Heterologous Expression of the Constitutive Disease Resistance 2 and 8 Genes from Poncirus trifoliata Restored the Hypersensitive Response and Resistance of Arabidopsis cdr1 Mutant to Bacterial Pathogen Pseudomonas syringae
Abstract
1. Introduction
2. Results
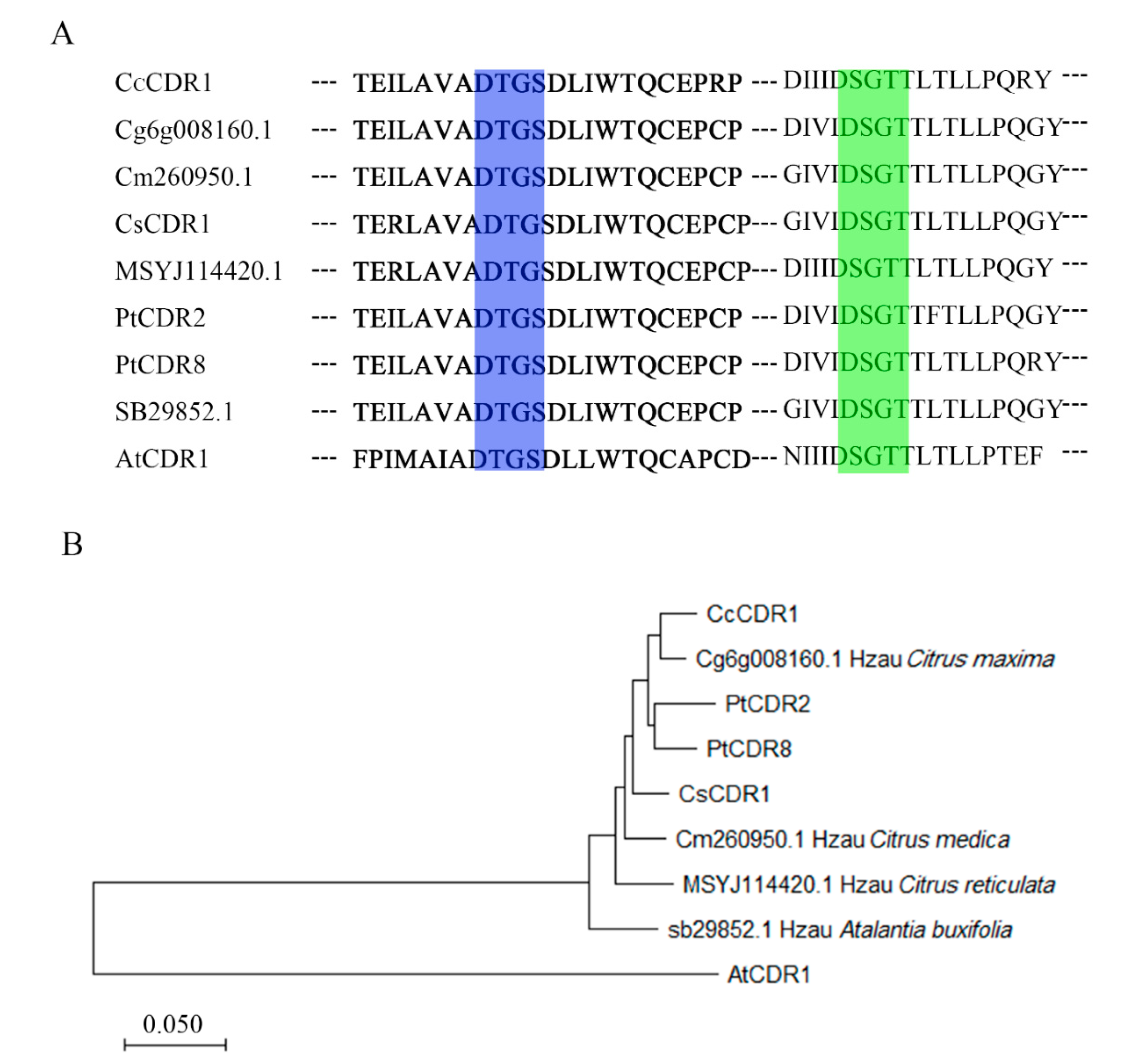
2.1. Cloning and Structure of Constitutive Disease Resistance 2 and 8 from Poncirus trifoliata
2.2. PtCDR2 and PtCDR8 Restored the Hypersensitive Response of Arabidopsis cdr1 Mutant to Pathogen
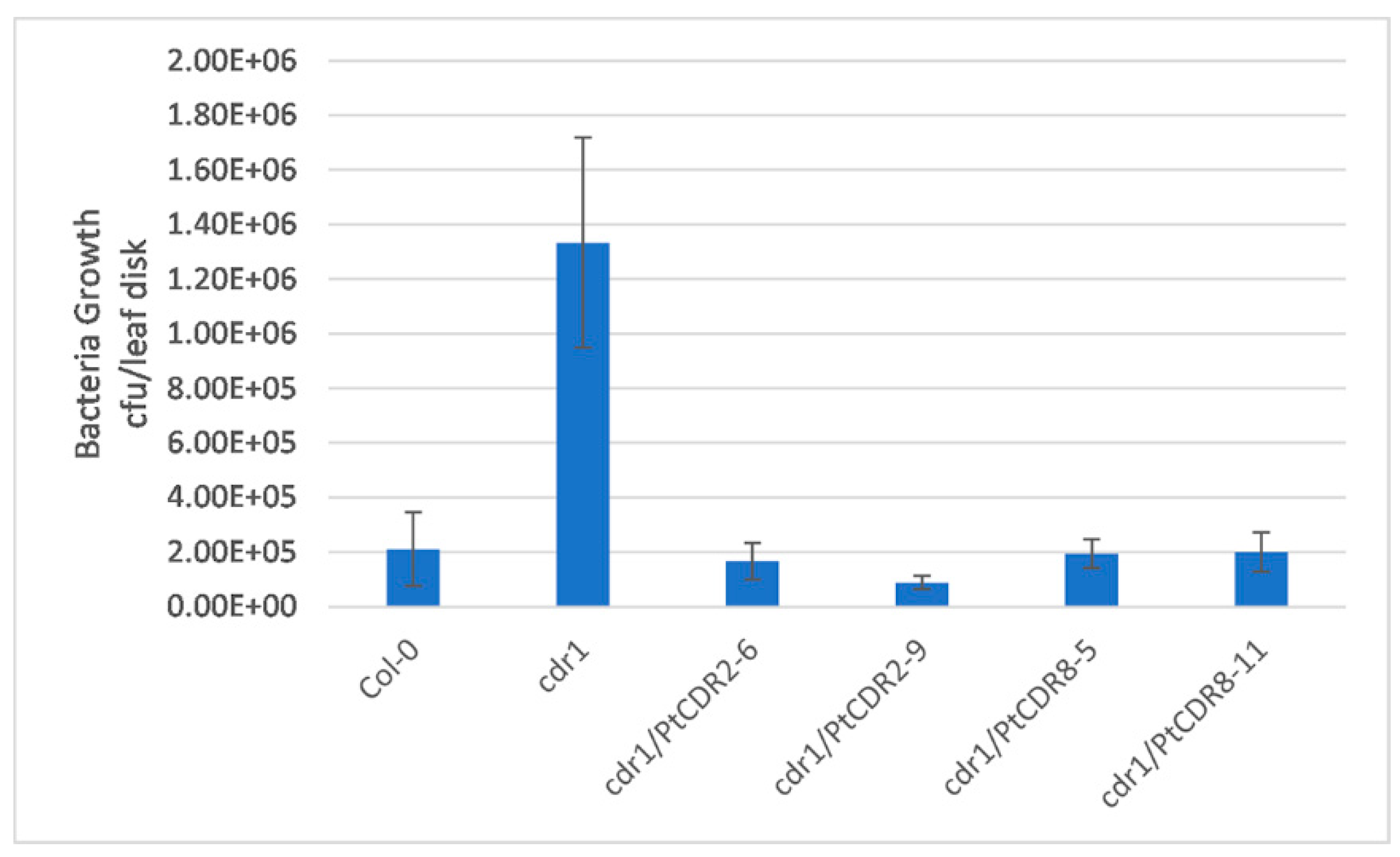
2.3. PtCDR2 and PtCDR8 Inhibited Pst DC3000 avrRpt2 Growth
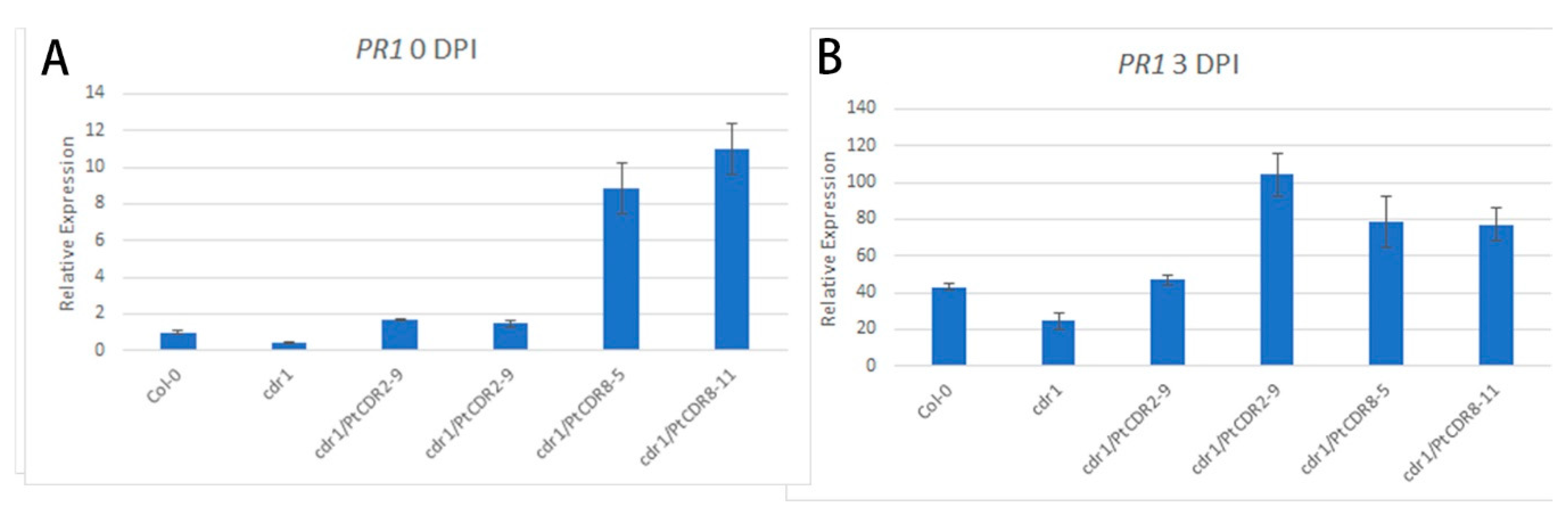
2.4. PtCDR2 and PtCDR8 Increased the Expression Level of PATHOGENESIS RELATED 1 (PR1) in the cdr1 Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Sequence Analysis
4.3. Gene Cloning, Construction of Expression Vectors, and Transformation of Arabidopsis cdr1 Mutant
4.4. Disease Assay
4.5. Gene Expression Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Bové, J.M. Huanglongbing or yellow shoot, a disease of Gondwanan origin: Will it destroy citrus worldwide? Phytoparasitica 2014, 42, 579–583. [Google Scholar] [CrossRef]
- Dala-Paula, B.M.; Plotto, A.; Bai, J.; Manthey, J.A.; Baldwin, E.A.; Ferrarezi, R.S.; Gloria, M.B.A. Effect of Huanglongbing or greening disease on orange juice quality, a review. Front. Plant Sci. 2018, 9, 1976. [Google Scholar] [CrossRef]
- Spreen, T.H.; Baldwin, J.P.; Futch, S.H. An economic assessment of the impact of Huanglongbing on citrus tree plantings in Florida. HortScience 2014, 49, 1052–1055. [Google Scholar] [CrossRef]
- Gottwald, T.R. Current epidemiological understanding of citrus Huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef]
- Li, J.Y.; Pang, Z.Q.; Trivedi, P.; Zhou, X.F.; Ying, X.B.; Jia, H.G.; Wang, N.A. ’Candidatus Liberibacter asiaticus’ encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol. Plant Microbe Interact. 2017, 30, 620–630. [Google Scholar] [CrossRef]
- Ying, X.; Wan, M.; Hu, L.; Zhang, J.; Li, H.; Lv, D. Identification of the virulence factors of Candidatus liberibacter asiaticus via heterologous expression in Nicotiana benthamiana using tobacco mosaic virus. Int. J. Mol. Sci. 2019, 20, 5575. [Google Scholar] [CrossRef]
- Jain, M.; Fleites, L.A.; Gabriel, D.W. Prophage-encoded peroxidase in ’Candidatus liberibacter asiaticus’ is a secreted effector that suppresses plant defenses. Mol. Plant Microbe Interact. 2015, 28, 1330–1337. [Google Scholar] [CrossRef]
- Li, H.; Ying, X.; Shang, L.; Redfern, B.; Kypraios, N.; Xie, X.; Xu, F.; Wang, S.; Zhang, J.; Jian, H.; et al. Heterologous expression of clibasia_03915/clibasia_04250 by tobacco mosaic virus resulted in phloem necrosis in the senescent leaves of Nicotiana benthamiana. Int. J. Mol. Sci. 2020, 21, 1414. [Google Scholar] [CrossRef]
- Pitino, M.; Armstrong, C.M.; Cano, L.M.; Duan, Y.P. Transient expression of Candidatus Liberibacter Asiaticus effector induces cell death in Nicotiana benthamiana. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Clark, K.; Franco, J.Y.; Schwizer, S.; Pang, Z.; Hawara, E.; Liebrand, T.W.H.; Pagliaccia, D.; Zeng, L.; Gurung, F.B.; Wang, P.; et al. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 2018, 9, 1718. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fan, Y.; Zhang, C.; Dai, M.; Wang, X.; Li, W. Nuclear import of a secreted "Candidatus Liberibacter asiaticus" protein is temperature dependent and contributes to pathogenicity in Nicotiana benthamiana. Front. Microbiol. 2019, 10, 1684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X.F.; Liu, X.L.; Fan, Y.Y.; Zhang, Y.Q.; Zhou, X.P.; Li, W.M. A novel ’Candidatus Liberibacter asiaticus’-encoded sec-dependent secretory protein suppresses programmed cell death in Nicotiana benthamiana. Int. J. Mol. Sci. 2019, 20, 5802. [Google Scholar] [CrossRef] [PubMed]
- Ramadugu, C.; Keremane, M.L.; Halbert, S.E.; Duan, Y.P.; Roose, M.L.; Stover, E.; Lee, R.F. Long-term field evaluation reveals Huanglongbing resistance in Citrus relatives. Plant Dis. 2016, 100, 1858–1869. [Google Scholar] [CrossRef] [PubMed]
- Boina, D.R.; Bloomquist, J.R. Chemical control of the Asian citrus psyllid and of Huanglongbing disease in citrus. Pest Manag. Sci. 2015, 71, 808–823. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, R.; Albrecht, U.; Padmanabhan, C.; Wang, A.; Coffey, M.D.; Girke, T.; Wang, Z.; Close, T.J.; Roose, M.; et al. Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus Huanglongbing disease. Mol. Plant 2013, 6, 301–310. [Google Scholar] [CrossRef]
- Hu, J.; Jiang, J.; Wang, N. Control of citrus Huanglongbing via trunk injection of plant defense activators and antibiotics. Phytopathology 2018, 108, 186–195. [Google Scholar] [CrossRef]
- Hoffman, M.T.; Doud, M.S.; Williams, L.; Zhang, M.Q.; Ding, F.; Stover, E.; Hall, D.; Zhang, S.; Jones, L.; Gooch, M.; et al. Heat treatment eliminates ’Candidatus Liberibacter asiaticus’ from infected citrus trees under controlled conditions. Phytopathology 2013, 103, 15–22. [Google Scholar] [CrossRef]
- Doud, M.M.; Wang, Y.; Hoffman, M.T.; Latza, C.L.; Luo, W.; Armstrong, C.M.; Gottwald, T.R.; Dai, L.; Luo, F.; Duan, Y. Solar thermotherapy reduces the titer of Candidatus Liberibacter asiaticus and enhances canopy growth by altering gene expression profiles in HLB-affected citrus plants. Hortic. Res. 2017, 4, 17054. [Google Scholar] [CrossRef]
- Citrus Statistics. Available online: http://flcitrusmutual.com/citrus-101/citrusstatistics.aspx (accessed on 1 December 2019).
- Deng, H.H.; Achor, D.; Exteberria, E.; Yu, Q.B.; Du, D.L.; Stanton, D.; Liang, G.; Gmitter, F.G. Phloem regeneration is a mechanism for Huanglongbing-tolerance of "Bearss" lemon and "LB8-9" Sugar Belle® mandarin. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Fan, J.; Chen, C.; Yu, Q.; Khalaf, A.; Achor, D.S.; Brlansky, R.H.; Moore, G.A.; Li, Z.G.; Gmitter, F.G., Jr. Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to ’Candidatus Liberibacter asiaticus’ infection. Mol. Plant Microbe Interact. 2012, 25, 1396–1407. [Google Scholar] [CrossRef]
- Stover, E.; McCollum, G. Incidence and severity of Huanglongbing and Candidatus Liberibacter asiaticus titer among field-infected citrus cultivars. HortScience 2011, 46, 1344. [Google Scholar] [CrossRef]
- Stover, E.; Inch, S.; Richardson, M.L.; Hall, D.G. Conventional citrus of some scion/rootstock combinations show field tolerance under high Huanglongbing disease pressure. HortScience 2016, 51, 127. [Google Scholar] [CrossRef]
- Killiny, N. Metabolite signature of the phloem sap of fourteen citrus varieties with different degrees of tolerance to Candidatus Liberibacter asiaticus. Physiol. Mol. Plant Pathol. 2017, 97, 20–29. [Google Scholar] [CrossRef]
- Folimonova, S.Y.; Robertson, C.J.; Garnsey, S.M.; Gowda, S.; Dawson, W.O. Examination of the responses of different genotypes of citrus to Huanglongbing (citrus greening) under different conditions. Phytopathology 2009, 99, 1346–1354. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, U.; Bowman, K.D. Tolerance of trifoliate citrus rootstock hybrids to Candidatus Liberibacter asiaticus. Sci. Hortic. 2012, 147, 71–80. [Google Scholar] [CrossRef]
- Albrecht, U.; Bowman, K.D. Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci. 2012, 185–186, 118–130. [Google Scholar] [CrossRef]
- Miles, G.P.; Stover, E.; Ramadugu, C.; Keremane, M.L.; Lee, R.F. Apparent tolerance to Huanglongbing in citrus and citrus-related germplasm. HortScience 2017, 52, 31. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a modified plant thionin enhances disease resistance to citrus canker and Huanglongbing (HLB). Front. Plant Sci. 2016, 7, 1078. [Google Scholar] [CrossRef]
- Zou, X.; Jiang, X.; Xu, L.; Lei, T.; Peng, A.; He, Y.; Yao, L.; Chen, S. Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to Huanglongbing. Plant Mol. Biol. 2017, 93, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Dutt, M.; Barthe, G.; Irey, M.; Grosser, J. Transgenic citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against Huanglongbing (HLB.; Citrus Greening). PLoS ONE 2015, 10, e0137134. [Google Scholar] [CrossRef] [PubMed]
- Kost, T.D.; Gessler, C.; Jansch, M.; Flachowsky, H.; Patocchi, A.; Broggini, G.A. Development of the first cisgenic apple with increased resistance to fire blight. PLoS ONE 2015, 10, e0143980. [Google Scholar] [CrossRef]
- Vanblaere, T.; Szankowski, I.; Schaart, J.; Schouten, H.; Flachowsky, H.; Broggini, G.A.; Gessler, C. The development of a cisgenic apple plant. J. Biotechnol. 2011, 154, 304–311. [Google Scholar] [CrossRef]
- Edenbrandt, A.K.; Gamborg, C.; Thorsen, B.J. Consumers’ preferences for bread: Transgenic, cisgenic, organic or pesticide-free? J. Agric. Econ. 2018, 69, 121–141. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.; Yu, X.; Stover, E.; Luo, F.; Duan, Y. Transcriptome Profiling of Huanglongbing (HLB) Tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 2016, 7, 933. [Google Scholar] [CrossRef]
- Simoes, I.; Faro, R.; Bur, D.; Faro, C. Characterization of recombinant CDR1, an Arabidopsis aspartic proteinase involved in disease resistance. J. Biol. Chem. 2007, 282, 31358–31365. [Google Scholar] [CrossRef]
- Simoes, I.; Faro, C. Structure and function of plant aspartic proteinases. Eur. J. Biochem. 2004, 271, 2067–2075. [Google Scholar] [CrossRef]
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988. [Google Scholar] [CrossRef]
- Prasad, B.D.; Creissen, G.; Lamb, C.; Chattoo, B.B. Overexpression of rice (Oryza sativa L.) OsCDR1 leads to constitutive activation of defense responses in rice and Arabidopsis. Mol. Plant Microbe Interact. 2009, 22, 1635–1644. [Google Scholar] [CrossRef]
- Prasad, B.D.; Creissen, G.; Lamb, C.; Chattoo, B.B. Heterologous expression and characterization of recombinant OsCDR1, a rice aspartic proteinase involved in disease resistance. Protein Expr. Purif. 2010, 72, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Roose, M.L.; Yu, Q.; Du, D.; Yu, Y.; Zhang, Y.; Deng, Z.; Stover, E.; Gmitter, F.G., Jr. Construction of high-density genetic maps and detection of QTLs associated with Huanglongbing tolerance in citrus. Front. Plant Sci. 2018, 9, 1694. [Google Scholar] [CrossRef] [PubMed]
- Boava, L.P.; Cristofani-Yaly, M.; Machado, M.A. Physiologic, anatomic, and gene expression changes in Citrus sunki, Poncirus trifoliata, and their hybrids after ’Candidatus Liberibacter asiaticus’ infection. Phytopathology 2017, 107, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Kumar, B.; Albrecht, U.; Du, D.; Huang, M.; Yu, Q.; Zhang, Y.; Duan, Y.P.; Bowman, K.D.; Gmitter, F.G., Jr.; et al. Genome resequencing and transcriptome profiling reveal structural diversity and expression patterns of constitutive disease resistance genes in Huanglongbing-tolerant Poncirus trifoliata and its hybrids. Hortic. Res. 2017, 4, 17064. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef]
- Yu, Q.; Chen, C.; Du, D.; Huang, M.; Yao, J.; Yu, F.; Brlansky, R.H.; Gmitter, F.G. Reprogramming of a defense signaling pathway in rough lemon and sweet orange is a critical element of the early response to ‘Candidatus Liberibacter asiaticus’. Hortic. Res. 2017, 4, 17063. [Google Scholar] [CrossRef]
- Pitino, M.; Allen, V.; Duan, Y. LasΔ5315 effector induces extreme starch accumulation and chlorosis as Ca. Liberibacter asiaticus infection in Nicotiana benthamiana. Front. Plant Sci. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Duan, Y.; Zhou, L.; Turechek, W.W.; Stover, E.; Powell, C.A. Screening molecules for control of citrus Huanglongbing using an optimized regeneration system for ’Candidatus Liberibacter asiaticus’-infected periwinkle (Catharanthus roseus) cuttings. Phytopathology 2010, 100, 239–245. [Google Scholar] [CrossRef]
- Piquerez, S.J.; Harvey, S.E.; Beynon, J.L.; Ntoukakis, V. Improving crop disease resistance: Lessons from research on Arabidopsis and tomato. Front. Plant Sci. 2014, 5, 671. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [PubMed]
- Omar, A.A.; Murata, M.M.; El-Shamy, H.A.; Graham, J.H.; Grosser, J.W. Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 2018, 27, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Francis, M.I.; Dawson, W.O.; Graham, J.H.; Orbovic, V.; Triplett, E.W.; Mou, Z. Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 2010, 128, 91–100. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotech. 2014, 32, 656–662. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y.; et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017, 49, 765–772. [Google Scholar] [CrossRef]
- Ying, X.B.; Dong, L.; Zhu, H.; Duan, C.G.; Du, Q.S.; Lv, D.Q.; Fang, Y.Y.; Garcia, J.A.; Fang, R.X.; Guo, H.S. RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell 2010, 22, 1358–1372. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, X.; Redfern, B.; Gmitter, F.G., Jr.; Deng, Z. Heterologous Expression of the Constitutive Disease Resistance 2 and 8 Genes from Poncirus trifoliata Restored the Hypersensitive Response and Resistance of Arabidopsis cdr1 Mutant to Bacterial Pathogen Pseudomonas syringae. Plants 2020, 9, 821. https://doi.org/10.3390/plants9070821
Ying X, Redfern B, Gmitter FG Jr., Deng Z. Heterologous Expression of the Constitutive Disease Resistance 2 and 8 Genes from Poncirus trifoliata Restored the Hypersensitive Response and Resistance of Arabidopsis cdr1 Mutant to Bacterial Pathogen Pseudomonas syringae. Plants. 2020; 9(7):821. https://doi.org/10.3390/plants9070821
Chicago/Turabian StyleYing, Xiaobao, Bryce Redfern, Frederick G. Gmitter, Jr., and Zhanao Deng. 2020. "Heterologous Expression of the Constitutive Disease Resistance 2 and 8 Genes from Poncirus trifoliata Restored the Hypersensitive Response and Resistance of Arabidopsis cdr1 Mutant to Bacterial Pathogen Pseudomonas syringae" Plants 9, no. 7: 821. https://doi.org/10.3390/plants9070821
APA StyleYing, X., Redfern, B., Gmitter, F. G., Jr., & Deng, Z. (2020). Heterologous Expression of the Constitutive Disease Resistance 2 and 8 Genes from Poncirus trifoliata Restored the Hypersensitive Response and Resistance of Arabidopsis cdr1 Mutant to Bacterial Pathogen Pseudomonas syringae. Plants, 9(7), 821. https://doi.org/10.3390/plants9070821




