Easy-to-Use InDel Markers for Genetic Mapping between Col-0 and Ler-0 Accessions of Arabidopsis thaliana
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. DNA Extraction
3.2. PCR and Agarose Gel Electrophoresis
Author Contributions
Funding
Conflicts of Interest
References
- Jander, G.; Norris, S.R.; Rounsley, S.D.; Bush, D.F.; Levin, I.M.; Last, R.L. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002, 129, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.L.; Cnudde, F.; Gerats, T. Forward genetics and map-based cloning approaches. Trends Plant Sci. 2003, 8, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.-L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, J.; Dai, X.; Zhang, D.; Zhao, Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 2016, 171, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, A.; Ausubel, F.M. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993, 4, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J.; Drenkarci, E.; Preuss, D.; Ausubel, F.M. Use of cleaved amplified polymorphic sequences (CAPS) as genetic markers in Arabidopsis thaliana. Methods Mol. Biol. 1998, 82, 173–182. [Google Scholar] [PubMed]
- Bell, C.J.; Ecker, J.R. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 1994, 19, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Neff, M.M.; Neff, J.D.; Chory, J.; Pepper, A.E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998, 14, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Li, L.; Peng, Z.; Wei, B.; Tang, S.; Ding, M.; Liu, J.; Zhang, F.; Zhao, Y.; Gu, H.; et al. A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. Plant J. 2010, 63, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Pacurar, D.I.; Pacurar, M.L.; Street, N.; Bussell, J.D.; Pop, T.I.; Gutierrez, L.; Bellini, C. A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J. Exp. Bot. 2012, 63, 2491–2501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Y.; Glazebrook, J.; Li, X. Identification of components in disease-resistance signaling in Arabidopsis by map-based cloning. Methods Mol. Biol. 2007, 354, 69–78. [Google Scholar] [PubMed]
- Xing, J.; Weng, Q.; Si, H.; Han, J.; Dong, J. Identification and molecular tagging of two Arabidopsis resistance genes to Botrytis cinerea. Front. Agric. China 2011, 5, 430–436. [Google Scholar] [CrossRef]
- Bryant, F.; Hughes, D.; Hassani-Pak, K.; Eastmond, P. Basic LEUCINE ZIPPER TRANSCRIPTION FACTOR67 transactivates DELAY OF GERMINATION1 to establish primary seed dormancy in Arabidopsis. Plant Cell 2019, 31, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, M. Technical report: Part 2. Basic requirements for designing optimal PCR primers. J. Clin. Lab. Anal. 1996, 10, 285–293. [Google Scholar] [CrossRef]
- Zapata, L.; Ding, J.; Willing, E.M.; Hartwig, B.; Bezdan, D.; Jiao, W.B.; Patel, V.; Velikkakam James, G.; Koornneef, M.; Ossowski, S.; et al. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. USA 2016, 113, 4052–4060. [Google Scholar] [CrossRef] [PubMed]
- Sahu, B.; Sumit, R.; Srivastava, S.; Bhattacharyya, M. Sequence based polymorphic (SBP) marker technology for targeted genomic regions: Its application in generating a molecular map of the Arabidopsis thaliana genome. BMC Genom. 2012, 13, 20. [Google Scholar] [CrossRef] [PubMed]
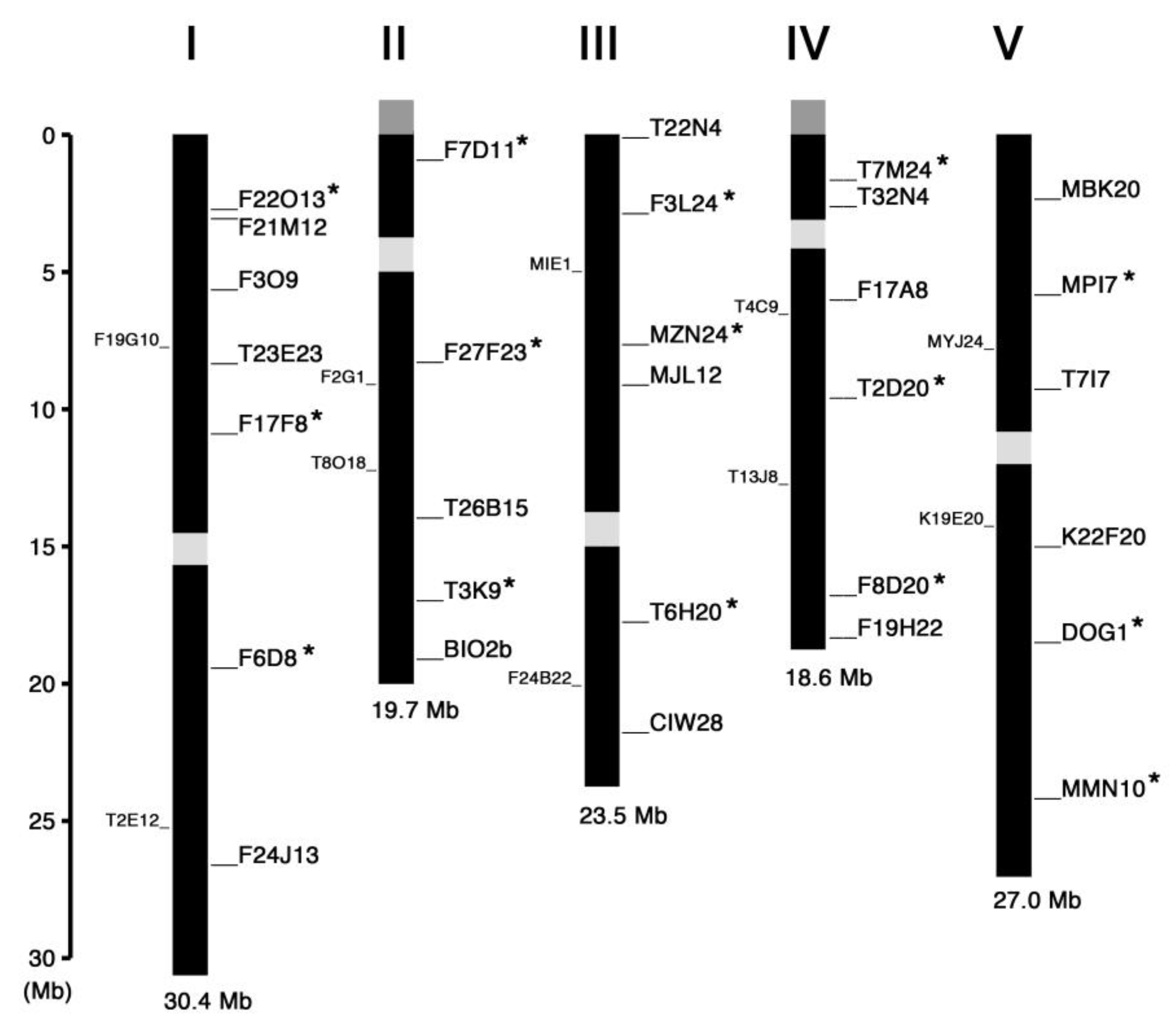
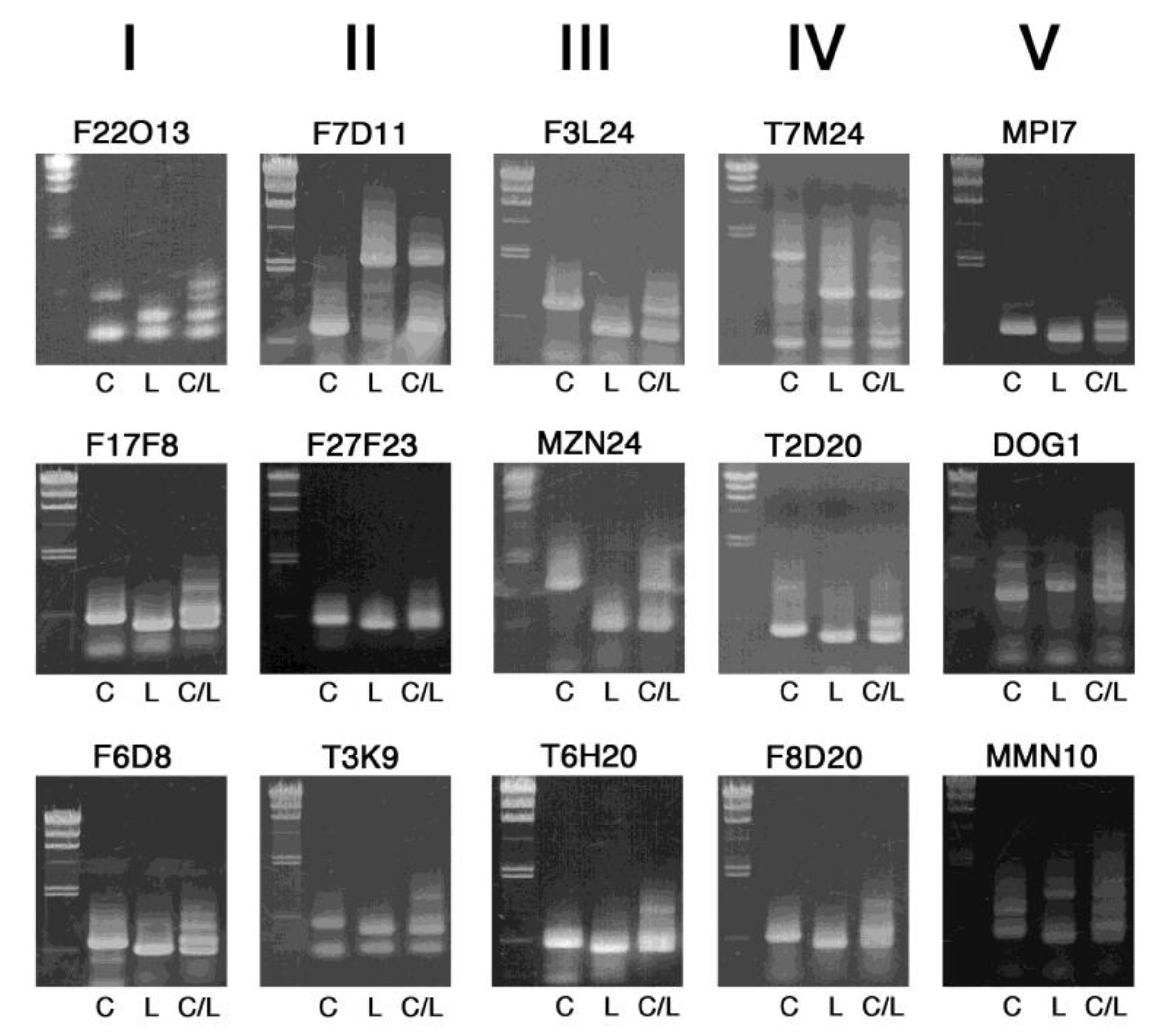
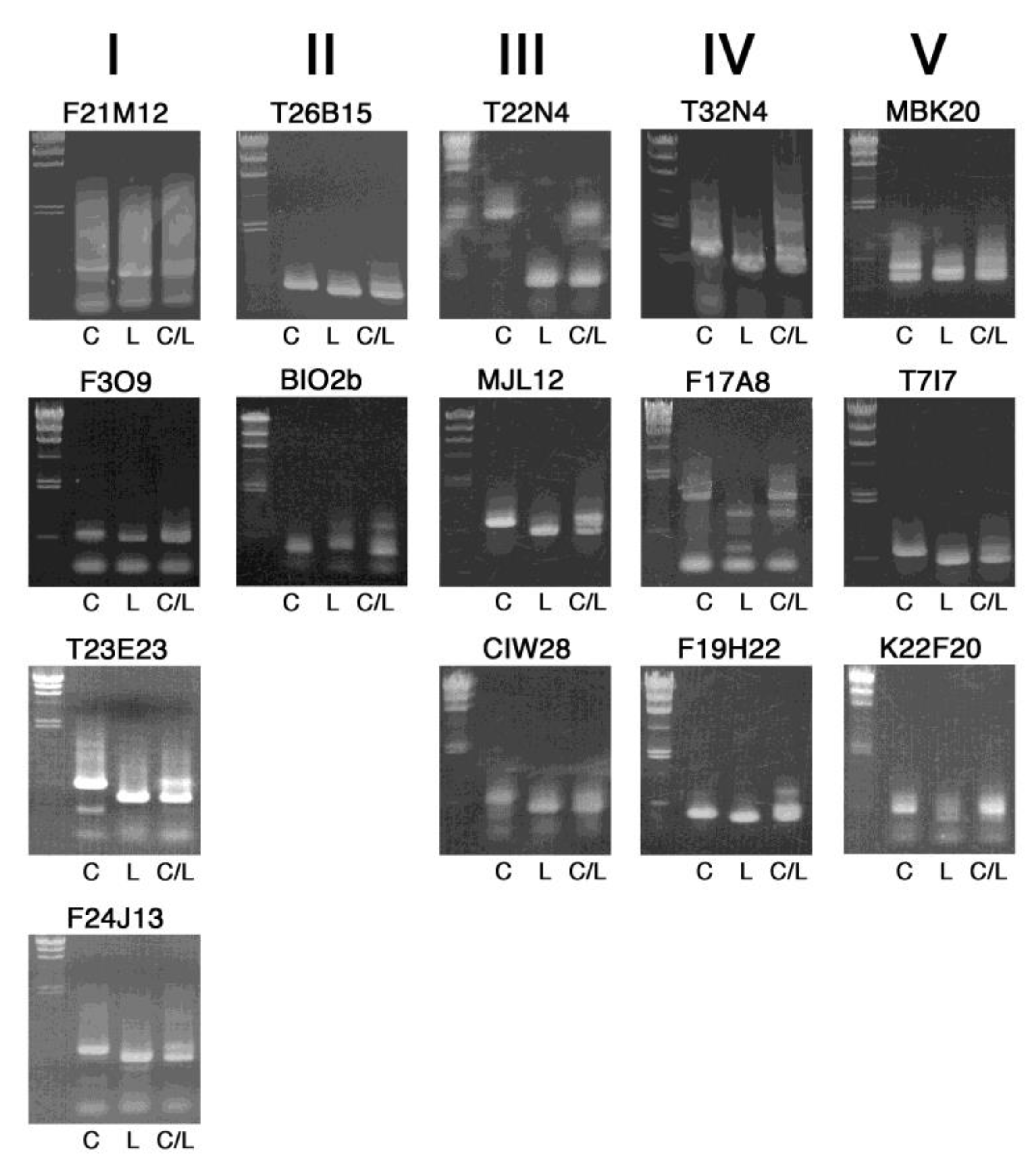
| Marker Name | Chr. | Position (Mbp) | Gene ID * | Length (bp) (Col-0/Ler) | Primer Sequence |
|---|---|---|---|---|---|
| F22O13 | 1 | 2.826 | At1g04667 | 460/210 | F: GTGTTGGGGAGAGCTTATAG |
| R: TCCACTTCCAACCATCAGAG | |||||
| F21M12 | 1 | 3.138 | At1g09700 (HYL1) | 580/500 | F: AAGACTCCATCTTGACACTG |
| R: CCTCAACCTACTGATCATTG | |||||
| F3O9 | 1 | 5.653 | At1g16530 (ASL9) | 610/530 | F: TTTTGGTCGGGTATGGAATG |
| R: CCAGAAGTTGCTCGTTAAAG | |||||
| T23E23 | 1 | 8.494 | At1g24000 | 540/420 | F: AAGGTCTTGTAGCGATCTAG |
| R: AACCCAACTGGCTCATTTTG | |||||
| F17F8 | 1 | 11.016 | At1g30930 | 460/360 | F: GGAAGAGGATTGACTCAAAG |
| R: CTACCGCTAGGACTTTCATG | |||||
| F6D8 | 1 | 19.621 | At1g52690 (LEA7) | 640/540 | F: GAGACACAGAGGAAGTGAAG |
| R: CTGACCAGCAAATTCTCAAG | |||||
| F24J13 | 1 | 26.624 | At1g70610 (ABCB26) | 600/500 | F: GCTACCCTTCAAGAGATGAG |
| R: TCGTAGAGTTGCAGCAAAAG | |||||
| F7D11 | 2 | 1.614 | At2g04622 | 670/580 | F: AGCGAACTTCGTTGATGTTC |
| R: CAATGTATATGCTCTTCTAGAG | |||||
| F27F23 | 2 | 8.410 | At2g19410 | 460/400 | F: TGACCAGTTGTACCAATGTG |
| R: GTCTGCGACAAAAAATACTG | |||||
| T26B15 | 2 | 13.825 | At2g32560 | 470/410 | F: AACACACTCTCTCTCTCTTG |
| R: AGGTCAAGAACCGACATTTG | |||||
| T3K9 | 2 | 17.107 | At2g40990 | 470/360 | F: TCATCGGAAGGAGCATTATG |
| R: AGGATGTTCCAGAGAGAATG | |||||
| BIO2b | 2 | 18.012 | At2g43360 (BIO2) | 390/460 | F: TGTACCTCCCTGAAGTTATG |
| R: TCTTGACCTCCTCTTCCATG | |||||
| T22N4 | 3 | 0.130 | At3g01345 | 2040/400 | F: TGACTGTTTGACTCCAAGTG |
| R: GTTACGAACCTCTGGTATTG | |||||
| F3L24 | 3 | 2.849 | At3g09270 (GSTU8) | 770/470 | F: TGAGCAATGATGGTTAGCAG |
| R: GAACGTAACTGCTTACGTAG | |||||
| MZN24 | 3 | 7.665 | At3g21750 (UGT71B1) | 1250/500 | F: ATCCGAACCGAAATCAACTG |
| R: GACTGAACGAGAGGAACATG | |||||
| MJL12 | 3 | 9.194 | At3g25240 | 650/490 | F: GGAGGCTAGAGACTCATATG |
| R: AGGGGATATTCGACTGAGAG | |||||
| T6H20 | 3 | 17.243 | At3g46820 (TOPP5) | 510/430 | F: AGTCCACCATGCATACAAAG |
| R: TGCATTGGTTTCTCTGCTTG | |||||
| CIW28 | 3 | 21.869 | At3g59140 (ABCC10) | 630/450 | F: GAGCACAAGTCTCTTACAAG |
| R: CCCTAAGTTTCACAAAGAATG | |||||
| T7M24 | 4 | 1.788 | At4g03826 | 1350/710 | F: TTTGGCGCTGTTGCCAATTG |
| R: TAATGCGCGAGGTGGATATG | |||||
| T32N4 | 4 | 2.549 | At4g04985 | 1180/900 | F: CTCAAGGTCGACATGATAATG |
| R: GTATAACGCGGGTCAATCTC | |||||
| F17A8 | 4 | 6.109 | At4g09670 | 1280/560 | F: TGCTCGAGAGACTTTTCGAG |
| R: CATAGACAGCCACACCAATG | |||||
| T2D20 | 4 | 9.652 | At4g17200 | 420/360 | F: TGGTCTTCTTATGCTCCAAG |
| R: AGAGGAAGCACACAGTATTG | |||||
| F8D20 | 4 | 16.924 | At4g35700 (DAZ3) | 540/460 | F: GGCGAGGATTGACTTAAATG |
| R: ACTGTTGCGATAATGCAGTG | |||||
| F19H22 | 4 | 18.133 | At4g38870 | 380/320 | F: GCGTTGTTGAGTGTAGCAAG |
| R: GAGATCGATCGTCATCTTTC | |||||
| MBK20 | 5 | 2.476 | At5g07770 (FH16) | 410/350 | F: AGAGACCCTTTTCTCTGTTG |
| R: GGAGCTTACCATCATATCAG | |||||
| MPI7 | 5 | 5.902 | At5g17860 (CAX7) | 570/490 | F: TCCAATTAGACCGCATATTAG |
| R: TTCGTTGCTTGAGACACTAG | |||||
| T7I7 | 5 | 9.271 | At5g26594 (RR24) | 650/500 | F: TGGCACCAAGAAGCAACTAG |
| R: TCCTAACTATCAACCAACTTG | |||||
| K22F20 | 5 | 15.027 | At5g00540 | 490/420 | F: ACCGCTACCATTTGTTCTTG |
| R: CCAACGTTCTTCCCTGTTAG | |||||
| DOG1 | 5 | 18.591 | At5g45830 (DOG1) | 1010/1300 | F: GCGTGTTTGTGTTTTGTGTG |
| R: ATCCGCTGTCTCAGGACATC | |||||
| MMN10 | 5 | 24.108 | At5g59840 (RABE1B) | 530/460 | F: TGAAGGATGACTCGTCTGTG |
| R: GATGGCTCTTTCACCACTAG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, T.; Nishii, Y.; Matsuo, H.; Takahashi, T. Easy-to-Use InDel Markers for Genetic Mapping between Col-0 and Ler-0 Accessions of Arabidopsis thaliana. Plants 2020, 9, 779. https://doi.org/10.3390/plants9060779
Tanaka T, Nishii Y, Matsuo H, Takahashi T. Easy-to-Use InDel Markers for Genetic Mapping between Col-0 and Ler-0 Accessions of Arabidopsis thaliana. Plants. 2020; 9(6):779. https://doi.org/10.3390/plants9060779
Chicago/Turabian StyleTanaka, Takahiro, Yuichi Nishii, Hirotoshi Matsuo, and Taku Takahashi. 2020. "Easy-to-Use InDel Markers for Genetic Mapping between Col-0 and Ler-0 Accessions of Arabidopsis thaliana" Plants 9, no. 6: 779. https://doi.org/10.3390/plants9060779
APA StyleTanaka, T., Nishii, Y., Matsuo, H., & Takahashi, T. (2020). Easy-to-Use InDel Markers for Genetic Mapping between Col-0 and Ler-0 Accessions of Arabidopsis thaliana. Plants, 9(6), 779. https://doi.org/10.3390/plants9060779






