Barley’s Second Spring as a Model Organism for Chloroplast Research
Abstract
:1. Barley, the Crop and the Model Species
1.1. A Brief History of Genome Manipulation in Barley
1.2. Early Studies and Milestones in Understanding of Chloroplast Biogenesis and Physiology in Barley
2. Arabidopsis thaliana as the Model Plant of Modern Times
3. The Genomes of Barley
3.1. The Nuclear Genome
3.2. The Exomes
4. Barley Genetic Resources: Natural and Induced Genetic Diversity
4.1. Natural Genetic Diversity
4.2. Induced Genetic Diversity: Random Chemical and Physical Mutagenesis
4.3. Induced Genetic Diversity: Genome Transformation and Insertional Mutagenesis
4.4. Induced Genetic Diversity: Gene Editing
5. Barley is Ready for a New Age of Functional Genomics Studies and Genetic Improvements
5.1. Plastid-to-Nucleus Retrograde Signalling
5.2. Photosynthesis and Yield
5.3. Optimization of Antenna Size in Crop Canopies
5.4. Increased Photosynthetic Electron Transport
5.5. Improving the Adaptation to Fluctuating Light: Dissipation of Excess Energy through Non-Photochemical Quenching
5.6. Transgenic Manipulation of the Calvin–Benson Cycle
5.7. Photorespiration and Photorespiratory Bypasses
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean Basin. In Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Oxford University Press: Oxford, UK, 2013; ISBN 9780191810046. [Google Scholar]
- Dawson, I.K.; Russell, J.; Powell, W.; Steffenson, B.; Thomas, W.T.B.; Waugh, R. Barley: A translational model for adaptation to climate change. New Phytol. 2015, 206, 913–931. [Google Scholar] [CrossRef] [PubMed]
- Stadler, L.J. Some genetic effect of X-rays in plants. J. Hered. 1930, 21, 3–20. [Google Scholar] [CrossRef]
- Afsson, Å.K.E.G. Studies on the genetic basis of chlorophyll formation and the mechanism of induced mutating. Hereditas 1938, 24, 33–93. [Google Scholar] [CrossRef]
- Afsson, A.K.E.G. Mutation experiments in barley. Hereditas 1941, 27, 225–242. [Google Scholar] [CrossRef]
- Smith, L. Effects of atomic bomb radiations and x-rays on seeds of cereals: A comparison of the effects of ionizing radiations from the “ test able” atomic bomb and from x-rays on seeds of barley, wheat and oats. J. Hered. 1950, 41, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, Å.; Key, J.M. The genetical effects of musterd gas substances and neutrons. Hereditas 1948, 34, 371–386. [Google Scholar] [CrossRef]
- Ehrenberg, L.; Gustafsson, Å.; Lundqvist, U.; Stenhagen, E.; Thorell, B. Chemically induced mutation and sterility in Barley. Acta Chem. Scand. 1956, 10, 492–494. [Google Scholar] [CrossRef]
- Bouma, J.; Ohnoutka, Z. Importance and application ot the mutant “Diamant” in spring barley breeding. Plant Mutat. Breed. Crop Improv. 1991, 1, 127–134. [Google Scholar]
- Forster, B.P. Mutation genetics of salt tolerance in barley: An assessment of Golden Promise and other semi-dwarf mutants. Euphytica 2001, 120, 317–328. [Google Scholar] [CrossRef]
- Henningsen, K.W.; Boynton, J.E. Macromolecular physiology of plastids. VII. The effect of a brief illumination on plastids of dark-grown barley leaves. J. Cell Sci. 1969, 5, 757–793. [Google Scholar]
- Kannanga, G.; Gamini, C. The formation of Ribulose Diphosphate Carboxylase Protein during chloroplast development in Barley. Plant Physiol. 1969, 44, 1533–1537. [Google Scholar] [CrossRef] [PubMed]
- Wellburn, A.R.; Robinson, D.C.; Wellburn, F.A.M. Chloroplast development in low light-grown barley seedlings. Planta 1982, 154, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Gollmer, I.; Batschauer, A. The light-dependent control of chloroplast development in barley (Hordeum vulgare L). J. Cell. Biochem. 1983, 23, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Fradkin, L.I.; Kolyago, V.M.; Nisenbaum, G.D.; Domanskaya, I.N. Disintegration and fractionation of Barley chloroplast membranes at different concentrations of digitonin and chloroplasts. Biokhimiya 1978, 43, 723–733. [Google Scholar]
- Nielsen, N.C.; Smillie, R.M.; Henningsen, K.W.; Von Wettstein, D.; French, C.S. Composition and function of thylakoid membranes from grana-rich and grana-deficient chloroplast mutants of barley. Plant Physiol. 1979, 63, 174–182. [Google Scholar] [CrossRef]
- Thornber, J.P.; Highkin, H.R. Composition of the photosynthetic apparatus of normal barley leaves and a mutant lacking chlorophyll b. Eur. J. Biochem. 1974, 41, 109–116. [Google Scholar] [CrossRef]
- Król, M.; Spangfort, M.D.; Huner, N.P.; Oquist, G.; Gustafsson, P.; Jansson, S. Chlorophyll a/b-binding proteins, pigment conversions, and early light-induced proteins in a chlorophyll b-less barley mutant. Plant Physiol. 1995, 107, 873–883. [Google Scholar] [CrossRef] [Green Version]
- Król, M.; Ivanov, A.G.; Jansson, S.; Kloppstech, K.; Huner, N.P.A. Greening under high light or cold temperature affects the level of xanthophyll-cycle pigments, early light-inducible proteins, and light-harvesting polypeptides in wild-type barley and the chlorina f2 mutant. Plant Physiol. 1999, 120, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Bossmann, B.; Knoetzel, J.; Jansson, S. Screening of chlorina mutants of barley (Hordeum vulgare L.) with antibodies against light-harvesting proteins of PS I and PS II: Absence of specific antenna proteins. Photosynth. Res. 1997, 52, 127–136. [Google Scholar] [CrossRef]
- Goodchild, D.J.; Highkin, H.R.; Boardman, N.K. The fine structure of chloroplasts in a barley mutant lacking chlorophyll B. Exp. Cell Res. 1966, 43, 684–688. [Google Scholar] [CrossRef]
- Von Wettstein, D.; Kahn, A.; Nielsen, O.F.; Gough, S. Genetic Regulation of Chlorophyll Synthesis Analyzed with Mutants in Barley. Science 1974, 184, 800–802. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.P.; Kim, C.; Lee, D.W.; Apel, K. TIGRINA d, required for regulating the biosynthesis of tetrapyrroles in barley, is an ortholog of the FLU gene of Arabidopsis thaliana. FEBS Lett. 2003, 553, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Meskauskiene, R.; Nater, M.; Goslings, D.; Kessler, F.; Op den Camp, R.; Apel, K. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2001, 98, 12826–12831. [Google Scholar] [CrossRef] [Green Version]
- Gustafsoon, Å. Drastic morphological mutation in Barley. Hereditas 1946, 32, 120–122. [Google Scholar] [CrossRef]
- Rzeznicka, K.; Walker, C.J.; Westergren, T.; Kannangara, C.G.; Von Wettstein, D.; Merchant, S.; Gough, S.P.; Hansson, M. Xantha-I encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 5886–5891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, P.E.; Willows, R.D.; Petersen, B.L.; Vothknecht, U.C.; Stummann, B.M.; Kannangara, C.G.; Von Wettstein, D.; Henningsen, K.W. Structural genes for Mg-chelatase subunits in barley: Xantha-f, -g and -h. Mol. Gen. Genet. 1996, 250, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.H.C.; French, C.S.; Koski, V.M. The Hill Reaction: Development of Chloroplast Activity During Greening of Etiolated Barley Leaves. Plant Physiol. 1952, 27, 212–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, D.; Laetsch, W.M. Structure and Function of Developing Barley Plastids. Plant Physiol. 1974, 54, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Hajdukiewicz, P.T.J.; Allison, L.A.; Maliga, P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997, 16, 4041–4048. [Google Scholar] [CrossRef] [Green Version]
- Siemenroth, A.; Wollgiehn, R.; Neumann, D.; Börner, T. Synthesis of ribosomal RNA in ribosome-deficient plastids of the mutant “albostrians” of Hordeum vulgare L. Planta 1981, 153, 547–555. [Google Scholar] [CrossRef]
- Hess, W.R.; Prombona, A.; Fieder, B.; Subramanian, A.R.; Börner, T. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: Evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 1993, 12, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, R.; Börner, T.; Knoth, R. Plastid ribosome deficiency in plastid mutants of Pelargonium and Hordeum. Genetics 1973, 74, 103–104. [Google Scholar]
- Börner, T.; Schumann, B.; Hagemann, R. Biochemical studies on a plastid ribosome-deficient mutant of Hordeum vulgare. In Genetics Biogenesis of Chloroplasts Mitochondria; Bücher, T., Neupert, W., Sebald, W., Werner, S., Eds.; Elsevier; North-Hill Medical Press: Amsterdam, The Netherlands, 1976; pp. 41–48. [Google Scholar]
- Börner, T. The discovery of plastid-to-nucleus retrograde signaling—a personal perspective. Protoplasma 2017, 254, 1845–1855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chilton, M.D.; Drummond, M.H.; Merlo, D.J.; Sciaky, D.; Montoya, A.L.; Gordon, M.P.; Nester, E.W. Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell 1977, 11, 263–271. [Google Scholar] [CrossRef]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X.; et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Li, C.; Sun, X.; Wang, P.; Zhao, S. Effects of ridge direction on growth and yield of tomato in solar greenhouse with diffuse film. Nongye Gongcheng Xuebao/Trans. Chinese Soc. Agric. Eng. 2017, 33, 242–248. [Google Scholar] [CrossRef]
- Kirst, H.; Gabilly, S.T.; Niyogi, K.K.; Lemaux, P.G.; Melis, A. Photosynthetic antenna engineering to improve crop yields. Planta 2017, 245, 1009–1020. [Google Scholar] [CrossRef]
- Pogson, B.J.; Ganguly, D.; Albrecht-Borth, V. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 1017–1024. [Google Scholar] [CrossRef] [Green Version]
- Boffey, S.A.; Selldén, G.; Leech, R.M. Influence of Cell Age on Chlorophyll Formation in Light-grown and Etiolated Wheat Seedlings. Plant Physiol. 1980, 65, 680–684. [Google Scholar] [CrossRef] [Green Version]
- Mullet, J.E. Chloroplast Development and Gene Expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 475–502. [Google Scholar] [CrossRef]
- Pogson, B.J.; Albrecht, V. Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiol. 2011, 155, 1545–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarvis, P.; López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.X.; Waugh, R.; Langridge, P.; Close, T.J.; Wise, R.P.; Graner, A.; Matsumoto, T.; Sato, K.; Schulman, A.; Ariyadasa, R.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–771. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monat, C.; Padmarasu, S.; Lux, T.; Wicker, T.; Gundlach, H.; Himmelbach, A.; Ens, J.; Li, C.; Muehlbauer, G.J.; Schulman, A.H.; et al. TRITEX: Chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 2019, 20, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, M.; Mascher, M.; Wright, J.; Padmarasu, S.; Himmelbach, A.; Heavens, D.; Milne, L.; Clavijo, B.; Stein, N.; Waugh, R. A Genome Assembly of the Barley “Transformation Reference” Cultivar Golden Promise. G3 Genes Genomes Genet. 2020, 10, 1823–1827. [Google Scholar] [CrossRef] [Green Version]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. BARLEX—The barley draft genome explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef] [Green Version]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016, 1533, 1–31. [Google Scholar] [CrossRef]
- Deng, W.; Nickle, D.C.; Learn, G.H.; Maust, B.; Mullins, J.I. ViroBLAST: A stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics 2007, 23, 2334–2336. [Google Scholar] [CrossRef]
- Tello-Ruiz, M.K.; Naithani, S.; Stein, J.C.; Gupta, P.; Campbell, M.; Olson, A.; Wei, S.; Preece, J.; Geniza, M.J.; Jiao, Y.; et al. Gramene 2018: Unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res. 2018, 46, D1181–D1189. [Google Scholar] [CrossRef] [Green Version]
- Spannagl, M.; Nussbaumer, T.; Bader, K.C.; Martis, M.M.; Seidel, M.; Kugler, K.G.; Gundlach, H.; Mayer, K.F.X. PGSB plantsDB: Updates to the database framework for comparative plant genome research. Nucleic Acids Res. 2016, 44, D1141–D1147. [Google Scholar] [CrossRef] [PubMed]
- Rapazote-Flores, P.; Bayer, M.; Milne, L.; Mayer, C.D.; Fuller, J.; Guo, W.; Hedley, P.E.; Morris, J.; Halpin, C.; Kam, J.; et al. BaRTv1.0: An improved barley reference transcript dataset to determine accurate changes in the barley transcriptome using RNA-seq. BMC Genom. 2019, 20, 968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascher, M.; Richmond, T.A.; Gerhardt, D.J.; Himmelbach, A.; Clissold, L.; Sampath, D.; Ayling, S.; Steuernagel, B.; Pfeifer, M.; D’Ascenzo, M.; et al. Barley whole exome capture: A tool for genomic research in the genus Hordeum and beyond. Plant J. 2013, 76, 494–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, J.; Mascher, M.; Dawson, I.K.; Kyriakidis, S.; Calixto, C.; Freund, F.; Bayer, M.; Milne, I.; Marshall-Griffiths, T.; Heinen, S.; et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 2016, 48, 1024–1030. [Google Scholar] [CrossRef] [Green Version]
- Bustos-Korts, D.; Dawson, I.K.; Russell, J.; Tondelli, A.; Guerra, D.; Ferrandi, C.; Strozzi, F.; Nicolazzi, E.L.; Molnar-Lang, M.; Ozkan, H.; et al. Exome sequences and multi-environment field trials elucidate the genetic basis of adaptation in barley. Plant J. 2019, 99, 1172–1191. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.; Hasibuzzaman, A.S.M.; Abdullah, H.M.; Kallol, M.M.H. Rediscovery of Genetic and Genomic Resources for Future Food Security. In Genetic and Genomic Resources and their Explotation for Unlocking Genetic Potential from the Wild Relatives; Springer: Singapore, 2020; pp. 193–210. [Google Scholar]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Caldwell, D.G.; McCallum, N.; Shaw, P.; Muehlbauer, G.J.; Marshall, D.F.; Waugh, R. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.). Plant J. 2004, 40, 143–150. [Google Scholar] [CrossRef]
- Talamè, V.; Bovina, R.; Sanguineti, M.C.; Tuberosa, R.; Lundqvist, U.; Salvi, S. TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant Biotechnol. J. 2008, 6, 477–485. [Google Scholar] [CrossRef]
- Szurman-Zubrzycka, M.E.; Zbieszczyk, J.; Marzec, M.; Jelonek, J.; Chmielewska, B.; Kurowska, M.M.; Krok, M.; Daszkowska-Golec, A.; Guzy-Wrobelska, J.; Gruszka, D.; et al. HorTILLUS—a rich and renewable source of induced mutations for forward/reverse genetics and pre-breeding programs in barley (Hordeum vulgare L.). Front. Plant Sci. 2018, 9, 216. [Google Scholar] [CrossRef]
- Schreiber, M.; Barakate, A.; Uzrek, N.; Macaulay, M.; Sourdille, A.; Morris, J.; Hedley, P.E.; Ramsay, L.; Waugh, R. A highly mutagenised barley (cv. Golden Promise) TILLING population coupled with strategies for screening-by-sequencing. Plant Methods 2019, 15, 99. [Google Scholar] [CrossRef]
- Gottwald, S.; Bauer, P.; Komatsuda, T.; Lundqvist, U.; Stein, N. TILLING in the two-rowed barley cultivar “Barke” reveals preferred sites of functional diversity in the gene HvHox1. BMC Res. Notes 2009, 2, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lababidi, S.; Mejlhede, N.; Rasmussen, S.K.; Backes, G.; Al-Said, W.; Baum, M.; Jahoor, A. Identification of barley mutants in the cultivar “lux” at the dhn loci through tilling. Plant Breed. 2009, 128, 332–336. [Google Scholar] [CrossRef]
- Kurowska, M.; Labocha-Pawłowska, A.; Gnizda, D.; Maluszynski, M.; Szarejko, I. Molecular analysis of point mutations in a barley genome exposed to MNU and Gamma rays. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2012, 738–739, 52–70. [Google Scholar] [CrossRef] [PubMed]
- Druka, A.; Franckowiak, J.; Lundqvist, U.; Bonar, N.; Alexander, J.; Houston, K.; Radovic, S.; Shahinnia, F.; Vendramin, V.; Morgante, M.; et al. Genetic dissection of barley morphology and development. Plant Physiol. 2011, 155, 617–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dockter, C.; Gruszka, D.; Braumann, I.; Druka, A.; Druka, I.; Franckowiak, J.; Gough, S.P.; Janeczko, A.; Kurowska, M.; Lundqvist, J.; et al. Induced variations in brassinosteroid genes define barley height and sturdiness, and expand the green revolution genetic toolkit. Plant Physiol. 2014, 166, 1912–1927. [Google Scholar] [CrossRef] [Green Version]
- Jost, M.; Taketa, S.; Mascher, M.; Himmelbach, A.; Yuo, T.; Shahinnia, F.; Rutten, T.; Druka, A.; Schmutzer, T.; Steuernagel, B.; et al. A homolog of blade-on-petiole 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiol. 2016, 171, 1113–1127. [Google Scholar] [CrossRef] [Green Version]
- Komatsuda, T.; Pourkheirandish, M.; He, C.; Azhaguvel, P.; Kanamori, K.; Perovic, D.; Stein, N.; Graner, A.; Wicker, T.; Tagiri, A.; et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 2007, 104, 1424–1429. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, L.; Comadran, J.; Druka, A.; Marshall, D.F.; Thomas, W.T.B.; MacAulay, M.; MacKenzie, K.; Simpson, C.; Fuller, J.; Bonar, N.; et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 2011, 43, 169–172. [Google Scholar] [CrossRef]
- Henikoff, S.; Till, B.J.; Comai, L. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 2004, 135, 630–636. [Google Scholar] [CrossRef] [Green Version]
- Waugh, R.; Leader, D.J.; McCallum, N.; Caldwell, D. Harvesting the potential of induced biological diversity. Trends Plant Sci. 2006, 11, 71–79. [Google Scholar] [CrossRef]
- Till, B.J.; Reynolds, S.H.; Greene, E.A.; Codomo, C.A.; Enns, L.C.; Johnson, J.E.; Burtler, C.; Odden, A.R.; Young, K.; Taylor, N.E.; et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003, 13, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, I.M.; Nagalakshmi, U.; Lieberman, M.C.; Ngo, K.J.; Krasileva, K.V.; Vasquez-Gross, H.; Akhunova, A.; Akhunov, E.; Dubcovsky, J.; Tai, T.H.; et al. Efficient genome-wide detection and cataloging of EMS-induced mutations using Exome capture and next-generation sequencing. Plant Cell 2014, 26, 1382–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, C.S.; Kronstad, W.E. Tissue culture and plant regeneration from immature embryo explants of Barley, Hordeum vulgare. Theor. Appl. Genet. 1986, 71, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Dahleen, L.S.; Bregitzer, P. An improved media system for high regeneration rates from barley immature embryo-derived callus cultures of commercial cultivars. Crop Sci. 2002, 42, 934–938. [Google Scholar] [CrossRef]
- Harwood, W.A. A protocol for high-throughput agrobacterium-mediated barley transformation. Methods Mol. Biol. 2014, 1099, 251–260. [Google Scholar] [CrossRef] [PubMed]
- McGrath, P.F.; Vincent, J.R.; Lei, C.H.; Pawlowski, W.P.; Torbert, K.A.; Gu, W.; Kaeppler, H.F.; Wan, Y.; Lemaux, P.G.; Rines, H.R.; et al. Coat protein-mediated resistance to isolates of barley yellow dwarf in oats and barley. Eur. J. Plant Pathol. 1997, 103, 695–710. [Google Scholar] [CrossRef]
- Hückelhoven, R. BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 2004, 9, 299–307. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Biselli, C.; Collins, N.C.; Consonni, G.; Stanca, A.M.; Schulze-Lefert, P.; Valè, G. The CC-NB-LRR-Type RDG2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death. PLoS ONE 2010, 5, e12599. [Google Scholar] [CrossRef] [Green Version]
- Eichmann, R.; Bischof, M.; Weis, C.; Shaw, J.; Lacomme, C.; Schweizer, P.; Duchkov, D.; Hensel, G.; Kumlehn, J.; Hückelhoven, R. Bax inhibitor-1 is required for full susceptibility of barley to powdery mildew. Mol. Plant-Microbe Interact. 2010, 23, 1217–1227. [Google Scholar] [CrossRef] [Green Version]
- Horvath, H.; Rostoks, N.; Brueggeman, R.; Steffenson, B.; Von Wettstein, D.; Kleinhofs, A. Genetically engineered stem rust resistance in barley using the Rpg1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 364–369. [Google Scholar] [CrossRef] [Green Version]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef]
- Seiler, C.; Harshavardhan, V.T.; Reddy, P.S.; Hensel, G.; Kumlehn, J.; Eschen-Lippold, L.; Rajesh, K.; Korzun, V.; Wobus, U.; Lee, J.; et al. Abscisic acid flux alterations result in differential abscisic acid signaling responses and impact assimilation efficiency in barley under terminal drought stress. Plant Physiol. 2014, 164, 1677–1696. [Google Scholar] [CrossRef] [PubMed]
- Soltész, A.; Vágújfalvi, A.; Rizza, F.; Kerepesi, I.; Galiba, G.; Cattivelli, L.; Coraggio, I.; Crosatti, C. The rice Osmyb4 gene enhances tolerance to frost and improves germination under unfavourable conditions in transgenic barley plants. J. Appl. Genet. 2012, 53, 133–143. [Google Scholar] [CrossRef]
- Soltész, A.; Smedley, M.; Vashegyi, I.; Galiba, G.; Harwood, W.; Vágújfalvi, A. Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J. Exp. Bot. 2013, 64, 1849–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovalchuk, N.; Jia, W.; Eini, O.; Morran, S.; Pyvovarenko, T.; Fletcher, S.; Bazanova, N.; Harris, J.; Beck-Oldach, K.; Shavrukov, Y.; et al. Optimization of TaDREB3 gene expression in transgenic barley using cold-inducible promoters. Plant Biotechnol. J. 2013, 11, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Murray, F.; Matthews, P.; Jacobsen, J.; Gubler, F. Increased expression of HvGAMYB in transgenic barley increases hydrolytic enzyme production by aleurone cells in response to gibberellin. J. Cereal Sci. 2006, 44, 317–322. [Google Scholar] [CrossRef]
- Kihara, M.; Okada, Y.; Kuroda, H.; Saeki, K.; Yoshigi, N.; Ito, K. Improvement of β-amylase thermostability in transgenic barley seeds and transgene stability in progeny. Mol. Breed. 2000, 6, 511–517. [Google Scholar] [CrossRef]
- Tull, D.; Phillipson, B.A.; Kramhøft, B.; Knudsen, S.; Olsen, O.; Svensson, B. Enhanced amylolytic activity in germinating barley through synthesis of a bacterial Alpha-amylase. J. Cereal Sci. 2003, 37, 71–80. [Google Scholar] [CrossRef]
- Jensen, L.G.; Olsen, O.; Kops, O.; Wolf, N.; Thomsen, K.K.; Von Wettstein, D. Transgenic barley expressing a protein-engineered, thermostable (1,3-1,4)-β-glucanase during germination. Proc. Natl. Acad. Sci. USA 1996, 93, 3487–3491. [Google Scholar] [CrossRef] [Green Version]
- Nuutila, A.M.; Ritala, A.; Skadsen, R.W.; Mannonen, L.; Kauppinen, V. Expression of fungal thermotolerant endo-1,4-β-glucanase in transgenic barley seeds during germination. Plant Mol. Biol. 1999, 41, 777–783. [Google Scholar] [CrossRef]
- Koprek, T.; McElroy, D.; Louwerse, J.; Williams-Carrier, R.; Lemaux, P.G. An efficient method for dispersing Ds elements in the barley genome as a tool for determining gene function. Plant J. 2000, 24, 253–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayliffe, M.A.; Pallotta, M.; Langridge, P.; Pryor, A.J. A barley activation tagging system. Plant Mol. Biol. 2007, 64, 329–347. [Google Scholar] [CrossRef] [PubMed]
- Lazarow, K.; Lütticke, S. An Ac/Ds-mediated gene trap system for functional genomics in barley. BMC Genom. 2009, 10, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryder, P.; McHale, M.; Fort, A.; Spillane, C. Generation of stable nulliplex autopolyploid lines of Arabidopsis thaliana using CRISPR/Cas9 genome editing. Plant Cell Rep. 2017, 36, 1005–1008. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA – Guided DNA Endonuclease S figs. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef] [Green Version]
- Holme, I.B.; Wendt, T.; Gil-Humanes, J.; Deleuran, L.C.; Starker, C.G.; Voytas, D.F.; Brinch-Pedersen, H. Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Mol. Biol. 2017, 95, 111–121. [Google Scholar] [CrossRef]
- Gasparis, S.; Kała, M.; Przyborowski, M.; Łyżnik, L.A.; Orczyk, W.; Nadolska-Orczyk, A. A simple and efficient CRISPR/Cas9 platform for induction of single and multiple, heritable mutations in barley (Hordeum vulgare L.). Plant Methods 2018, 14, 111. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Galli, M.; Ordon, J.; Stuttmann, J.; Kogel, K.H.; Imani, J. Further analysis of barley MORC1 using a highly efficient RNA-guided Cas9 gene-editing system. Plant Biotechnol. J. 2018, 16, 1892–1903. [Google Scholar] [CrossRef] [Green Version]
- Gasparis, S.; Przyborowski, M.; Kała, M.; Nadolska-Orczyk, A. Knockout of the HvCKX1 or HvCKX3 Gene in Barley (Hordeum vulgare L.) by RNA-Guided Cas9 Nuclease Affects the Regulation of Cytokinin Metabolism and Root Morphology. Cells 2019, 8, 782. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Zhong, X.; Li, Q.; Lan, J.; Tang, H.; Qi, P.; Ma, J.; Wang, J.; Chen, G.; Pu, Z.; et al. Mutation of the D-hordein gene by RNA-guided Cas9 targeted editing reducing the grain size and changing grain compositions in barley. Food Chem. 2020, 311, 125892. [Google Scholar] [CrossRef] [PubMed]
- Saski, C.; Tomkins, J.; Lee, S.-B.; Daniell, H.; Fjellheim, S.; Rognli, O.A.; Guda, C.; Jansen, R.K.; Luo, H.; Clarke, J.L. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 2007, 115, 571–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, J.; Rogowska-Wrzesinska, A.; Jensen, O.N. Functional proteomics of barley and barley chloroplasts-strategies, methods and perspectives. Front. Plant Sci. 2013, 4, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pogson, B.J.; Woo, N.S.; Förster, B.; Small, I.D. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008, 3, 602–609. [Google Scholar] [CrossRef]
- Wagner, D.; Przybyla, D.; Op Den Camp, R.; Kim, C.; Landgraf, F.; Keun, P.L.; Würsch, M.; Laloi, C.; Nater, M.; Hideg, E.; et al. The genetic basis of singlet oxygen-induced stress response of Arabidopsis thaliana. Science 2004, 306, 1183–1185. [Google Scholar] [CrossRef]
- Maruta, T.; Noshi, M.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 2012, 287, 11717–11729. [Google Scholar] [CrossRef] [Green Version]
- Pfalz, J.; Liebers, M.; Hirth, M.; Grübler, B.; Holtzegel, U.; Schröter, Y.; Dietzel, L.; Pfannschmidt, T. Environmental control of plant nuclear gene expression by chloroplast redox signals. Front. Plant Sci. 2012, 3, 257. [Google Scholar] [CrossRef] [Green Version]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Savchenko, T.; Baidoo, E.E.K.; Chehab, W.E.; Hayden, D.M.; Tolstikov, V.; Corwin, J.A.; Kliebenstein, D.J.; Keasling, J.D.; Dehesh, K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 2012, 149, 1525–1535. [Google Scholar] [CrossRef] [Green Version]
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540. [Google Scholar] [CrossRef] [Green Version]
- D’Alessandro, S.; Havaux, M. Sensing β-carotene oxidation in photosystem II to master plant stress tolerance. New Phytol. 2019, 223, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Chi, W.; Feng, P.; Ma, J.; Zhang, L. Metabolites and chloroplast retrograde signaling. Curr. Opin. Plant Biol. 2015, 25, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Tian, L. Recent advances in understanding carotenoid-derived signaling molecules in regulating plant growth and development. Front. Plant Sci. 2015, 6, 790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the Languages of the Chloroplast: Retrograde Signaling and Beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53. [Google Scholar] [CrossRef] [PubMed]
- Kleine, T.; Leister, D. Retrograde signaling: Organelles go networking. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- de Souza, A.; Wang, J.-Z.; Dehesh, K. Retrograde Signals: Integrators of Interorganellar Communication and Orchestrators of Plant Development. Annu. Rev. Plant Biol. 2017, 68, 85–108. [Google Scholar] [CrossRef]
- Susek, R.E.; Ausubel, F.M.; Chory, J. Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 1993, 74, 787–799. [Google Scholar] [CrossRef] [Green Version]
- Larkin, R.M.; Stefano, G.; Ruckle, M.E.; Stavoe, A.K.; Sinkler, C.A.; Brandizzi, F.; Malmstrom, C.M.; Osteryoung, K.W.; Chory, J. REDUCED CHLOROPLAST COVERAGE genes from Arabidopsis thaliana help to establish the size of the chloroplast compartment. Proc. Natl. Acad. Sci. USA 2016, 113, E1116–E1125. [Google Scholar] [CrossRef] [Green Version]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from Chloroplasts Converge to Regulate Nuclear Gene Expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Tadini, L.; Pesaresi, P.; Kleine, T.; Rossi, F.; Guljamow, A.; Sommer, F.; Mühlhaus, T.; Schroda, M.; Masiero, S.; Pribil, M.; et al. Gun1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 2016, 170, 1817–1830. [Google Scholar] [CrossRef] [Green Version]
- Tadini, L.; Peracchio, C.; Trotta, A.; Colombo, M.; Mancini, I.; Jeran, N.; Costa, A.; Faoro, F.; Marsoni, M.; Vannini, C.; et al. GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J. 2020, 101, 1198–1220. [Google Scholar] [CrossRef]
- Wu, G.Z.; Meyer, E.H.; Richter, A.S.; Schuster, M.; Ling, Q.; Schöttler, M.A.; Walther, D.; Zoschke, R.; Grimm, B.; Jarvis, R.P.; et al. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 2019, 5, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Börner, T.; Meister, A. Chlorophyll and carotenoid content of ribosome-deficient plastids. Photosynthetica 1980, 14, 589–593. [Google Scholar]
- Yaronskaya, E.; Ziemann, V.; Walter, G.; Averina, N.; Börner, T.; Grimm, B. Metabolic control of the tetrapyrrole biosynthetic pathway for porphyrin distribution in the barley mutant albostrians. Plant J. 2003, 35, 512–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feierabend, J.; Mikus, M. Occurrence of a High Temperature Sensitivity of Chloroplast Ribosome Formation in Several Higher Plants. Plant Physiol. 1977, 59, 863–867. [Google Scholar] [CrossRef]
- Woodson, J.D.; Perez-Ruiz, J.M.; Chory, J. Heme synthesis by plastid ferrochelatase i regulates nuclear gene expression in plants. Curr. Biol. 2011, 21, 897–903. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Hensel, G.; Mascher, M.; Melzer, M.; Budhagatapalli, N.; Rutten, T.; Himmelbach, A.; Beier, S.; Korzun, V.; Kumlehn, J.; et al. Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell 2019, 31, 1430–1445. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Dale, B.E. Global potential bioethanol production from wasted crops and crop residues. Biomass Bioenergy 2004, 26, 361–375. [Google Scholar] [CrossRef]
- Gu, J.; Zhou, Z.; Li, Z.; Chen, Y.; Wang, Z.; Zhang, H.; Yang, J. Photosynthetic properties and potentials for improvement of photosynthesis in pale green leaf rice under high light conditions. Front. Plant Sci. 2017, 8, 1082. [Google Scholar] [CrossRef] [Green Version]
- Chida, H.; Nakazawa, A.; Akazaki, H.; Hirano, T.; Suruga, K.; Ogawa, M.; Satoh, T.; Kadokura, K.; Yamada, S.; Hakamata, W.; et al. Expression of the algal cytochrome c6 gene in Arabidopsis enhances photosynthesis and growth. Plant Cell Physiol. 2007, 48, 948–957. [Google Scholar] [CrossRef]
- Yadav, S.K.; Khatri, K.; Rathore, M.S.; Jha, B. Introgression of UfCyt c 6, a thylakoid lumen protein from a green seaweed Ulva fasciata Delile enhanced photosynthesis and growth in tobacco. Mol. Biol. Rep. 2018, 45, 1745–1758. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, P.; Hertle, A.; Pribil, M.; Kleine, T.; Wagner, R.; Strissel, H.; Lhnatowicz, A.; Bonardi, V.; Scharfenberg, M.; Schneider, A.; et al. Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 2009, 21, 2402–2423. [Google Scholar] [CrossRef] [Green Version]
- Simkin, A.J.; McAusland, L.; Lawson, T.; Raines, C.A. Overexpression of the rieskeFeS protein increases electron transport rates and biomass yield. Plant Physiol. 2017, 175, 134–145. [Google Scholar] [CrossRef] [Green Version]
- Kromdijk, J.; Głowacka, K.; Leonelli, L.; Gabilly, S.T.; Iwai, M.; Niyogi, K.K.; Long, S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 2016, 354, 857–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simkin, A.J.; McAusland, L.; Headland, L.R.; Lawson, T.; Raines, C.A. Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco. J. Exp. Bot. 2015, 66, 4075–4090. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Lopez-Calcagno, P.E.; Davey, P.A.; Headland, L.R.; Lawson, T.; Timm, S.; Bauwe, H.; Raines, C.A. Simultaneous stimulation of sedoheptulose 1,7-bisphosphatase, fructose 1,6-bisphophate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnol. J. 2017, 15, 805–816. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, S.; Lawson, T.; Zakhleniuk, O.V.; Lloyd, J.C.; Raines, C.A. Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 2005, 138, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Ding, F.; Wang, M.; Zhang, S.; Ai, X. Changes in SBPase activity influence photosynthetic capacity, growth, and tolerance to chilling stress in transgenic tomato plants. Sci. Rep. 2016, 6, 32741. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, K.; Suzuki, N.; Iwamae, T.; Inui, M.; Yukawa, H. Increased fructose 1,6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J. Exp. Bot. 2012, 63, 3001–3009. [Google Scholar] [CrossRef]
- Yamori, W.; Masumoto, C.; Fukayama, H.; Makino, A. Rubisco activase is a key regulator of non-steady-state photosynthesis at any leaf temperature and, to a lesser extent, of steady-state photosynthesis at high temperature. Plant J. 2012, 71, 871–880. [Google Scholar] [CrossRef]
- Sanz-Barrio, R.; Corral-Martinez, P.; Ancin, M.; Segui-Simarro, J.M.; Farran, I. Overexpression of plastidial thioredoxin f leads to enhanced starch accumulation in tobacco leaves. Plant Biotechnol. J. 2013, 11, 618–627. [Google Scholar] [CrossRef] [Green Version]
- Farran, I.; Fernandez-San Millan, A.; Ancin, M.; Larraya, L.; Veramendi, J. Increased bioethanol production from commercial tobacco cultivars overexpressing thioredoxin f grown under field conditions. Mol. Breed. 2014, 34, 457–469. [Google Scholar] [CrossRef] [Green Version]
- Toivola, J.; Nikkanen, L.; Dahlström, K.M.; Salminen, T.A.; Lepistö, A.; Vignols, F.; Rintamäki, E. Overexpression of chloroplast NADPH-dependent thioredoxin reductase in Arabidopsis enhances leaf growth and elucidates in vivo function of reductase and thioredoxin domains. Front. Plant Sci. 2013, 4, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikkanen, L.; Toivola, J.; Rintamäki, E. Crosstalk between chloroplast thioredoxin systems in regulation of photosynthesis. Plant Cell Environ. 2016, 39, 1691–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timm, S.; Florian, A.; Arrivault, S.; Stitt, M.; Fernie, A.R.; Bauwe, H. Glycine decarboxylase controls photosynthesis and plant growth. FEBS Lett. 2012, 586, 3692–3697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timm, S.; Wittmiß, M.; Gamlien, S.; Ewald, R.; Florian, A.; Frank, M.; Wirtz, M.; Hell, R.; Fernie, A.R.; Bauwe, H. Mitochondrial dihydrolipoyl dehydrogenase activity shapes photosynthesis and photorespiration of Arabidopsis Thaliana. Plant Cell 2015, 27, 1968–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Calcagno, P.E.; Fisk, S.; Brown, K.L.; Bull, S.E.; South, P.F.; Raines, C.A. Overexpressing the H-protein of the glycine cleavage system increases biomass yield in glasshouse and field-grown transgenic tobacco plants. Plant Biotechnol. J. 2019, 7, 141–151. [Google Scholar] [CrossRef]
- Kebeish, R.; Niessen, M.; Thiruveedhi, K.; Bari, R.; Hirsch, H.J.; Rosenkranz, R.; Stäbler, N.; Schönfeld, B.; Kreuzaler, F.; Peterhänsel, C. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat. Biotechnol. 2007, 25, 593–599. [Google Scholar] [CrossRef]
- South, P.F.; Cavanagh, A.P.; Liu, H.W.; Ort, D.R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 2019, 367, 45. [Google Scholar] [CrossRef] [Green Version]
- Jansson, C.; Wullschleger, S.D.; Kalluri, U.C.; Tuskan, G.A. Phytosequestration: Carbon Biosequestration by Plants and the Prospects of Genetic Engineering. Bioscience 2010, 60, 685–696. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.-G.; Long, S.P.; Ort, D.R. Improving Photosynthetic Efficiency for Greater Yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blankenship, R.E.; Chen, M. Spectral expansion and antenna reduction can enhance photosynthesis for energy production. Curr. Opin. Chem. Biol. 2013, 17, 457–461. [Google Scholar] [CrossRef]
- Nakajima, Y.; Ueda, R. Improvement of photosynthesis in dense microalgal suspension by reduction of light harvesting pigments. J. Appl. Phycol. 1997, 9, 503–510. [Google Scholar] [CrossRef]
- Beckmann, J.; Lehr, F.; Finazzi, G.; Hankamer, B.; Posten, C.; Wobbe, L.; Kruse, O. Improvement of light to biomass conversion by de-regulation of light-harvesting protein translation in Chlamydomonas reinhardtii. J. Biotechnol. 2009, 142, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Perrine, Z.; Negi, S.; Sayre, R.T. Optimization of photosynthetic light energy utilization by microalgae. Algal Res. 2012, 1, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Demmig-Adams, B.; Adams, W.W. Chlorophyll and carotenoid composition in leaves of Euonymus kiautschovicus acclimated to different degrees of light stress in the field. Aust. J. Plant Physiol. 1996, 23, 649–659. [Google Scholar] [CrossRef]
- Li, X.P.; Müller-Moulé, P.; Gilmore, A.M.; Niyogi, K.K. PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl. Acad. Sci. USA 2002, 99, 15222–15227. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Bueno, M.L.; Johnson, M.P.; Zia, A.; Ruban, A.V.; Horton, P. The Lhcb protein and xanthophyll composition of the light harvesting antenna controls the ΔpH-dependency of non-photochemical quenching in Arabidopsis thaliana. FEBS Lett. 2008, 582, 1477–1482. [Google Scholar] [CrossRef] [Green Version]
- Driever, S.M.; Simkin, A.J.; Alotaibi, S.; Fisk, S.J.; Madgwick, P.J.; Sparks, C.A.; Jones, H.D.; Lawson, T.; Parry, M.A.J.; Raines, C.A. Increased sbpase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160384. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.R.; Khaleda, L.; Jung, I.J.; Kim, J.Y.; Lee, S.Y.; Cha, J.Y.; Kim, W.Y. Overexpression of chloroplast-localized NADPH-dependent thioredoxin reductase C (NTRC) enhances tolerance to photo-oxidative and drought stresses in Arabidopsis thaliana. J. Plant Biol. 2017, 60, 175–180. [Google Scholar] [CrossRef]
- Kisaki, T.; Tolbert, N.E. Glycine as a substrate for photorespiration. Plant Cell Physiol. 1970, 11, 247–258. [Google Scholar] [CrossRef]
- Kisaki, T.; Imai, A.; Tolbert, N.E. Intracellular localization of enzymes related to photorespiration in green leaves. Plant Cell Physiol. 1971, 12, 267–273. [Google Scholar] [CrossRef]
- Eisenhut, M.; Ruth, W.; Haimovich, M.; Bauwe, H.; Kaplan, A.; Hagemann, M. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. USA 2008, 105, 17199–17204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, G.; Motokawa, Y.; Yoshida, T.; Hiraga, K. Glycine cleavage system: Reaction mechanism, physiological significance, and hyperglycinemia. Proc. Japan Acad. Ser. B Phys. Biol. Sci. 2008, 84, 246–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelitch, I.; Schultes, N.P.; Peterson, R.B.; Brown, P.; Brutnell, T.P. High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol. 2009, 149, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, C.; Kern, R.; Hüge, J.; Stal, L.J.; Tsuji, Y.; Kopka, J.; Shiraiwa, Y.; Bauwe, H.; Hagemann, M. Cyanobacterial lactate oxidases serve as essential partners in N2 fixation and evolved into photorespiratory glycolate oxidases in plants. Plant Cell 2011, 23, 2978–2990. [Google Scholar] [CrossRef] [Green Version]

| Tool | Description/Application | URL | Reference |
|---|---|---|---|
| BARLEX | The Barley Genome Explorer permits visual inspection of BAC overlaps, and comparisons of BACs and provides useful information on genes and markers | http://barlex.barleysequence.org | [49] |
| EnsemblPlants | A genome browser that incorporates genomic data from diverse organisms, including numerous plant species. It enables users to compare genome-scale datasets with the aid of a single collection of interfaces | http://plants.ensembl.org | [50] |
| IPK Barley BLAST Server | Barley BLAST server for genome-scale homology-based searches | http://webblast.ipk-gatersleben.de/barley | [51] |
| Golden Promise Genome | GMAP and BLAST server for barley (cv. Golden Promise) genome comparisons, including mapping of transcripts | https://ics.hutton.ac.uk/gmapper/ | [48] |
| Gramene | Integrated data resource for comparative functional genomics in crops and model plant species | http://www.gramene.org | [52] |
| PlantsDB | Provides data and information resources for individual plant species and a platform for integrative and comparative plant genome research. | http://pgsb.helmholtz-muenchen.de/plant/ | [53] |
| BaRTv1.0 | Barley Reference Transcript Dataset provides access to 177,240 barley-expressed transcripts covering 60,444 genes | https://ics.hutton.ac.uk/barleyrtd/ | [54] |
| Collections of natural genetic diversity | ||
| Gene Bank | Country | URL |
| PGRC Plant Gene Resources of Canada, Saskatoon Research Centre, Agriculture and Agri-Food Canada) | Canada | https://pgrc.agr.gc.ca/index_e.html |
| NSGC The National Small Grains Collection is part of the National Plant Germplasm System (NPGS) of the United States Department of Agriculture - Agricultural Research Service (USDA-ARS) | USA | https://www.ars.usda.gov/pacific-west-area/aberdeen-id/small-grains-and-potato-germplasm-research/docs/barley-wheat-genetic-stocks-collections-1/ |
| ICARDA International Centre for Agricultural Research in the Dry Areas | Global | https://grs.icarda.org/ |
| IPK Leibniz Institute of Plant Genetics and Crop Plant Research | Germany | http://gbis.ipk-gatersleben.de |
| WHEALBI WHEAt and barley Legacy for Breeding Improvement | France | http://wheat-urgi.versailles.inra.fr/Projects/Achieved-projects/Whealbi |
| NORDGEN Nordic Genetic Resources Centre | Sweden | https://www.nordgen.org/bgs/ |
| GRU Germplasm Resource Unit, John Innes Centre | UK | https://www.seedstor.ac.uk/ |
| NARO NIAS, National Institute of Agrobiological Sciences | Japan | https://www.gene.affrc.go.jp/databases_en.php |
| Online platforms for barley germplasm searches | ||
| Name | Description | URL |
| GENESIS | An online platform containing information about plant genetic resources for food and agriculture, conserved in gene banks worldwide | https://www.genesys-pgr.org/ |
| SINGER (The system-wide Information Network for Genetic Resources) | An online catalogue of crop collections together with their locations | https://www.gbif.org/dataset/85818aea-f762-11e1-a439-00145eb45e9a |
| EURISCO (The European Search Catalogue for Plant Genetic Resources) | Information on more than 2 million crop plant accessions and their wild relatives, preserved ex situ by almost 400 institutes in Europe and beyond | https://www.ecpgr.cgiar.org/resources/germplasm-databases/eurisco-catalogue/ |
| Induced Mutant Populations | ||
|---|---|---|
| Cultivar | Mutagen | Reference |
| Optic | EMS | [60] |
| Barke | EMS | [64] |
| Morex | NaN3 | [61] |
| Lux | NaN3 | [65] |
| DH-930-36 | MNU | [66] |
| DH-930-36 | Gamma rays | [66] |
| Sebastian | NaN3+MNU | [62] |
| Golden Promise | EMS | [63] |
| Target | Efficiency Gain | Strategy | Outcome | References |
|---|---|---|---|---|
| Retrograde signalling | ||||
| 1. Investigating the existence of a GUN1-dependent retrograde signalling pathway in barley | Not expected | Knock-out of the HORVU.MOREX.r2.5HG0366860.1 gene through gene editing | Molecular details of the retrograde signalling pathway involved in chloroplast biogenesis with possible repercussions for the control of leaf life-cycle | [122] |
| Light phase of photosynthesis | ||||
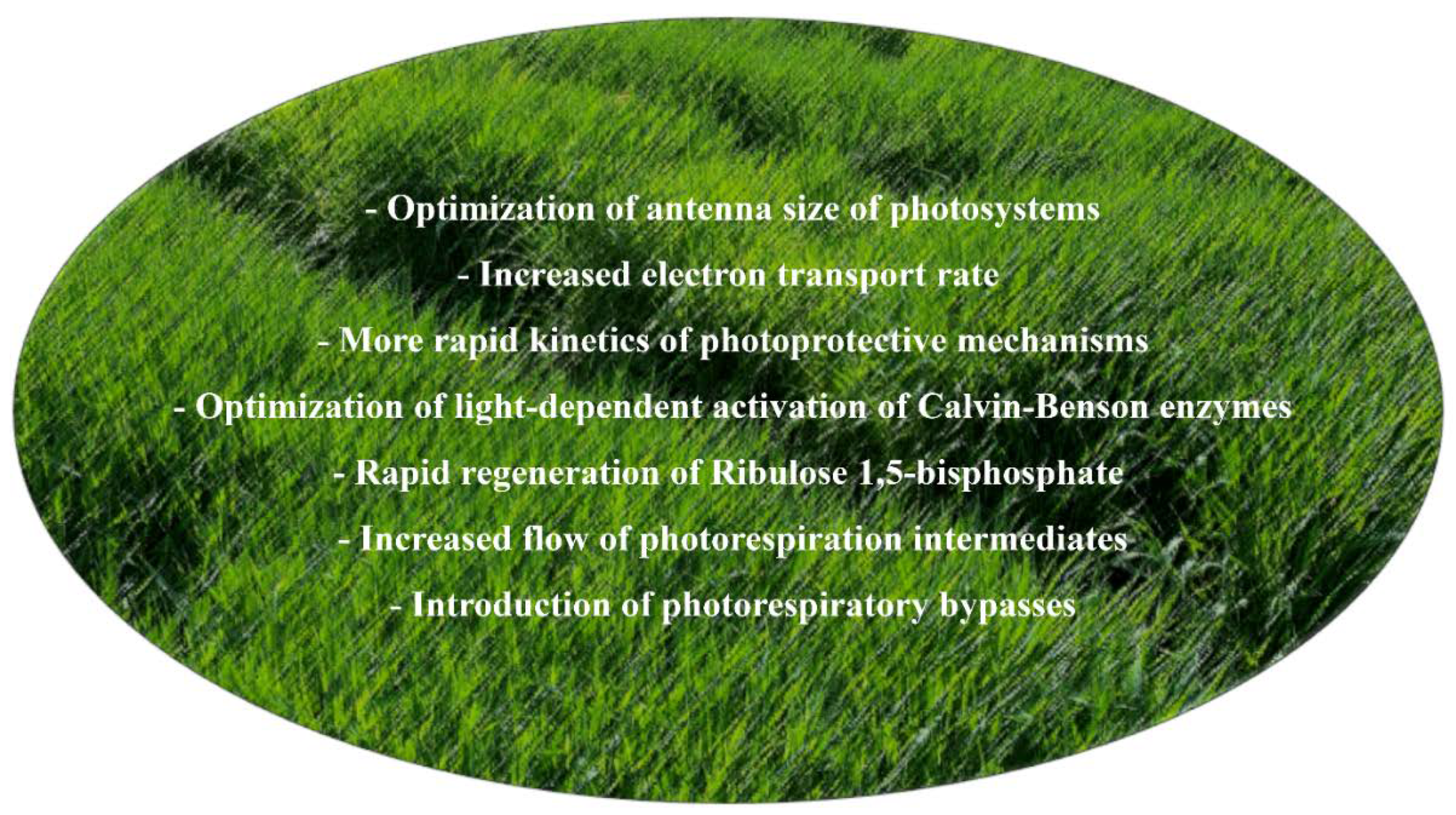
| 1. Optimisation of the antenna size | 20–50% | Reduction of photosystem antenna size obtained by either reducing chlorophyll production or decreasing levels of the photosystem antenna proteins by gene editing or introgression of induced mutations. Identification of allelic variants by allele mining and TILLING. | More uniform photosynthetic performance throughout the crop canopy and prevention of photo-oxidative damage in the upper layers of the canopy. Increases in land–surface reflectivity to offset greenhouse gas warming. | [39,132] |
| 2. Increased photosynthetic electron transport | 30–70% | Increased accumulation of electron carriers, such as cytochrome c6, plastocyanin and Rieske proteins, by transgenic approaches. Identification of allelic variants by allele mining and TILLING. | Increased electron transport rate through the thylakoid membranes | [133,134,135,136] |
| 3. Fine-Tuning of NPQ | 30% | Increased accumulation of VDE, ZEP and PsbS by transgenic approaches. Identification of allelic variants by allele mining and TILLING. | More rapid induction and relaxation of heat dissipation at PSII. | [137] |
| Dark phase of photosynthesis | ||||
| 1. Increasing the abundance of different enzymes of the Calvin–Benson cycle | >30% | Increased accumulation of SBPase and FBPA enzymes by transgenic approaches. Identification of allelic variants by allele mining and TILLING. | Optimization of ribulose 1,5-bisphosphate (RuBP) regeneration. | [138,139,140,141,142] |
| 2. Increasing the efficiency of light activation of Calvin–Benson enzymes | >20% | Increased accumulation of Rubisco activase, TRX f and NTRC by transgenic approaches. Identification of allelic variants by allele mining and TILLING. | More efficient light-dependent activation of Calvin–Benson enzyme optimises CO2 fixation. | [143,144,145,146,147] |
| Photorespiration | ||||
| 1. Increasing the photorespiration flow of intermediates | >15% | Increased accumulation of H- and L-proteins by transgenic approaches. | Reduced accumulation of photorespiration intermediates and increased CO2 assimilation rate | [148,149,150] |
| 2. Synthetic bypasses to photorespiration | >20% | Introduction of natural and synthetic glycolate catabolic pathways in the chloroplast | Increased CO2 assimilation rates | [151,152] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotasperti, L.; Sansoni, F.; Mizzotti, C.; Tadini, L.; Pesaresi, P. Barley’s Second Spring as a Model Organism for Chloroplast Research. Plants 2020, 9, 803. https://doi.org/10.3390/plants9070803
Rotasperti L, Sansoni F, Mizzotti C, Tadini L, Pesaresi P. Barley’s Second Spring as a Model Organism for Chloroplast Research. Plants. 2020; 9(7):803. https://doi.org/10.3390/plants9070803
Chicago/Turabian StyleRotasperti, Lisa, Francesca Sansoni, Chiara Mizzotti, Luca Tadini, and Paolo Pesaresi. 2020. "Barley’s Second Spring as a Model Organism for Chloroplast Research" Plants 9, no. 7: 803. https://doi.org/10.3390/plants9070803
APA StyleRotasperti, L., Sansoni, F., Mizzotti, C., Tadini, L., & Pesaresi, P. (2020). Barley’s Second Spring as a Model Organism for Chloroplast Research. Plants, 9(7), 803. https://doi.org/10.3390/plants9070803








