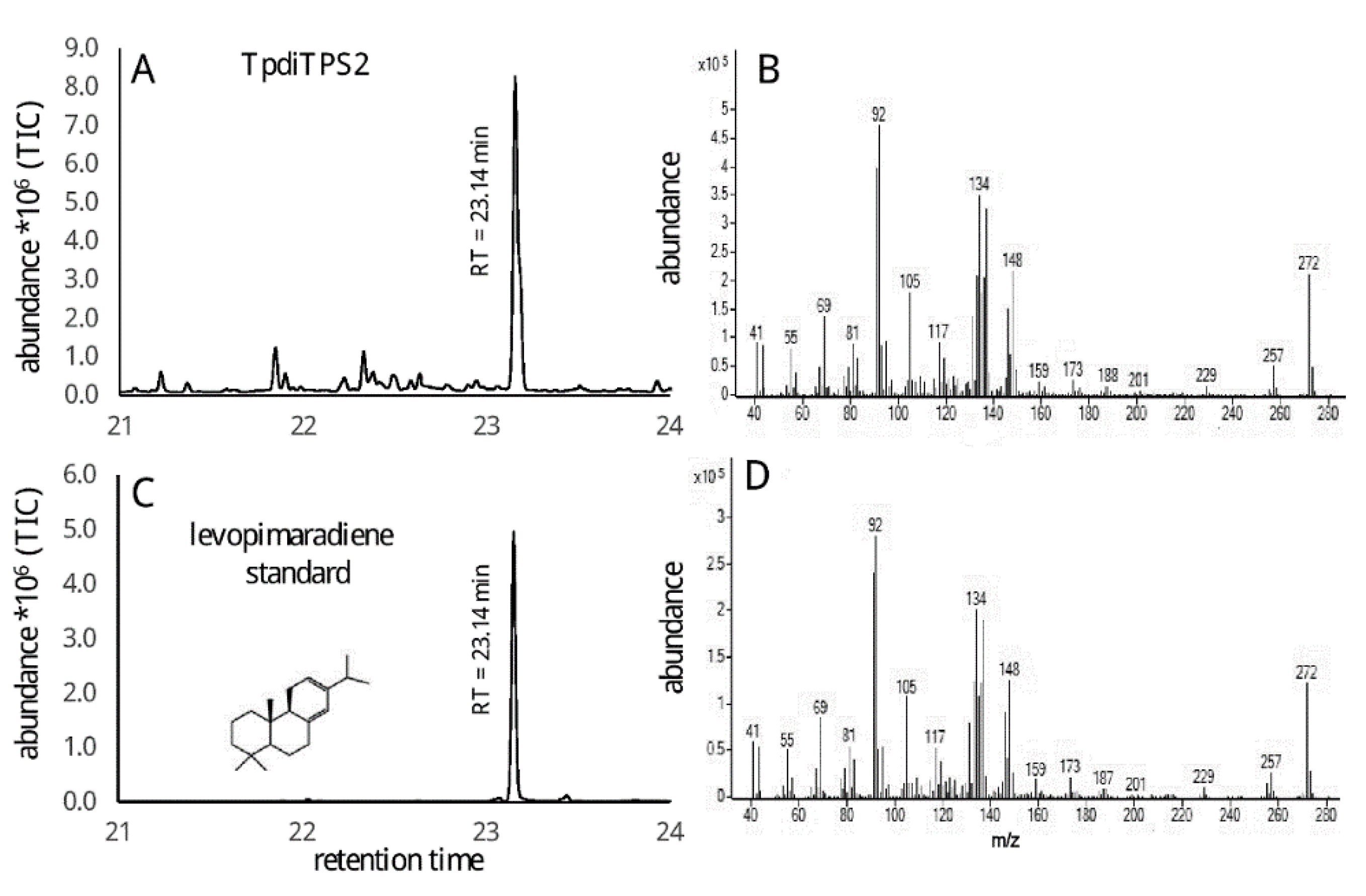
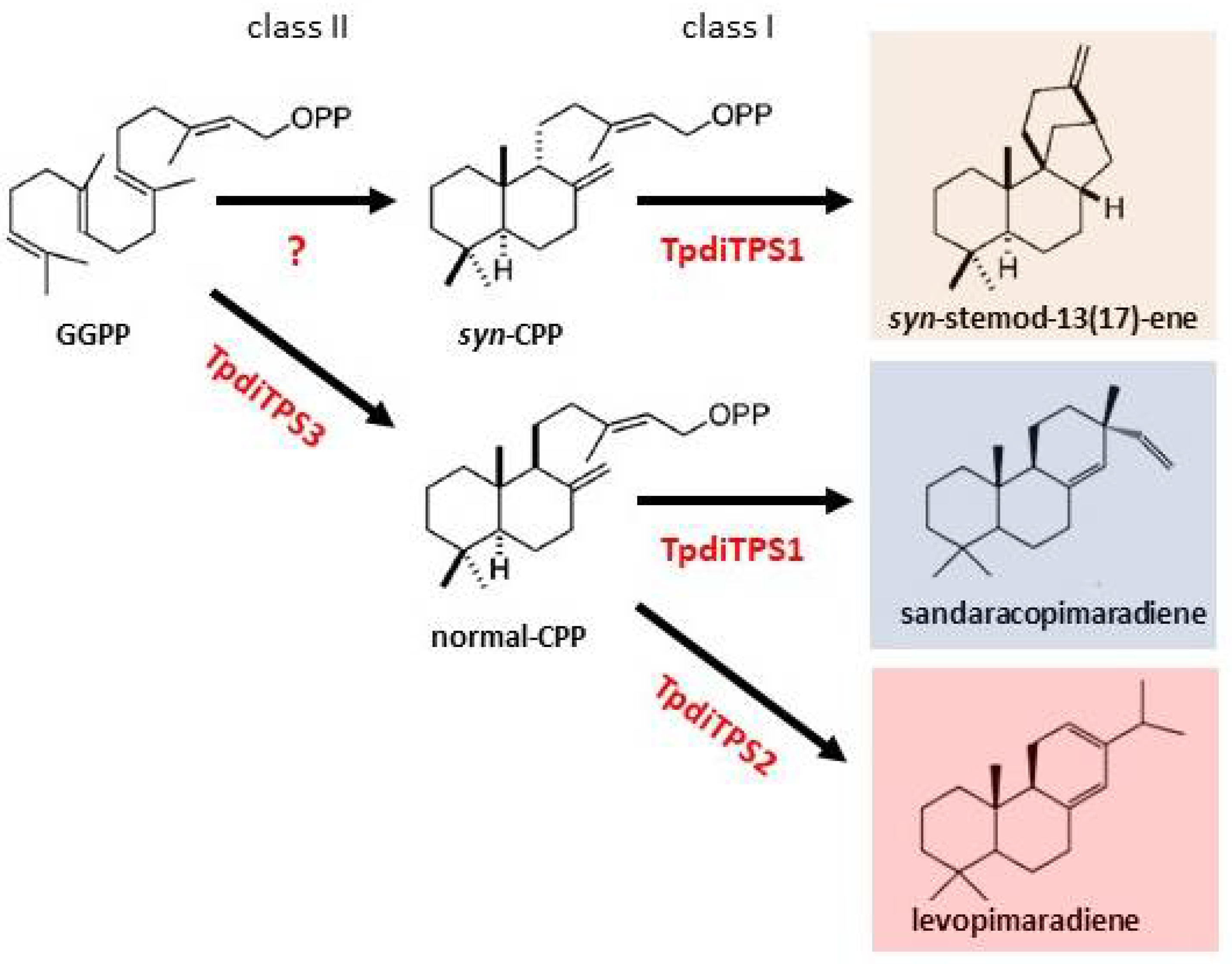
2.2. Assessment of Potential Mono-Functional Class I Diterpene Synthase Activity in TpdiTPS1 and 2
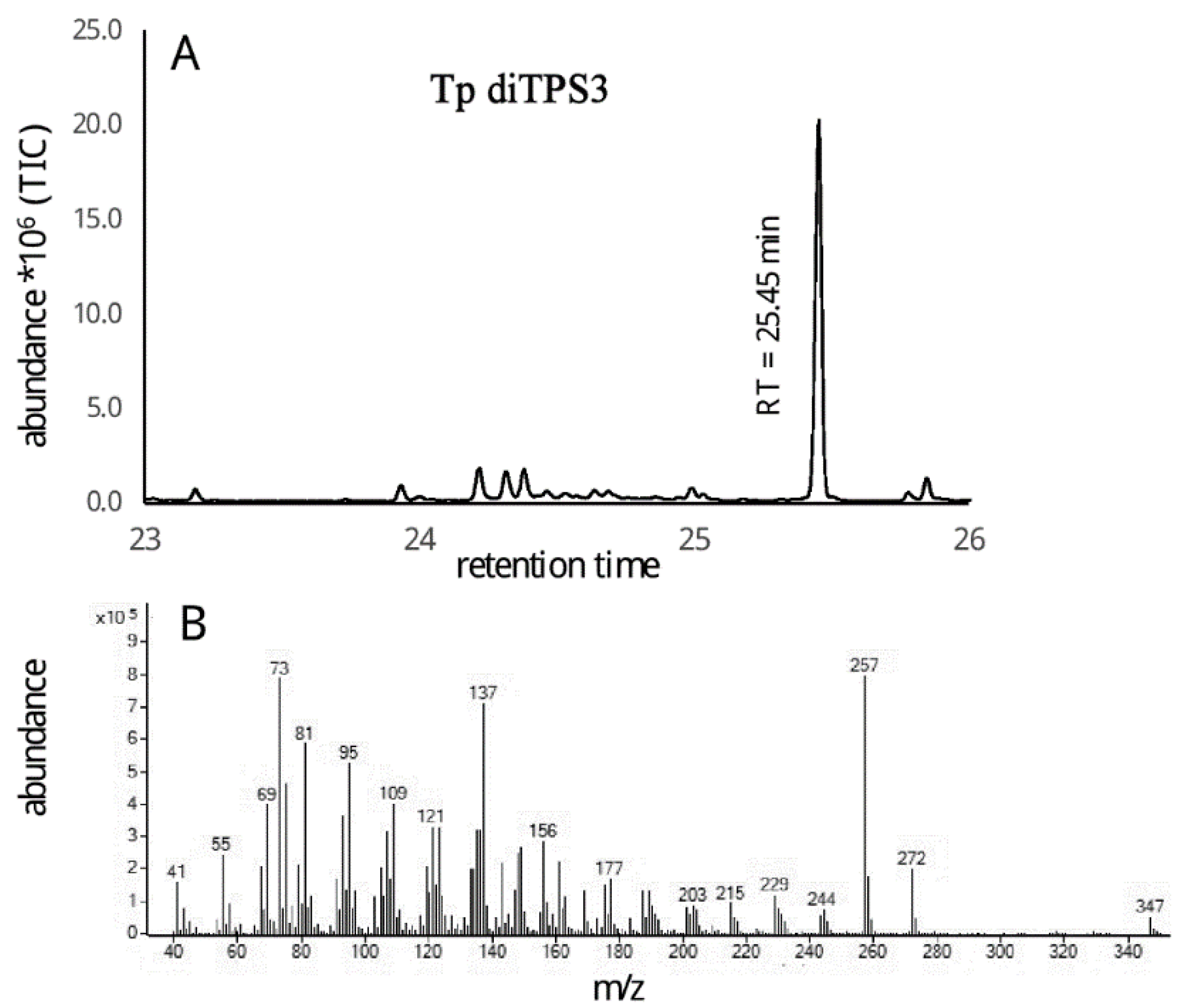
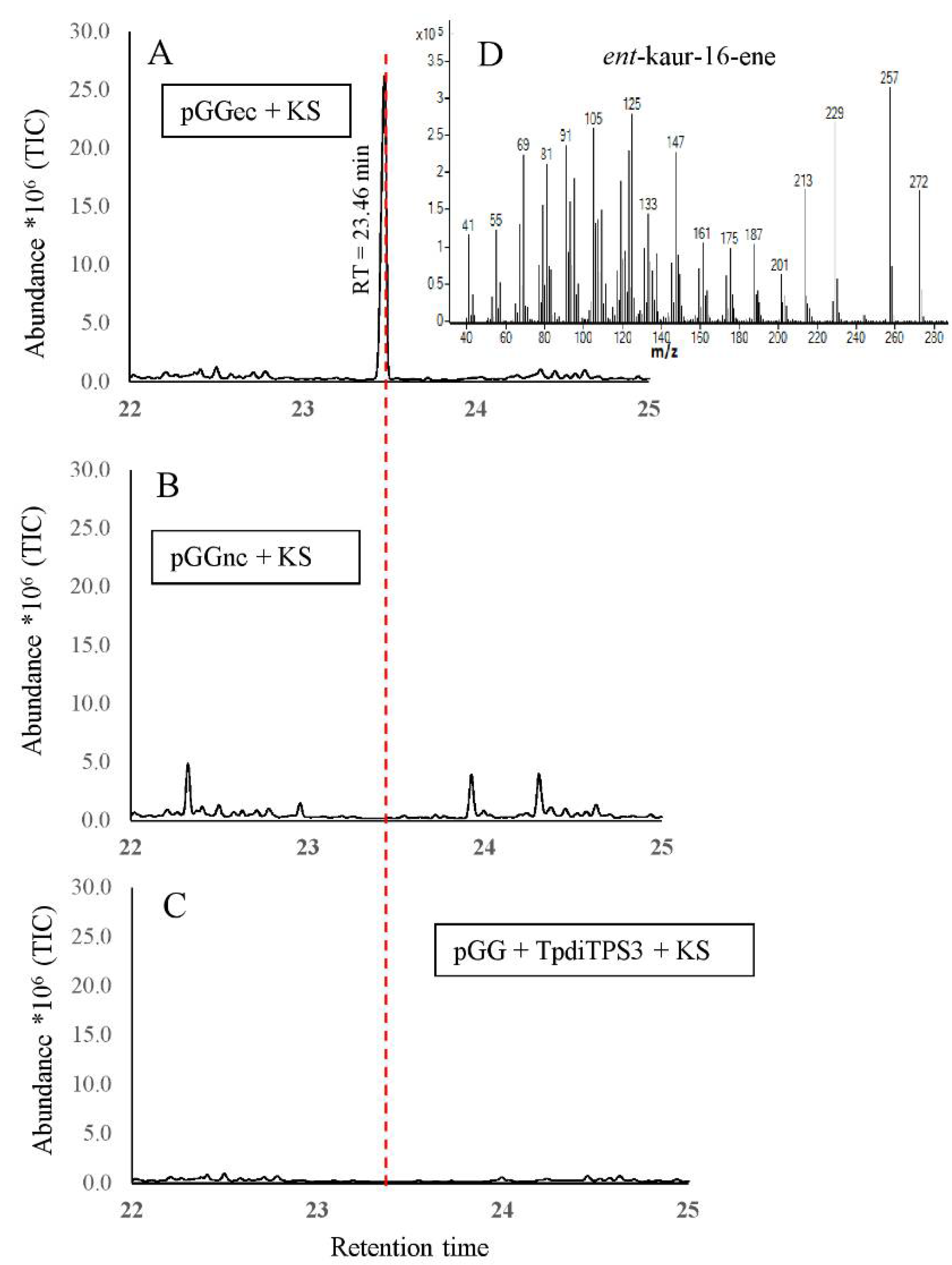
2.3. Monofunctional Class II TpdiTPS3 Synthesize the Intermediate CDP
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Selection of Candidate Genes
4.3. Cloning of diTPS cDNAs
4.4. Transformation and In Vivo Coexpression
4.5. Isolation of Recombinant Protein Products
4.6. Silica and Alumina Chromatography
4.7. GC-MS Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gonzalez, J.S. Growth, Properties and Uses of Western Red Cedar; Forintek Canada Corp.: Vancouver, BC, Canada, 2004. [Google Scholar]
- Gregory, C.; McBeath, A.; Filipescu, C. An Economic Assessment of the Western Redcedar Industry in British Columbia; Technical Report, Information report FI-X-017; Natural Resources Canada, Canadian Forest Service, Canadian Wood Fibre Centre: Victoria, BC, Canada, 2018; p. 38. ISBN 9780660240947. [Google Scholar]
- Hebda, R.J.; Mathewes, R.W. Holocene History of Cedar and Native Indian Cultures of the North American Pacific Coast. Science 1984, 225, 711–713. [Google Scholar] [CrossRef] [PubMed]
- Minore, D. Western Redcedar—A Literature Review; General Technical Report; US Department of Agriculture, Forest Service, Pacific Northwest Forest and Range Experiment Station: Portland, OR, USA, 1983; p. 75. [Google Scholar]
- Sturrock, R.N.; Braybrooks, A.V.; Reece, P. Decay of Living Western Redcedar: A Literature Review; Technical Report, Information report FI-X-014; Natural Resources Canada, Canadian Forest Service, Canadian Wood Fibre Centre: Victoria, BC, Canada, 2017; p. 41. [Google Scholar]
- International Association of Wood Anatomists. Multilingual Glossary of Terms Used in Wood Anatomy (2d. rev.). 1964, pp. 1–26. Available online: http://www.jwrs.org/kenkyu/wa_wp/glossary/IAWA_glossary.pdf (accessed on 12 August 2020).
- Spicer, R. Symplasmic networks in secondary vascular tissues: Parenchyma distribution and activity supporting long-distance transport. J. Exp. Bot. 2014, 65, 1829–1848. [Google Scholar] [CrossRef] [PubMed]
- Spicer, R.; Holbrook, N.M. Parenchyma cell respiration and survival in secondary xylem: Does metabolic activity decline with cell age? Plant Cell Environ. 2007, 30, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Spicer, R. Senescence in Secondary Xylem: Heartwood Formation as an Active Developmental Program. Physiol. Ecol. Vasc. Transp. Plants 2005, 457–475. [Google Scholar] [CrossRef]
- Bannan, M.W. Vertical Resin Ducts in the Secondary Wood of the Abietineae. New Phytol. 1936, 35, 11–46. [Google Scholar] [CrossRef]
- Franceschi, V.R.; Krekling, T.; Christiansen, E. Application of methyl jasmonate on Picea abies (Pinaceae) stems induces defense-related responses in phloem and xylem. Am. J. Bot. 2002, 89, 578–586. [Google Scholar] [CrossRef]
- Martin, D.; Tholl, D.; Gershenzon, J.; Bohlmann, J. Methyl jasmonate induces traumatic resin ducts, terpenoid resin biosynthesis, and terpenoid accumulation in developing xylem of Norway spruce stems. Plant Physiol. 2002, 129, 1003–1018. [Google Scholar] [CrossRef]
- McKay, S.A.B.; Hunter, W.L.; Godard, K.A.; Wang, S.X.; Martin, D.M.; Bohlmann, J.; Plant, A.L. Insect attack and wounding induce traumatic resin duct development and gene expression of (−)-pinene synthase in Sitka spruce. Plant Physiol. 2003, 133, 368–378. [Google Scholar] [CrossRef]
- Phillips, M.A.; Croteau, R.B. Resin-based defenses in conifers. Trends Plant Sci. 1999, 4, 184–190. [Google Scholar] [CrossRef]
- Hudgins, J.W.; Franceschi, V.R. Methyl jasmonate-induced ethylene production is responsible for conifer phloem defense responses and reprogramming of stem cambial zone for traumatic resin duct formation. Plant Physiol. 2004, 135, 2134–2149. [Google Scholar] [CrossRef]
- Shain, L.; Mackay, J.F.G. Seasonal fluctuation in respiration of aging xylem in relation to heartwood formation in Pinus radiata. Can. J. Bot. 1973, 51, 737–741. [Google Scholar] [CrossRef]
- Taylor, A.M.; Gartner, B.L.; Morrell, J.J. Heartwood formation and natural durability—A review. Wood Fiber Sci. 2002, 34, 587–611. [Google Scholar]
- Bamber, R.K. Heartwood, its function and formation. Wood Sci. Technol. 1976, 10, 1–8. [Google Scholar] [CrossRef]
- Morris, P.I.; Stirling, R. Western red cedar extractives associated with durability in ground contact. Wood Sci. Technol. 2012, 46, 991–1002. [Google Scholar] [CrossRef]
- Daniels, C.R.; Russell, J.H. Analysis of western redcedar (Thuja plicata Donn) heartwood components by HPLC as a possible screening tool for trees with enhanced natural durability. J. Chromatogr. Sci. 2007, 45, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Barton, G.M.; Macdonald, B. The chemistry and utilization of western red cedar. Can. For. Serv. 1971, 1023, 31. [Google Scholar]
- Roff, J.W.; Atkinson, J.M. Toxicity tests of water soluble phenolic fraction (thujaplicin-free) of western red cedar. Can. J. Bot. 1954, 32, 308–309. [Google Scholar] [CrossRef]
- Gardner, J.A.F.; Barton, G.M.; Maclean, H. The Polyoxyphenols of Western Red Cedar (Thuja Plicata Donn.): I. Isolation and Preliminary Characterization of Plicatic Acid. Can. J. Chem. 1959, 37, 1703–1709. [Google Scholar] [CrossRef]
- Stirling, R.; Morris, P.I. Potential contributions of lignans to decay resistance in western red cedar. Wood Sci. Technol. 2016, 50, 399–412. [Google Scholar] [CrossRef]
- Buckland, D.C. Investigations of Decay in Western Red Cedar in British Columbia. Can. J. Res. 1946, 24c, 158–181. [Google Scholar] [CrossRef]
- Taylor, A.M.; Gartner, B.L.; Morrell, J.J.; Tsunoda, K. Effects of heartwood extractive fractions of Thuja plicata and Chamaecyparis nootkatensis on wood degradation by termites or fungi. J. Wood Sci. 2006, 52, 147–153. [Google Scholar] [CrossRef]
- Von Rudloff, E.; Nair, G.V. The Sesquiterpene Alcohols of the Heartwood of Thuja Occidentals L. Can. J. Chem. 1964, 42, 421–425. [Google Scholar] [CrossRef]
- Fang, J.M.; Chen, Y.C.; Wang, B.W.; Cheng, Y.S. Terpenes from heartwood of Juniperus chinensis. Phytochemistry 1996, 41, 1361–1365. [Google Scholar] [CrossRef]
- Ro, D.K.; Arimura, G.I.; Lau, S.Y.W.; Piers, E.; Bohlmann, J. Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc. Natl. Acad. Sci. USA 2005, 102, 8060–8065. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Huffaker, A.; Sims, J.W.; Christensen, S.A.; Lu, X.; Okada, K.; Peters, R.J. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014, 79, 659–678. [Google Scholar] [CrossRef] [PubMed]
- Keeling, C.I.; Bohlmann, J. Diterpene resin acids in conifers. Phytochemistry 2006, 67, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Jassbi, A.R.; Gase, K.; Hettenhausen, C.; Schmidt, A.; Baldwin, I.T. Silencing geranylgeranyl diphosphate synthase in Nicotiana attenuata dramatically impairs resistance to tobacco hornworm. Plant Physiol. 2008, 146, 974–986. [Google Scholar] [CrossRef]
- Vaughan, M.M.; Wang, Q.; Webster, F.X.; Kiemle, D.; Hong, Y.J.; Tantillo, D.J.; Coates, R.M.; Wray, A.T.; Askew, W.; O’Donnell, C.; et al. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 2013, 25, 1108–1125. [Google Scholar] [CrossRef]
- Peters, R.J. Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep. 2010, 27, 1521–1530. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Zerbe, P.; Hamberger, B.; Yuen, M.M.S.; Chiang, A.; Sandhu, H.K.; Madilao, L.L.; Nguyen, A.; Hamberger, B.; Bach, S.S.; Bohlmann, J. Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 2013, 162, 1073–1091. [Google Scholar] [CrossRef] [PubMed]
- Prisic, S.; Xu, J.; Coates, R.M.; Peters, R.J. Probing the role of the DXDD motif in class II diterpene cyclases. ChemBioChem 2007, 8, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Hoshino, T. Functional analysis of the DXDDTA motif in squalene-hopene cyclase by site-directed mutagenesis experiments: Initiation site of the polycyclization reaction and stabilization site of the carbocation intermediate of the initially cyclized A-ring. Biosci. Biotechnol. Biochem. 1999, 63, 2189–2198. [Google Scholar] [CrossRef]
- Peters, R.J.; Ravn, M.M.; Coates, R.M.; Croteau, R.B. Bifunctional abietadiene synthase: Free diffusive transfer of the (+)-copalyl diphosphate intermediate between two distinct active sites. J. Am. Chem. Soc. 2001, 123, 8974–8978. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Honzatko, R.B.; Peters, R.J. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat. Prod. Rep. 2012, 29, 1153–1175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Peters, R.J. Electrostatic effects on (di)terpene synthase product outcome. Chem. Commun. 2011, 47, 4074–4080. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wilderman, P.R.; Peters, R.J. Following evolution’s lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. USA 2007, 104, 7387–7401. [Google Scholar] [CrossRef]
- Criswell, J.; Potter, K.; Shephard, F.; Beale, M.H.; Peters, R.J. A single residue change leads to a hydroxylated product from the class II diterpene cyclization catalyzed by abietadiene synthase. Org. Lett. 2012, 14, 5828–5831. [Google Scholar] [CrossRef]
- Wilderman, P.R.; Peters, R.J. A single residue switch converts abietadiene synthase into a pimaradiene specific cyclase. J. Am. Chem. Soc. 2007, 129, 15736–15737. [Google Scholar] [CrossRef]
- Keeling, C.I.; Weisshaar, S.; Lin, R.P.C.; Bohlmann, J. Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc. Natl. Acad. Sci. USA 2008, 105, 1085–1089. [Google Scholar] [CrossRef]
- Funk, C.; Croteau, R. Diterpenoid Resin Acid Biosynthesis in Conifers: Characterization of Two Cytochrome P450-Dependent Monooxygenases and an Aldehyde Dehydrogenase Involved in Abietic Acid Biosynthesis. Arch. Biochem. Biophys. 1994, 308, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Hamberger, B.; Ohnishi, T.; Hamberger, B.; Séguin, A.; Bohlmann, J. Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol. 2011, 157, 1677–1695. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.; Lewinsohn, E.; Vogel, B.S.; Steele, C.L.; Croteau, R. Regulation of Oleoresinosis in Grand Fir (Abies grandis) (Coordinate Induction of Monoterpene and Diterpene Cyclases and Two Cytochrome P450-Dependent Diterpenoid Hydroxylases by Stem Wounding). Plant Physiol. 1994, 106, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Bathe, U.; Tissier, A. Cytochrome P450 enzymes: A driving force of plant diterpene diversity. Phytochemistry 2019, 161, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Geisler, K.; Jensen, N.B.; Yuen, M.M.S.; Madilao, L.; Bohlmann, J. Modularity of conifer diterpene resin acid biosynthesis: P450 enzymes of different CYP720B clades use alternative substrates and converge on the same products. Plant Physiol. 2016, 171, 152–164. [Google Scholar] [CrossRef]
- Zerbe, P.; Bohlmann, J. Plant diterpene synthases: Exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 2015, 33, 419–428. [Google Scholar] [CrossRef]
- Harman-Ware, A.E.; Sykes, R.; Peter, G.F.; Davis, M. Determination of terpenoid content in pine by organic solvent extraction and fast-GC analysis. Front. Energy Res. 2016, 4, 1–9. [Google Scholar] [CrossRef]
- Jiang, Z.; Kempinski, C.; Chappell, J. Extraction and Analysis of Terpenes/Terpenoids. Curr. Protoc. Plant Biol. 2016, 1, 345–358. [Google Scholar] [CrossRef]
- Lewinsohn, E.; Savage, T.J.; Gijzen, M.; Croteau, R. Simultaneous analysis of monoterpenes and diterpenoids of conifer oleoresin. Phytochem. Anal. 1993, 4, 220–225. [Google Scholar] [CrossRef]
- Porter, L.J. The resin and fatty acid content of living Pinus radiata wood. N. Z. J. Sci. 1969, 12, 687–693. [Google Scholar]
- Hemingway, R.W.; HILLIS, W.E. Changes in fats and resins of Pinus radiata associated with heartwood formation. Appita 1971, 24, 439. [Google Scholar]
- Lloyd, J.A. Distribution of extractives in Pinus radiata earlywood and latewood. N. Z. J. For. Sci. 1978, 8, 288–294. [Google Scholar]
- Davis, E.M.; Croteau, R. Cyclization Enzymes in the Biosynthesis of Monoterpenes, Sesquiterpenes, and Diterpenes. Top. Curr. Chem. 2000, 209, 53–95. [Google Scholar]
- Christianson, D. Structural Biology and Chemistry of the Terpenoid Cyclases. Chem. Rev. 2006, 106, 3412–3442. [Google Scholar] [CrossRef] [PubMed]
- Cyr, A.; Wilderman, P.R.; Determan, M.; Peters, R.J. A modular approach for facile biosynthesis of labdane-related diterpenes. J. Am. Chem. Soc. 2007, 129, 6684–6685. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hillwig, M.L.; Tiernan, M.S.; Peters, R.J. Probing Labdane-Related Diterpenoid Biosynthesis in the Fungal Genus Aspergillus. J. Nat. Prod. 2017, 80, 328–333. [Google Scholar] [CrossRef]
- Martin, D.M.; Fäldt, J.; Bohlmann, J.; Martin, D.M.; Faldt, J.; Bohlmann, J. Functional Characterization of Nine Norway Spruce TPS Genes and Evolution of Gymnosperm Terpene Synthases of the TPS-d Subfamily. Am. Soc. Plant Biol. (ASPB) 2019, 135, 1908–1927. [Google Scholar] [CrossRef]
- Peters, R.J.; Flory, J.E.; Jetter, R.; Ravn, M.M.; Lee, H.J.; Coates, R.M.; Croteau, R.B. Abietadiene synthase from grand fir (Abies grandis): Characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry 2000, 39, 15592–15602. [Google Scholar]
- Peters, R.J.; Croteau, R.B. Abietadiene synthase catalysis: Conserved residues involved in protonation-initiated cyclization of geranylgeranyl diphosphate to (+)-copalyl diphosphate. Biochemistry 2002, 41, 1836–1842. [Google Scholar]
- Wu, Y.; Zhou, K.; Toyomasu, T.; Sugawara, C.; Oku, M.; Abe, S.; Usui, M.; Mitsuhashi, W.; Chono, M.; Chandler, P.M.; et al. Functional characterization of wheat copalyl diphosphate synthases sheds light on the early evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 2012, 84, 40–46. [Google Scholar] [CrossRef]
- Ma, L.T.; Lee, Y.R.; Tsao, N.W.; Wang, S.Y.; Zerbe, P.; Chu, F.H. Biochemical characterization of diterpene synthases of Taiwania cryptomerioides expands the known functional space of specialized diterpene metabolism in gymnosperms. Plant J. 2019, 100, 1254–1272. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.M.; Ma, L.T.; Ding, Y.; Schmelz, E.A.; Zerbe, P. Functional characterization of two class ii diterpene synthases indicates additional specialized diterpenoid pathways in maize (Zea mays). Front. Plant Sci. 2018, 871, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Hillwig, M.L.; Huango, L.; Cui, G.; Wang, X.; Kong, J.; Yang, B.; Peters, R.J. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett. 2009, 11, 5170–5173. [Google Scholar] [CrossRef] [PubMed]
- Vogel, B.S.; Wildung, M.R.; Vogel, G.; Croteau, R. Abietadiene Synthase from Grand Fir (Abies grandis). J. Biol. Chem. 1996, 271, 23262–23268. [Google Scholar] [PubMed]
- Schepmann, H.G.; Pang, J.; Matsuda, S.P.T. Cloning and characterization of Ginkgo biloba levopimaradiene synthase, which catalyzes the first committed step in ginkgolide biosynthesis. Arch. Biochem. Biophys. 2001, 392, 263–269. [Google Scholar] [CrossRef]
- Ro, D.K.; Bohlmann, J. Diterpene resin acid biosynthesis in loblolly pine (Pinus taeda): Functional characterization of abietadiene/levopimaradiene synthase (PtTPS-LAS) cDNA and subcellular targeting of PtTPS-LAS and abietadienol/abietadienal oxidase (PtAO, CYP720B1). Phytochemistry 2006, 67, 1572–1578. [Google Scholar] [CrossRef]
- Morrone, D.; Hillwig, M.L.; Mead, M.E.; Lowry, L.; Fulton, D.B.; Peters, R.J. Evident and latent plasticity across the rice diterpene synthase family with potential implications for the evolution of diterpenoid metabolism in the cereals. Biochem. J. 2011, 435, 589–595. [Google Scholar] [CrossRef]
- Hall, D.E.; Zerbe, P.; Jancsik, S.; Quesada, A.L.; Dullat, H.; Madilao, L.L.; Yuen, M.; Bohlmann, J. Evolution of conifer diterpene synthases: Diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol. 2013, 161, 600–616. [Google Scholar] [CrossRef]
- Keeling, C.I.; Weisshaar, S.; Ralph, S.G.; Jancsik, S.; Hamberger, B.; Dullat, H.K.; Bohlmann, J. Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biol. 2011, 11, 43. [Google Scholar] [CrossRef]
- Morrone, D.; Jin, Y.; Xu, M.; Choi, S.Y.; Coates, R.M.; Peters, R.J. An unexpected diterpene cyclase from rice: Functional identification of a stemodene synthase. Arch. Biochem. Biophys. 2006, 448, 133–140. [Google Scholar] [CrossRef]
- Keeling, C.I.; Madilao, L.L.; Zerbe, P.; Dullat, H.K.; Bohlmann, J. The primary diterpene synthase products of Picea abies levopimaradiene/abietadiene synthase (PaLAS) are epimers of a thermally unstable diterpenol. J. Biol. Chem. 2011, 286, 21145–21153. [Google Scholar] [CrossRef] [PubMed]
- Nault, J. Radial distribution of thujaplicins in old growth and second growth western red cedar (Thuja plicata Donn). Wood Sci. Technol. 1988, 22, 73–80. [Google Scholar] [CrossRef]
- Schultz, T.P.; Nicholas, D.D. Naturally durable heartwood: Evidence for a proposed dual defensive function of the extractives. Phytochemistry 2000, 54, 47–52. [Google Scholar] [CrossRef]
- Erdtman, H.; Gripenberg, J. Antibiotic Substances from the Heart Wood of Thuja plicata Donn. Nature 1948, 161, 719. [Google Scholar] [CrossRef] [PubMed]
- Stirling, R.; Morris, P.I. New Perspectives on the Role of Extractives in the Durability of Western Redcedar. Proc. Can. Wood Preserv. Assoc. 2011, 32, 12. [Google Scholar]
- Swan, E.P.; Jiang, K.S. Formation of heartwood extractives in Western Red Cedar. TAPPI 1970, 53, 844–846. [Google Scholar]
- Iwamoto, M.; Ohtsu, H.; Tokuda, H.; Nishino, H.; Matsunaga, S.; Tanaka, R. Anti-tumor promoting diterpenes from the stem bark of Thuja standishii (Cupressaceae). Bioorg. Med. Chem. 2001, 9, 1911–1921. [Google Scholar] [CrossRef]
- Azémard, C.; Ménager, M.; Vieillescazes, C. On the tracks of sandarac, review and chemical analysis. Environ. Sci. Pollut. Res. 2017, 24, 27746–27754. [Google Scholar] [CrossRef]
- Sugimoto, N.; Kuroyanagi, M.; Kato, T.; Sato, K.; Tada, A.; Yamazaki, T.; Tanamoto, K. Identification of the main constituents in sandarac resin, a natural gum base. J. Food Hyg. Soc. Jpn. 2006, 47, 76–79. [Google Scholar] [CrossRef]
- Edwards, O.E.; Nicolson, A.; Rodger, M.N. The Structure of Sandaracopimaric Acid. Can. J. Chem. 1960, 38, 663–667. [Google Scholar] [CrossRef]
- Simoneit, B.R.T.; Cox, R.E.; Oros, D.R.; Otto, A. Terpenoid compositions of resins from Callitris species (Cupressaceae). Molecules 2018, 23, 3384. [Google Scholar] [CrossRef]
- Micales, J.A.; Han, J.S.; Davis, J.L.; Young, R.A. Chemical Composition and Fungitoxic Activities of Pine Cone Extractives. In Mycotoxins, Wood Decay, Plant Stress, Biocorrosion, and General Biodeterioration; Springer: Boston, MA, USA, 1994; Volume 4, pp. 317–332. [Google Scholar]
- Ejike, C.E.C.C.; Gong, M.; Udenigwe, C.C. Phytoalexins from the Poaceae: Biosynthesis, function and prospects in food preservation. Food Res. Int. 2013, 52, 167–177. [Google Scholar] [CrossRef]
- Peters, R.J. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 2006, 67, 2307–2317. [Google Scholar] [CrossRef]
- Koga, J.; Shimura, M.; Oshima, K.; Ogawa, N.; Yamauchi, T.; Ogasawara, N. Phytocassanes A, B, C and D, novel diterpene phytoalexins from rice, Oryza sativa L. Tetrahedron 1995, 51, 7907–7918. [Google Scholar] [CrossRef]
- Koga, J.; Ogawa, N.; Yamauchi, T.; Klkuchi, M.; Ogasawara, N.; Shimura, M. Functional moiety for the antifungal activity of phytocassane E, a diterpene phytoalexin from rice. Phytochemistry 1997, 44, 249–253. [Google Scholar] [CrossRef]
- Fukuta, M.; Xuan, T.D.; Deba, F.; Tawata, S.; Khanh, T.D.; Chung, I.M. Comparative efficacies in vitro of antibacterial, fungicidal, antioxidant, and herbicidal activities of momilatones A and B. J. Plant Interact. 2007, 2, 245–251. [Google Scholar] [CrossRef]
- Keeling, C.I.; Bohlmann, J. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 2006, 170, 657–675. [Google Scholar] [CrossRef]
- Aranda, P.S.; LaJoie, D.M.; Jorcyk, C.L. Bleach Gel. Natl. Inst. Health 2012, 33, 366–369. [Google Scholar]
- Li, M.Z.; Elledge, S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 2007, 4, 251–256. [Google Scholar] [CrossRef]
- Morrone, D.; Lowry, L.; Determan, M.K.; Hershey, D.M.; Xu, M.; Peters, R.J. Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 2010, 85, 1893–1906. [Google Scholar] [CrossRef]
- Reiling, K.K.; Yoshikuni, Y.; Martin, V.J.J.; Newman, J.; Bohlmann, J.; Keasling, J.D. Mono and diterpene production in Escherichia coli. Biotechnol. Bioeng. 2004, 87, 200–212. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tasnim, S.; Gries, R.; Mattsson, J. Identification of Three Monofunctional Diterpene Synthases with Specific Enzyme Activities Expressed during Heartwood Formation in Western Redcedar (Thuja plicata) Trees. Plants 2020, 9, 1018. https://doi.org/10.3390/plants9081018
Tasnim S, Gries R, Mattsson J. Identification of Three Monofunctional Diterpene Synthases with Specific Enzyme Activities Expressed during Heartwood Formation in Western Redcedar (Thuja plicata) Trees. Plants. 2020; 9(8):1018. https://doi.org/10.3390/plants9081018
Chicago/Turabian StyleTasnim, Sifat, Regine Gries, and Jim Mattsson. 2020. "Identification of Three Monofunctional Diterpene Synthases with Specific Enzyme Activities Expressed during Heartwood Formation in Western Redcedar (Thuja plicata) Trees" Plants 9, no. 8: 1018. https://doi.org/10.3390/plants9081018
APA StyleTasnim, S., Gries, R., & Mattsson, J. (2020). Identification of Three Monofunctional Diterpene Synthases with Specific Enzyme Activities Expressed during Heartwood Formation in Western Redcedar (Thuja plicata) Trees. Plants, 9(8), 1018. https://doi.org/10.3390/plants9081018