Advances in Plant Regeneration: Shake, Rattle and Roll
Abstract
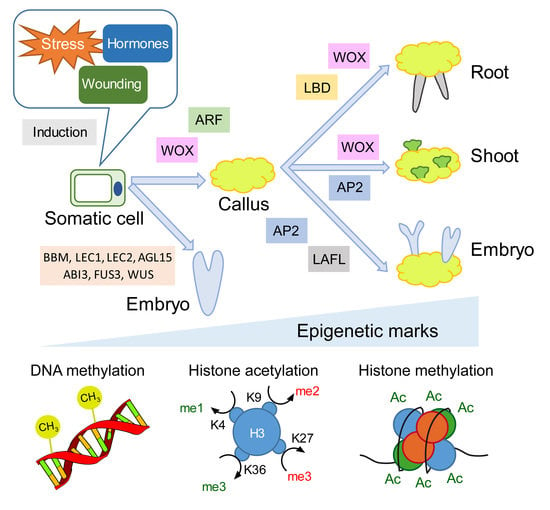
1. Introduction
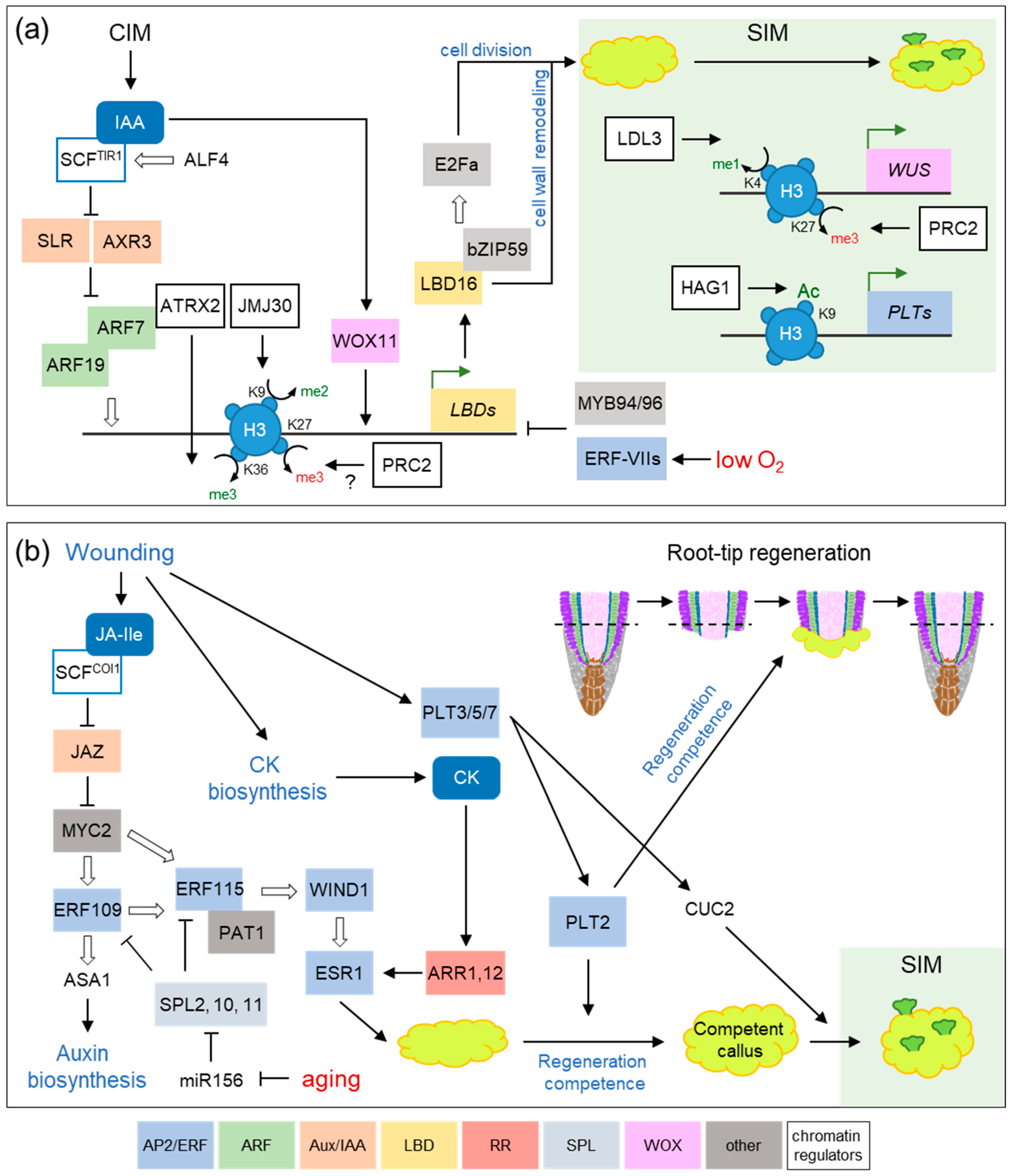
2. Transcription Factor Networks and Epigenetic Regulators during Hormone-Induced Callus Formation
3. Wound Signaling Regulates Tissue Regeneration through Conserved Gene Regulatory Networks
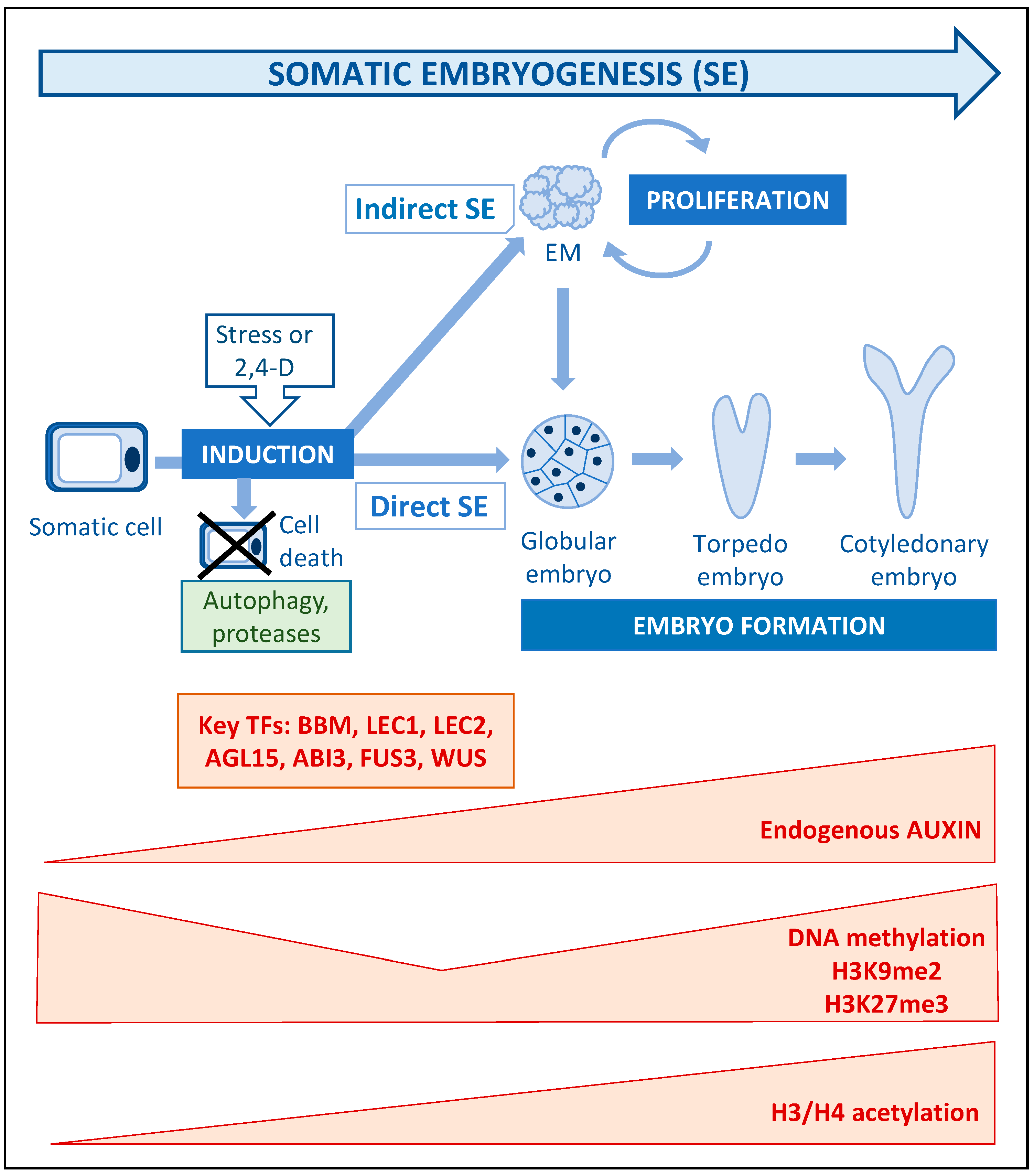
4. Somatic Embryogenesis: Stress, Auxin and Epigenetic Modifications as Key Players of Cell Totipotency Expression
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xu, L.; Huang, H. Genetic and epigenetic controls of plant regeneration. Curr. Top. Dev. Biol. 2014, 108, 1–33. [Google Scholar] [PubMed]
- Skoog, F.; Miller, C.O. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 1957, 11, 118–130. [Google Scholar] [PubMed]
- Sugimoto, K.; Jiao, Y.; Meyerowitz, E.M. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 2010, 18, 463–471. [Google Scholar] [CrossRef]
- DiDonato, R.J.; Arbuckle, E.; Buker, S.; Sheets, J.; Tobar, J.; Totong, R.; Grisafi, P.; Fink, G.R.; Celenza, J.L. Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J. 2004, 37, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, R.; Melnyk, C.W.; Christ, G.; Winkler, M.; Kirchsteiner, K.; Salehin, M.; Mergner, J.; Niemeyer, M.; Schwechheimer, C.; Calderón Villalobos, L.I.A.; et al. The Arabidopsis ALF 4 protein is a regulator of SCF E3 ligases. EMBO J. 2018, 37, 255–268. [Google Scholar] [CrossRef]
- Kim, Y.; Park, C.; Cha, S.; Han, M.; Ryu, K.-S.; Suh, J.-Y. Determinants of PB1 domain interactions in auxin response factor ARF5 and repressor IAA17. J. Mol. Biol. 2020. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A molecular framework for auxin-controlled homeostasis of shoot stem cells in arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef]
- Lee, K.; Park, O.S.; Seo, P.J. Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Liu, J.; Hu, X.; Qin, P.; Prasad, K.; Hu, Y.; Xu, L. The WOX11–LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 2018, 59, 739–748. [Google Scholar] [CrossRef]
- Jing, T.; Ardiansyah, R.; Xu, Q.; Xing, Q.; Müller-Xing, R. Reprogramming of cell fate during root regeneration by transcriptional and epigenetic networks. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Lee, K.; Seo, P.J. dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 2018, 23, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Lee, U.S.; Wagner, D. Tug of war: Adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 2016, 34, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Knowles, S.M.; Webb, C.J.; Celaya, R.B.; Cha, C.; Siu, J.P.; Tobin, E.M. The jumonji C domain-containing protein JMJ30 regulates period length in the arabidopsis circadian clock. Plant Physiol. 2011, 155, 906–915. [Google Scholar] [CrossRef] [PubMed]
- Gan, E.S.; Xu, Y.; Wong, J.Y.; Geraldine Goh, J.; Sun, B.; Wee, W.Y.; Huang, J.; Ito, T. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 2014, 5, 1–13. [Google Scholar] [CrossRef]
- Lee, K.; Park, O.-S.; Seo, P.J. JMJ30-mediated demethylation of H3K9me3 drives tissue identity changes to promote callus formation in Arabidopsis. Plant J. 2018, 95, 961–975. [Google Scholar] [CrossRef]
- Wiles, E.T.; Selker, E.U. H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2017, 43, 31–37. [Google Scholar] [CrossRef]
- He, C.; Chen, X.; Huang, H.; Xu, L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured arabidopsis tissues. PLoS Genet. 2012, 8, e1002911. [Google Scholar] [CrossRef]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef]
- Zhang, T.Q.; Lian, H.; Zhou, C.M.; Xu, L.; Jiao, Y.; Wang, J.W. A two-stepmodel for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 2017, 29, 1073–1087. [Google Scholar] [CrossRef]
- Liu, Y.; Tian, T.; Zhang, K.; You, Q.; Yan, H.; Zhao, N.; Yi, X.; Xu, W.; Su, Z. PCSD: A plant chromatin state database. Nucleic Acids Res. 2018, 46, D1157–D1167. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Minucci, S. A comprehensive review of Lysine-Specific Demethylase 1 and its roles in cancer. Epigenomics 2017, 9, 1123–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Yang, S.; Liu, X.; Wu, K. Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front. Plant Sci. 2015, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Martignago, D.; Bernardini, B.; Polticelli, F.; Salvi, D.; Cona, A.; Angelini, R.; Tavladoraki, P. The four FAD-dependent histone demethylases of arabidopsis are differently involved in the control of flowering time. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Hung, F.Y.; Chen, F.F.; Li, C.; Chen, C.; Lai, Y.C.; Chen, J.H.; Cui, Y.; Wu, K. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018, 46, 10669–10681. [Google Scholar] [CrossRef]
- Ishihara, H.; Sugimoto, K.; Tarr, P.T.; Temman, H.; Kadokura, S.; Inui, Y.; Sakamoto, T.; Sasaki, T.; Aida, M.; Suzuki, T.; et al. Primed histone demethylation regulates shoot regenerative competency. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Adamo, A.; Sesé, B.; Boue, S.; Castaño, J.; Paramonov, I.; Barrero, M.J.; Belmonte, J.C.I. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 2011, 13, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Kornet, N.; Scheres, B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 2009, 21, 1070–1079. [Google Scholar] [CrossRef]
- Kim, J.; Yang, W.; Forner, J.; Lohmann, J.U.; Noh, B.; Noh, Y. Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.K.; Li, H.J.; Metzger, E.; Schule, R.; Leiter, A.B. CtBP and associated LSD1 are required for transcriptional activation by NeuroD1 in gastrointestinal endocrine cells. Mol. Cell. Biol. 2014, 34, 2308–2317. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cao, H.; Zhang, Q.; Wang, H.; Xin, W.; Xu, E.; Zhang, S.; Yu, R.; Yu, D.; Hu, Y. Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nat. Plants 2018, 4, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cao, H.; Xu, E.; Zhang, S.; Hu, Y. Genome-wide identification of arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation. Plant Cell Physiol. 2018, 59, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liu, N.; Wang, L.; Li, J.; Zheng, X.; Xiang, F.; Liu, Z. MYB94 and MYB96 additively inhibit callus formation via directly repressing LBD29 expression in Arabidopsis thaliana. Plant Sci. 2020, 293, 110323. [Google Scholar] [CrossRef] [PubMed]
- Shang, B.; Xu, C.; Zhang, X.; Cao, H.; Xin, W.; Hu, Y. Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 5101–5106. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, T.; Okushima, Y.; Nagata, N.; Kojima, M.; Sakakibara, H.; Umeda, M. Synthesis of Very-Long-Chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 2013, 11, e1001531. [Google Scholar] [CrossRef]
- Grison, M.S.; Brocard, L.; Fouillen, L.; Nicolas, W.; Wewer, V.; Dörmann, P.; Nacir, H.; Benitez-Alfonso, Y.; Claverol, S.; Germain, V.; et al. Specific membrane lipid composition is important for plasmodesmata function in arabidopsis. Plant Cell 2015, 27, 1228–1250. [Google Scholar] [CrossRef]
- Yan, D.; Yadav, S.R.; Paterlini, A.; Nicolas, W.J.; Petit, J.D.; Brocard, L.; Belevich, I.; Grison, M.S.; Vaten, A.; Karami, L.; et al. Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat. Plants 2019, 5, 604–615. [Google Scholar] [CrossRef]
- Lup, S.D.; Tian, X.; Xu, J.; Pérez-Pérez, J.M. Wound signaling of regenerative cell reprogramming. Plant Sci. 2016, 250, 178–187. [Google Scholar] [CrossRef]
- Sena, G.; Wang, X.; Liu, H.Y.; Hofhuis, H.; Birnbaum, K.D. Organ regeneration does not require a functional stem cell niche in plants. Nature 2009, 457, 1150–1153. [Google Scholar] [CrossRef]
- Efroni, I.; Mello, A.; Nawy, T.; Ip, P.L.; Rahni, R.; Delrose, N.; Powers, A.; Satija, R.; Birnbaum, K.D. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 2016, 165, 1721–1733. [Google Scholar] [CrossRef]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K.; et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 2011, 21, 508–514. [Google Scholar] [CrossRef]
- Iwase, A.; Harashima, H.; Ikeuchi, M.; Rymen, B.; Ohnuma, M.; Komaki, S.; Morohashi, K.; Kurata, T.; Nakata, M.; Ohme-Takagi, M.; et al. WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in arabidopsis. Plant Cell 2017, 29, 54–69. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Rymen, B.; Lambolez, A.; Kojima, M.; Takebayashi, Y.; Heyman, J.; Watanabe, S.; Seo, M.; De Veylder, L.; et al. Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 2017, 175, 1158–1174. [Google Scholar] [CrossRef]
- Bustillo-Avendaño, E.; Ibáñez, S.; Sanz, O.; Barros, J.A.S.; Gude, I.; Perianez-Rodriguez, J.; Micol, J.L.; del Pozo, J.C.; Moreno-Risueno, M.A.; Pérez-Pérez, J.M. Regulation of hormonal control, cell reprogramming, and patterning during de novo root organogenesis. Plant Physiol. 2018, 176, 1709–1727. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dai, X.; Li, J.; Liu, N.; Liu, X.; Li, S.; Xiang, F. The Type-B cytokinin response regulator ARR1 inhibits shoot regeneration in an ARR12-dependent manner in Arabidopsis. Plant Cell 2020, tpc.00022.2019. [Google Scholar] [CrossRef]
- Heyman, J.; Cools, T.; Vandenbussche, F.; Heyndrickx, K.S.; Van Leene, J.; Vercauteren, I.; Vanderauwera, S.; Vandepoele, K.; De Jaeger, G.; Van Der Straeten, D.; et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 2013, 342, 860–863. [Google Scholar] [CrossRef] [PubMed]
- Heyman, J.; Cools, T.; Canher, B.; Shavialenka, S.; Traas, J.; Vercauteren, I.; Van Den Daele, H.; Persiau, G.; De Jaeger, G.; Sugimoto, K.; et al. The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence. Nat. Plants 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Rymen, B.; Kawamura, A.; Lambolez, A.; Inagaki, S.; Takebayashi, A.; Iwase, A.; Sakamoto, Y.; Sako, K.; Favero, D.S.; Ikeuchi, M.; et al. Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun. Biol. 2019, 404. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Borg, M.; Berger, F.; Chen, Z. The atypical histone variant H3.15 promotes callus formation in Arabidopsis thaliana. Development 2020, 147, dev184895. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Morishita, M.; Higuchi, Y.; Ichikawa, S.; Ishikawa, T.; Nishiyama, T.; Kabeya, Y.; Hiwatashi, Y.; Kurata, T.; Kubo, M.; et al. Physcomitrella STEMIN transcription factor introduces stem cell formation with epigenetic reprogramming. Nat. Plants 2019, 5, 681–690. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transciption. J. Exp. Bot. 2016, 68, erw443. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant regeneration: Cellular origins and molecular mechanisms. Development 2016, 143, 1442–1451. [Google Scholar] [CrossRef]
- Zhang, T.Q.; Lian, H.; Tang, H.; Dolezal, K.; Zhou, C.M.; Yu, S.; Chen, J.H.; Chen, Q.; Liu, H.; Ljung, K.; et al. An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell. 2015, 27, 349–360. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Massoumi, M.; Krens, F.A.; Visser, R.G.F.; De Klerk, G.-J.M. Azacytidine and miR156 promote rooting in adult but not in juvenile Arabidopsis tissues. J. Plant Physiol. 2017, 208, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Shang, G.D.; Pan, Y.; Xu, Z.G.; Zhou, C.M.; Mao, Y.B.; Bao, N.; Sun, L.; Xu, T.; Wang, J.W. AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 2020, 32, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Kareem, A.; Durgaprasad, K.; Sugimoto, K.; Du, Y.; Pulianmackal, A.J.; Trivedi, Z.B.; Abhayadev, P.V.; Pinon, V.; Meyerowitz, E.M.; Scheres, B.; et al. PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 2015, 25, 1017–1030. [Google Scholar] [CrossRef]
- Durgaprasad, K.; Roy, M.V.; Venugopal, A.; Kareem, A.; Raj, K.; Willemsen, V.; Mähönen, A.P.; Scheres, B.; Prasad, K. Gradient expression of transcription factor imposes a boundary on organ regeneration potential in plants. Cell Rep. 2019, 29, 453–463.e3. [Google Scholar] [CrossRef] [PubMed]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, D.; Shanmukhan, A.P.; Kareem, A.; Aiyaz, M.; Varapparambathu, V.; Toms, A.; Kerstens, M.; Valsakumar, D.; Landge, A.N.; Shaji, A.; et al. A coherent feed-forward loop drives vascular regeneration in damaged aerial organs of plants growing in a normal developmental context. Development 2020, 147, dev185710. [Google Scholar] [CrossRef]
- Germana, M.; Lambardi, M. In Vitro Embryogenesis in Higher Plants, 1st ed.; Humana Press-Springer: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Loyola-Vargas, V.; Ochoa-Alejo, N. Somatic Embryogenesis: Fundamental Aspects and Applications, 1st ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Díaz-Sala, C. Molecular dissection of the regenerative capacity of forest tree species: Special focus on conifers. Front. Plant Sci. 2019, 9, 1943. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Jain, S.M.; Gupta, P.K. Step Wise Protocols for Somatic Embryogenesis of Important Woody Plants, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Nic-Can, G.I.; Loyola-Vargas, V.M. The role of the auxins during somatic embryogenesis. In Somatic Embryogenesis: Fundamental Aspects and Applications, 1st ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 171–182. [Google Scholar] [CrossRef]
- Pais, M.S. Somatic embryogenesis induction in woody species: The future after OMICs data assessment. Front. Plant Sci. 2019, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Testillano, P.S. Microspore embryogenesis: Targeting the determinant factors of stress-induced cell reprogramming for crop improvement. J. Exp. Bot. 2019, 70, 2965–2978. [Google Scholar] [CrossRef]
- Bárány, I.; González-Melendi, P.; Fadón, B.; Mitykó, J.; Risueño, M.C.; Testillano, P.S. Microspore-derived embryogenesis in pepper (Capsicum annuum L.): Subcellular rearrangements through development. Biol. Cell 2005, 97, 709–722. [Google Scholar] [CrossRef]
- Custers, J.B.M.; Cordewener, J.H.G.; Nöllen, Y.; Dons, H.J.M.; Van Lockeren Campagne, M.M. Temperature controls both gametophytic and sporophytic development in microspore cultures of Brassica napus. Plant Cell Rep. 1994, 13, 267–271. [Google Scholar] [CrossRef]
- Prem, D.; Solís, M.T.; Bárány, I.; Rodríguez-Sanz, H.; Risueño, M.C.; Testillano, P.S. A new microspore embryogenesis system under low temperature which mimics zygotic embryogenesis initials, expresses auxin and efficiently regenerates doubled-haploid plants in Brassica napus. BMC Plant Biol. 2012, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The resurgence of haploids in higher plants. Trends Plant Sci. 2007, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Maluszynski, M.; Kasha, K.; Forster, B.; Szarejko, I. Doubled Haploid Production in Crop Plants: A Manual, 1st ed.; Springer: New York, NY, USA, 2003. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Bárány, I.; Prem, D.; Coronado, M.; Risueño, M.; Testillano, P. NO, ROS, and cell death associated with caspase-like activity increase in stress-induced microspore embryogenesis of barley. J. Exp. Bot. 2012, 63, 2007–2024. [Google Scholar] [CrossRef] [PubMed]
- Satpute, G.K.; Long, H.; Seguí-Simarro, J.M.; Risueño, M.C.; Testillano, P.S. Cell architecture during gametophytic and embryogenic microspore development in Brassica napus L. Acta Physiol. Plant. 2005, 27, 665–674. [Google Scholar] [CrossRef]
- Bárány, I.; Berenguer, E.; Solís, M.-T.; Pérez-Pérez, Y.; Santamaría, M.E.; Crespo, J.L.; Risueño, M.C.; Díaz, I.; Testillano, P.S. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 2018, 69, 1387–1402. [Google Scholar] [CrossRef]
- Berenguer, E.; Solís, M.-T.; Pérez-Pérez, Y.; Testillano, P.S. Proteases with caspase 3-like activity participate in cell death during stress-induced microspore embryogenesis of Brassica napus. EuroBiotech J. 2019, 3, 152–159. [Google Scholar] [CrossRef]
- Berenguer, E.; Minina, E.; Bárány, I.; Carneros, E.; Bozhkov, P.; Testillano, P.S. Suppression of metacaspase and autophagy-dependent cell death improves stress-induced microspore embryogenesis in Brassica napus. Plant Cell Physiol. 2020. First Revision. [Google Scholar]
- Pérez-Pérez, Y.; Bárány, I.; Berenguer, E.; Carneros, E.; Risueño, M.C.; Testillano, P.S. Modulation of autophagy and protease activities by small bioactive compounds to reduce cell death and improve stress-induced microspore embryogenesis initiation in rapeseed and barley. Plant Signal. Behav. 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef]
- Méndez-Hernández, H.A.; Ledezma-Rodríguez, M.; Avilez-Montalvo, R.N.; Juárez-Gómez, Y.L.; Skeete, A.; Avilez-Montalvo, J.; De-La-Peña, C.; Loyola-Vargas, V.M. Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 2019, 10, 77. [Google Scholar] [CrossRef]
- Rodríguez-Sanz, H.; Solís, M.; López, M.; Gómez-Cadenas, A.; Risueño, M.; Testillano, P. Auxin biosynthesis, accumulation, action and transport are involved in stress-induced microspore embryogenesis initiation and progression in Brassica napus. Plant Cell Physiol. 2015, 56. [Google Scholar] [CrossRef]
- Wójcik, A.M.; Wójcikowska, B.; Gaj, M.D. Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. Int. J. Mol. Sci. 2020, 21, 1333. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, Y.; El-Tantawy, A.-A.; Solís, M.T.; Risueño, M.C.; Testillano, P.S. Stress-induced microspore embryogenesis requires endogenous auxin synthesis and polar transport in barley. Front. Plant Sci. 2019, 10, 1200. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Gaj, M.D. Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 2017, 36, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, X.; Yuan, D.; Jin, F.; Zhang, Y.; Xu, J. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Corredoira, E.; Cano, V.; Bárány, I.; Solís, M.T.; Rodríguez, H.; Vieitez, A.M.; Risueño, M.C.; Testillano, P.S. Initiation of leaf somatic embryogenesis involves high pectin esterification, auxin accumulation and DNA demethylation in Quercus alba. J. Plant Physiol. 2017, 213, 42–54. [Google Scholar] [CrossRef]
- Grafi, G. Epigenetics in plant development and response to stress. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 351–352. [Google Scholar] [CrossRef]
- El-Tantawy, A.A.; Solís, M.T.; Risueño, M.C.; Testillano, P.S. Changes in DNA methylation levels and nuclear distribution patterns after microspore reprogramming to embryogenesis in barley. Cytogenet. Genome Res. 2014, 143, 200–208. [Google Scholar] [CrossRef]
- Solís, M.; Rodríguez-Serrano, M.; Meijón, M.; Canal, M.; Cifuentes, A.; Risueño, M.; Testillano, P. DNA methylation dynamics and MET1a-like gene expression changes during stress-induced pollen reprogramming to embryogenesis. J. Exp. Bot. 2012, 63, 6431–6444. [Google Scholar] [CrossRef]
- Berenguer, E.; Bárány, I.; Solís, M.-T.; Pérez-Pérez, Y.; Risueño, M.C.; Testillano, P.S. Inhibition of histone H3K9 methylation by BIX-01294 promotes stress-induced microspore totipotency and enhances embryogenesis initiation. Front. Plant Sci. 2017, 8, 1161. [Google Scholar] [CrossRef]
- De-la-Peña, C.; Nic-Can, G.I.; Galaz-Ávalos, R.M.; Avilez-Montalvo, R.; Loyola-Vargas, V.M. The role of chromatin modifications in somatic embryogenesis in plants. Front. Plant Sci. 2015, 6, 635. [Google Scholar] [CrossRef]
- Rodríguez-Sanz, H.; Moreno-Romero, J.; Solís, M.T.; Köhler, C.; Risueño, M.C.; Testillano, P.S. Changes in histone methylation and acetylation during microspore reprogramming to embryogenesis occur concomitantly with BnHKMT and BnHAT expression and are associated with cell totipotency, proliferation, and differentiation in Brassica napus. Cytogenet. Genome Res. 2014, 143, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Mozgová, I.; Muñoz-Viana, R.; Hennig, L. PRC2 represses hormone-induced somatic embryogenesis in vegetative tissue of Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006562. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Montalvo, P.; Sáenz-Carbonell, L.; De-la-Peña, C. 5-azacytidine: A promoter of epigenetic changes in the quest to improve plant somatic embryogenesis. Int. J. Mol. Sci. 2018, 19, 3182. [Google Scholar] [CrossRef] [PubMed]
- Solís, M.T.; El-Tantawy, A.A.; Cano, V.; Risueño, M.C.; Testillano, P.S. 5-azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Soriano, M.; Cordewener, J.; Muiño, J.M.; Riksen, T.; Fukuok, H.; Angenent, G.C.; Boutilier, K. The histone deacetylase inhibitor trichostatin a promotes totipotency in the male gametophyte. Plant Cell 2014, 26, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Botor, M.; Morończyk, J.; Wójcik, A.M.; Nodzyński, T.; Karcz, J.; Gaj, M.D. Trichostatin a triggers an embryogenic transition in arabidopsis explants via an auxin-related pathway. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Ji, L.; Mathioni, S.M.; Johnson, S.; Tucker, D.; Bewick, A.J.; Kim, K.D.; Daron, J.; Slotkin, R.K.; Jackson, S.A.; Parrott, W.A.; et al. Genome-wide reinforcement of DNA methylation occurs during somatic embryogenesis in soybean. Plant Cell 2019, 31, 2315–2331. [Google Scholar] [CrossRef]
- Su, Y.H.; Tang, L.P.; Zhao, X.Y.; Zhang, X.S. Plant cell totipotency: Insights into cellular reprogramming. J. Integr. Plant Biol. 2020, jipb.12972. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 2019, 70, 377–406. [Google Scholar] [CrossRef]
- Iwafuchi-Doi, M.; Zaret, K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014, 28, 2679–2692. [Google Scholar] [CrossRef] [PubMed]
- Mohyeldin, A.; Garzón-Muvdi, T.; Quiñones-Hinojosa, A. Oxygen in stem cell biology: A critical component of the stem cell niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; Van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Conde, J.V.; Berckhan, S.; Prasad, G.; Mendiondo, G.M.; Holdsworth, M.J. Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 2015, 169, 23–31. [Google Scholar] [CrossRef]
- Weits, D.A.; Kunkowska, A.B.; Kamps, N.C.W.; Portz, K.M.S.; Packbier, N.K.; Nemec Venza, Z.; Gaillochet, C.; Lohmann, J.U.; Pedersen, O.; van Dongen, J.T.; et al. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 2019, 569, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Lombardi, L.; Iacopino, S.; Pencik, A.; Novak, O.; Perata, P.; Giuntoli, B.; Licausi, F. Endogenous Hypoxia in Lateral Root Primordia Controls Root Architecture by Antagonizing Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 538–551. [Google Scholar] [CrossRef] [PubMed]
| Genes | Abbreviations | Function in Plant Regeneration | Molecular Function |
|---|---|---|---|
| ABA INSENSITIVE 3 | ABI3 | Quantitatively regulates BBM-mediated somatic embryogenesis. Acts as a positive regulator | Dof-type transcription factor |
| ABERRANT LATERAL ROOT FORMATION 4 | ALF4 | Formative divisions of XPP cells during LR formation. Callus formation upon CIM induction | SCFTIR1 regulation |
| AGAMOUS LIKE 15 | AGL15 | Activates auxin biosynthesis, leading to totipotency acquisition and SE initiation | MADS domain transcription factor |
| ANTHRANILATE SYNTHASE α1 | ASA1 | Tryptophan biosynthesis | Oxo-acid-lyase enzyme |
| ARABIDOPSIS RESPONSE REGULATOR 1 and 12 | ARR1 and 12 | Involved in CK-mediated ESR1 induction in order to promote shoot regeneration | Type-B Arabidopsis response regulator transcription factors |
| ARABIDOPSIS TRITHORAX-RELATED 2 | ATXR2 | Positively regulates LBD16 and LBD29 expression upon CIM induction | Histone lysine methyltransferase |
| AUXIN RESISTANT 3 | AXR3, IAA17 | Transcriptional repressor upon low auxin levels. Controls stem cell maintenance | Aux/IAA corepressor |
| AUXIN RESPONSE FACTOR 7 and 19 | ARF7 and 19 | LR formation / Positively regulates LBD16 and LBD29 expression upon CIM induction | Auxin-responsive transcription factor |
| BABY BOOM | BBM, PLT4, AIL5 | Its ectopic expression can also directly reprogram somatic cells and induce SE in the absence of exogenous stimuli | AP2/ERF transcription factor |
| BASIC REGION/LEUCINE ZIPPER MOTIF 59 | bZIP59 | Interacts with LBD16 upon CIM induction | bZIP transcription factor |
| E2 PROMOTER BINDING FACTOR a | E2Fa | DNA replication | E2F transcription factor |
| ENHANCER OF SHOOT REGENERATION 1 | ESR1 | Induces the expression of key shoot regulators (CUC1, RAP2.6L, ESR2, WUS, and STM) to promote shoot regeneration | AP2/ERF transcription factor |
| ETHYLENE RESPONSE FACTOR 109 | ERF109 | Up-regulates ERF115 expression. Up-regulates ASA1 expression, probably involved in the auxin biosynthetic pathway | AP2/ERF transcription factor |
| ETHYLENE RESPONSE FACTOR 115 | ERF115 | Acts as as a rate-limiting factor for quiescent center (QC) cell division after DNA damaging stress. Involved in WIND1 up-regulation upon wound signaling | AP2/ERF transcription factor |
| FUSCA 3 | FUS3 | Involved in embryo development. Essential for successful SE | B3 domain-containing transcription factor |
| GENERAL CONTROL NONREPRESSED 5 | GCN5, HAG1 | Root stem cell niche maintenance. Callus pluripotency and shoot induction upon SIM | Histone acetyltransferase |
| JASMONATE-ZIM DOMAIN PROTEINS | JAZ PROTEINS | Represses de novo root formation in Arabidopsis leaf explants. Their destabilization allows the action of positive regulators | Jasmonate zinc-finger inflorescence meristem domain transcription factor |
| JUMONJI C DOMAIN-CONTAINING 30 | JMJ30, JMJD5 | Positively regulates LBD16 and LBD29 expression upon CIM induction | Histone lysine demethylase |
| LATERAL ORGAN BOUNDARIES DOMAIN 16, 17, 18 and 29 | LBD16, 17, 18 and 29 | Callus formation upon CIM induction | LOB-domain transcription factor |
| LEAFY COTYLEDON 1 | LEC1 | Its ectopic expression can also directly reprogram somatic cells and induce SE in the absence of exogenous stimuli | B3 domain-containing transcription factor |
| LEAFY COTYLEDON 2 | LEC2 | Its ectopic expression can also directly reprogram somatic cells and induce SE in the absence of exogenous stimuli | B3 domain-containing transcription factor |
| LYSINE-SPECIFIC DEMETHYLASE 1-LIKE 3 | LDL3 | Presumably removes H3K4me2 during callus formation. It may allow the genes for shoot initiation to be expressed after SIM treatment | Histone lysine demethylase |
| microRNA156 | miRNA156 | Reduces SPL2, 10 and 11 expression, promoting AR formation | microRNA molecule |
| MONOPTEROS | MP, ARF5 | Hypophysis specification during embryogenesis | Auxin-responsive transcription factor |
| MYB94 and 96 | MYB94 and 96 | Regulates LBD29 expression upon CIM induction | MYB transcription factors |
| MYC2 | MYC2 | Acts upstream of ERF109 as a positive regulator | bHLH transcription factor |
| PHYTOCHROME A SIGNAL TRANSDUCTION 1 | PAT1 | Acts as a partner of ERF115 and induces WIND1 expression | GRAS transcription factor |
| PIN-FORMED 1 | PIN1 | Auxin transport | Auxin efflux facilitator |
| PLETHORA 3, 5 and 7 | PLT3, 5 and 7 | Induce the expression of genes involved in regeneration competence acquisition (PLT2) and differentiation factors (i.e., CUC2) | AP2/ERF transcription factor |
| POLYCOMB REPRESSIVE COMPLEX 2 | PRC2 | Di- and tri-methylation of Lys27 on histone H3. PRC2 activity blocks hormone-mediated SE | Histone lysine methyltransferase |
| RWP-RK DOMAIN-CONTAINING 4 | RKD4, GRD | Induces early embryo-specific genes when overexpressed in seedlings. Its ectopic expression can also directly reprogram somatic cells and induce SE in the absence of exogenous stimuli | RWP-RK-type transcription factor |
| SOLITARY ROOT 1 | SLR1, IAA14 | Formative divisions of XPP cells during LR formation | Aux/IAA corepressor |
| SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2, 10 and 11 | SPL2, 10 and 11 | Their up-regulation is linked to a decrease in wound-induced ARs, presumably due to the repression of ABR1, ERF109, ERF115 and RAP2.6L, among others | SPL transcription factor |
| TAA-RELATED 2 | TAR2 | Auxin biosynthesis | Tryptophan aminotransferase enzyme |
| TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 | TAA1 | Auxin biosynthesis | Tryptophan aminotransferase enzyme |
| WOUND INDUCED DEDIFFERENTIATION 1 | WIND1, RAP2.4 | Establishes and maintains dedifferentiated cell status | AP2/ERF transcription factor |
| WUSCHEL | WUS | Shoot induction upon SIM | Homeobox transcription factor |
| WUSCHEL RELATED HOMEOBOX 11 and 12 | WOX11 and 12 | Positively regulates LBD16 and LBD29 expression upon CIM induction | Homeobox transcription factor |
| YUCCA 1 and 4 | YUC1 and 4 | Auxin biosynthesis | Flavin-containing monooxygenase enzymes |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibáñez, S.; Carneros, E.; Testillano, P.S.; Pérez-Pérez, J.M. Advances in Plant Regeneration: Shake, Rattle and Roll. Plants 2020, 9, 897. https://doi.org/10.3390/plants9070897
Ibáñez S, Carneros E, Testillano PS, Pérez-Pérez JM. Advances in Plant Regeneration: Shake, Rattle and Roll. Plants. 2020; 9(7):897. https://doi.org/10.3390/plants9070897
Chicago/Turabian StyleIbáñez, Sergio, Elena Carneros, Pilar S. Testillano, and José Manuel Pérez-Pérez. 2020. "Advances in Plant Regeneration: Shake, Rattle and Roll" Plants 9, no. 7: 897. https://doi.org/10.3390/plants9070897
APA StyleIbáñez, S., Carneros, E., Testillano, P. S., & Pérez-Pérez, J. M. (2020). Advances in Plant Regeneration: Shake, Rattle and Roll. Plants, 9(7), 897. https://doi.org/10.3390/plants9070897