Paternity Assignment in White Guinea Yam (Dioscorea Rotundata) Half-Sib Progenies from Polycross Mating Design Using SNP Markers
Abstract
1. Introduction
2. Results
2.1. Heterozygosity and Genetic Diversity
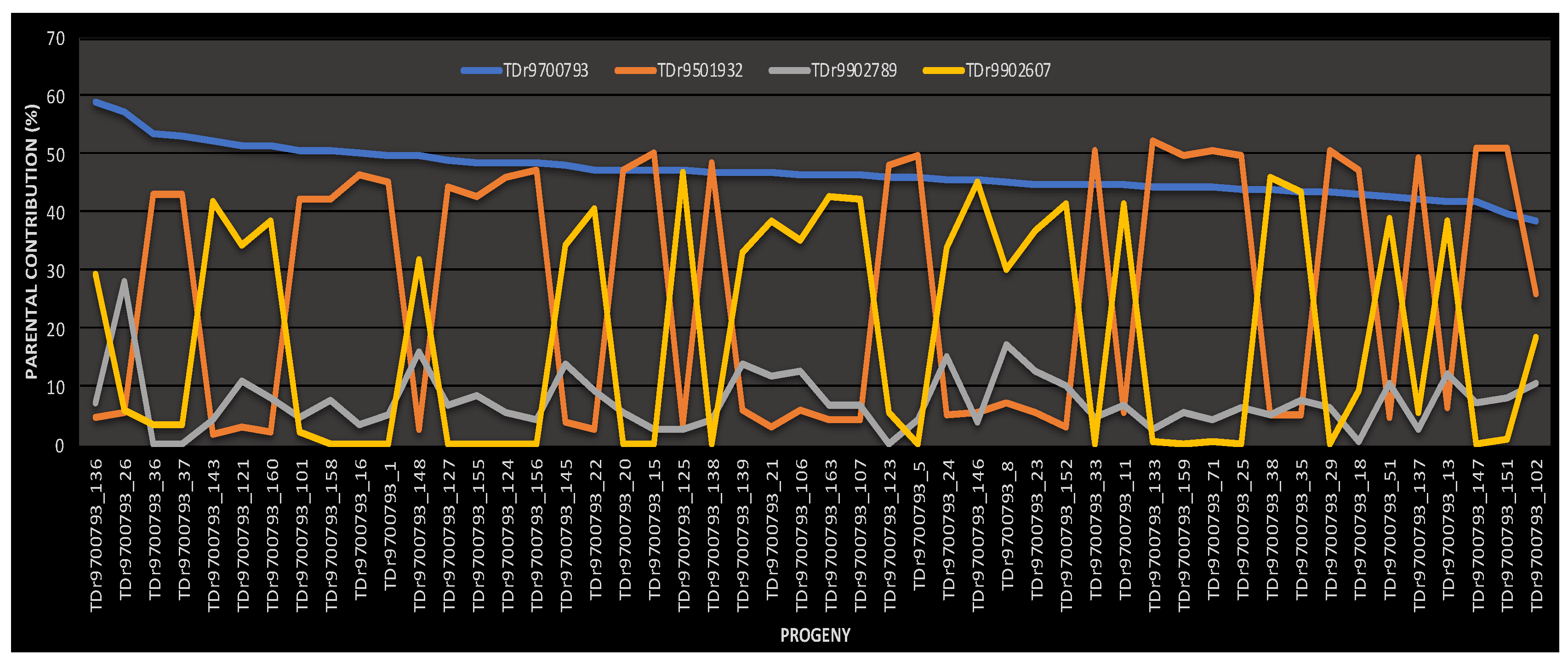
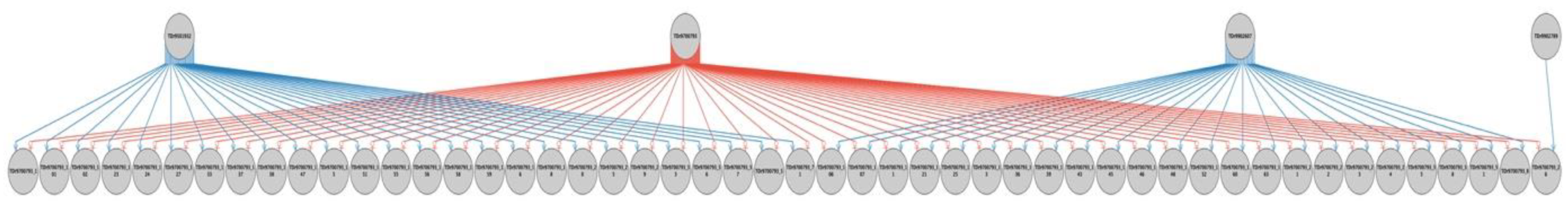
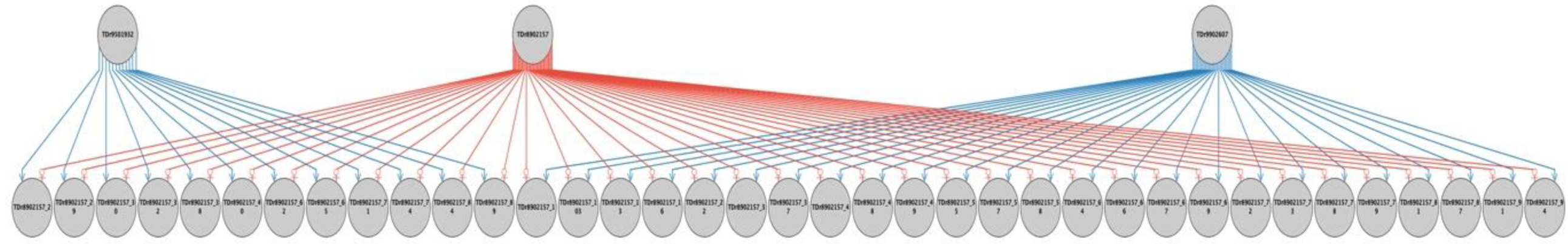
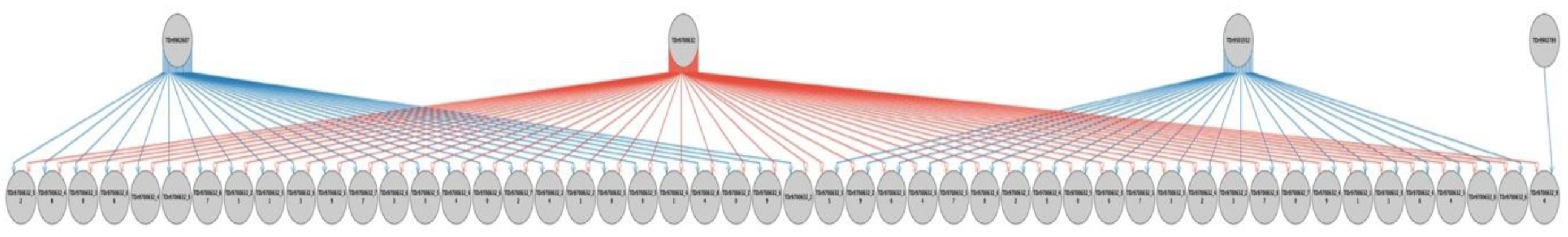
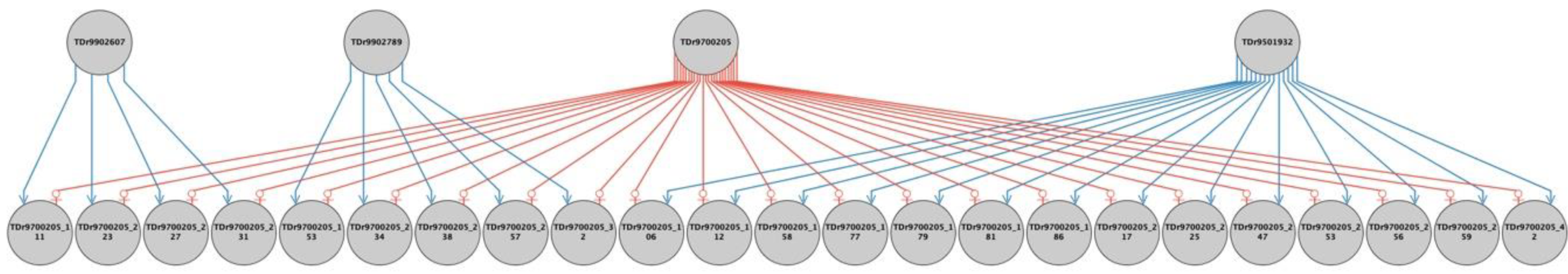
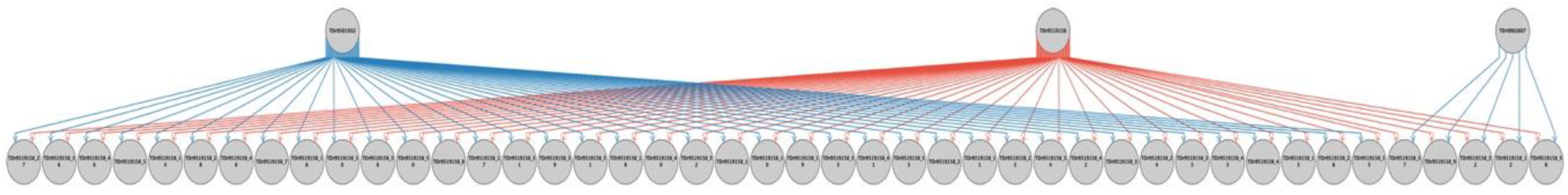
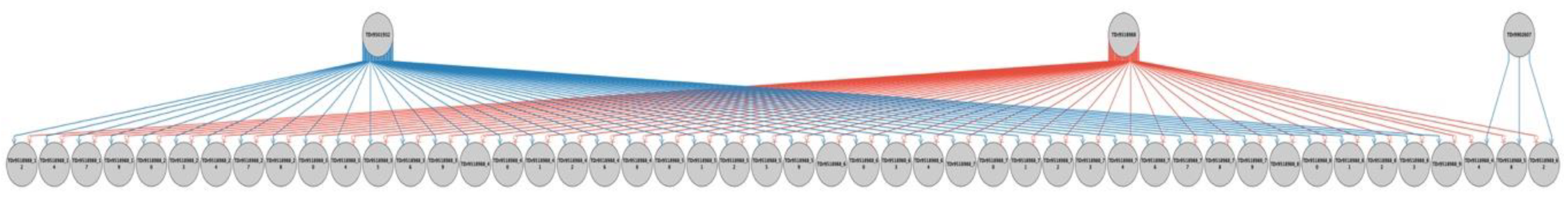
2.2. Parental Reconstruction and Gametic Composition in Half-Sib Progenies
2.3. Numbers and Patterns of SNP Mutations in Half-Sib Progenies of White Yam
3. Discussion
3.1. Allelic Diversity and Parental Reconstruction
3.2. Numbers and Patterns of SNP Mutations in Half-Sib Progenies of White Yam
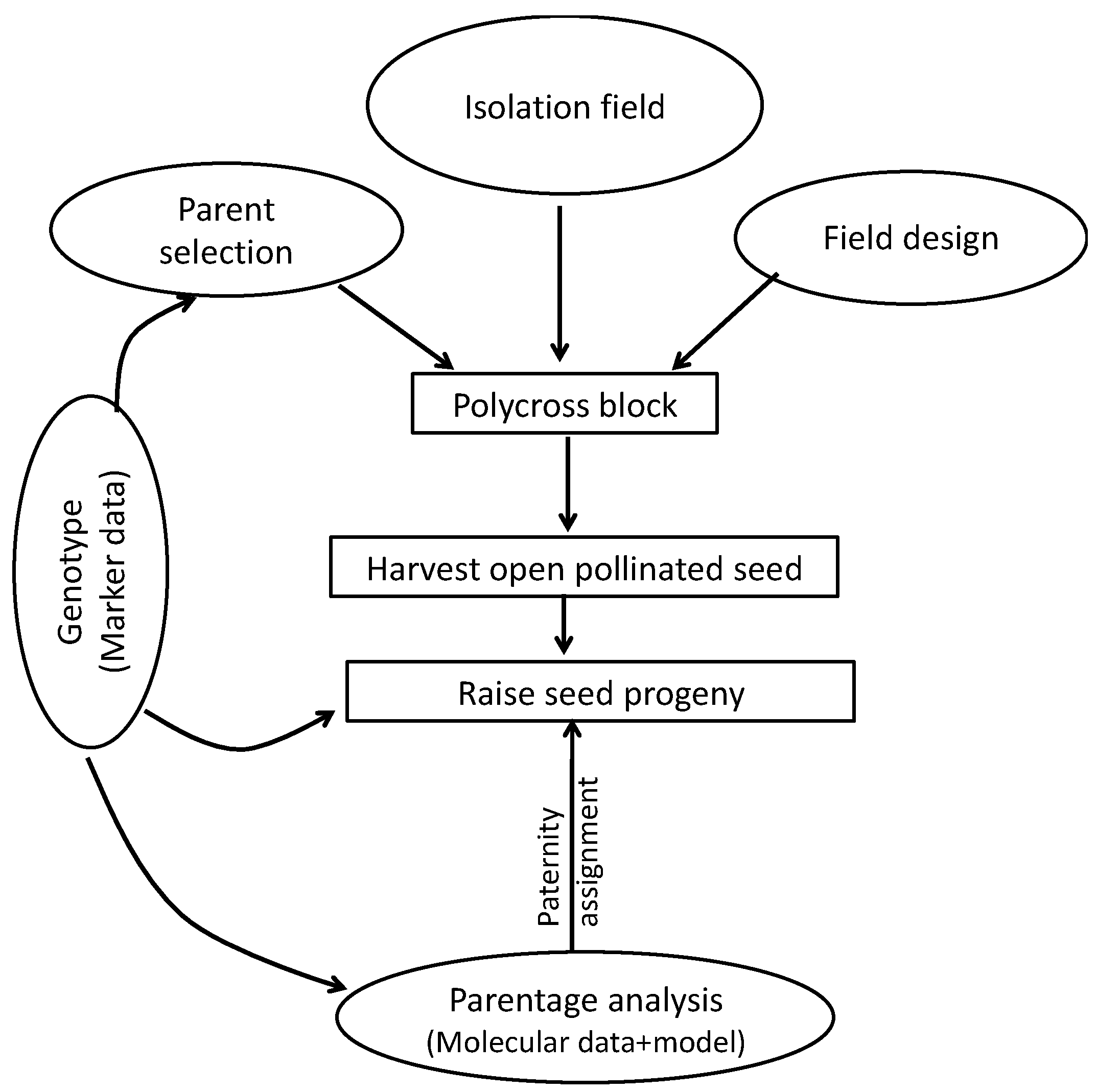
4. Materials and Methods
4.1. Experimental Materials
4.2. Genotyping
4.3. Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CTAB | cetyl trimethyl ammonium bromide |
| DArT | diversity array technology |
| DNA | deoxyribonucleic acid |
| IITA | international institute of tropical agriculture |
| ISRIC | international soil reference and information centre |
| MAT | months after transplanting |
| SNP | single nucleotide polymorphism |
| VCF | variant call format |
| WAS | weeks after planting |
References
- Govaerts, R.; Wilkin, P.; Saunders, R.M.K. World Checklist of Dioscoreales: Yams and Their Allies; The Board of Trustees of the Royal Botanic Gardens: London, UK, 2007; pp. 1–65. [Google Scholar]
- Bai, K.V.; Ekanayake, I.J. Taxonomy, morphology and floral biology. In Food Yams: Advances in Research; Orkwor, G.C., Asiedu, R., Ekanayake, I.J., Eds.; NRCRI and IITA: Ibadan, Nigeria, 1998; pp. 13–38. [Google Scholar]
- Lebot, V. Tropical Root and Tuber Crops: Cassava, Sweet Potato, Yams and Aroids; CABI: Wallingford, UK, 2009; 413p. [Google Scholar]
- Mignouna, H.D.; Abang, M.M.; Onasanya, A.; Agindotan, B.; Asiedu, R. Identification and potential use of RAPD markers linked to Yam mosaic virus resistance in white yam (Dioscorea rotundata Poir). Ann. Appl. Biol. 2002, 140, 163–169. [Google Scholar] [CrossRef]
- Scarcelli, N.; Daïnou, O.; Agbangla, C.; Tostain, S.; Pham, J.L. Segregation patterns of isozyme loci and microsatellite markers show the diploidy of African yam Dioscorea rotundata (2n = 40). Theor. Appl. Gen. 2005, 111, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Sybenga, J. Cytogenetics in Plant Breeding. In Monograph on Theoretical and Applied Genetics 17; Springer Science and Business Media: Berlin, Germany, 2012; p. 469. [Google Scholar]
- Stebbins, G. Types of polyploids: Their classification and significance. Adv. Genet. 1947, 1, 403–429. [Google Scholar] [PubMed]
- Stift, M.; Berenos, C.; Kuperus, P.; van Tienderen, P.H. Segregation models for disomic, tetrasomic and intermediate inheritance in tetraploids: A general procedure applied to Rorippa (yellow cress) microsatellite data. Genetics 2008, 179, 2113–2123. [Google Scholar] [CrossRef]
- Obidiegwu, J.E.; Akpabio, E.M. The geography of yam cultivation in southern Nigeria: Exploring its social meanings and cultural functions. J. Ethn. Foods 2017, 4, 28–35. [Google Scholar] [CrossRef]
- Alabi, T.R.; Adebola, P.O.; Asfaw, A.; De Koeyer, D.; Lopez-Montes, A.; Asiedu, R. Spatial multivariate cluster analysis for defining target population of environments in West Africa for yam breeding. Int. J. Appl. Geo. Res. 2019, 10, 1–30. [Google Scholar] [CrossRef]
- Darkwa, K.; Olasanmi, B.; Asiedu, R.; Asfaw, A. Review of empirical and emerging methods and tools for yam (Dioscorea spp.) improvement: Status and prospects. Plant Breed. 2019, 1–24. [Google Scholar] [CrossRef]
- Lebot, V.; Abraham, K.; Kaoh, J.; Rogers, C.; Molisalé, T. Development of anthracnose resistant hybrids of the Greater yam (Dioscorea alata L.) and interspecific hybrids with D. nummularia Lam. Gen. Res. Crop Evol. 2019, 66, 871–883. [Google Scholar] [CrossRef]
- Norman, P.E.; Asfaw, A.; Tongoona, P.B.; Danquah, A.; Danquah, E.Y.; De Koeyer, D.; Asiedu, R. Pollination success in some white yam genotypes under polycross and nested mating designs. Int. J. Biol. Sci. Appl. 2018, 5, 19–28. [Google Scholar]
- Jones, A.G.; Small, C.M.; Paczolt, K.A.; Ratterman, N.L. A practical guide to methods of parentage analysis. Mol. Ecol. Res. 2010, 10, 6–30. [Google Scholar] [CrossRef]
- Telfer, E.J.; Stovold, G.T.; Li, Y.; Silva-Junior, O.B.; Grattapaglia, D.G.; Dungey, H.S. Parentage reconstruction in Eucalyptus nitens using SNPs and microsatellite markers: A comparative analysis of marker data power and robustness. PLoS ONE 2015, 10, e0130601. [Google Scholar] [CrossRef]
- Herbinger, C.M.; Doyle, R.W.; Pitman, E.R.; Paquet, D.; Mesa, K.A.; Morris, D.B.; Wright, J.M.; Cook, D. DNA fingerprint based analysis of paternal and maternal effects on offspring growth and survival in communally reared rainbow trout. Aquaculture 1995, 137, 245–256. [Google Scholar] [CrossRef]
- Zoundjihékpon, J.; Hamon, S.; Tio-Touré, B.; Hamon, P. First controlled progenies checked by isozyme markers in cultivated yams Dioscorea cayenensis-rotundata. Theor. Appl. Genet. 1994, 88, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Sartie, A.; Asiedu, R. Development of mapping populations for genetic analysis in yams (Dioscorea rotundata Poir and Dioscorea alata L). Afr. J. Biotech. 2011, 10, 3040–3050. [Google Scholar]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H.; Weising, K.; Rotter, B. DNA fingerprinting in botany: Past, present, future. Investig. Genet. 2014, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Shete, S.; Tiwari, H.; Elston, R.C. On estimating the heterozygosity and polymorphism information content value. Theor. Popul. Biol. 2000, 57, 265–271. [Google Scholar] [CrossRef]
- Pruvost, N.B.M.; Hoffmann, A.; Reyer, H.U. Gamete production patterns, ploidy, and population genetics reveal evolutionary significant units in hybrid water frogs (Pelophylax exculentus). Ecol. Evol. 2013, 3, 2933–2948. [Google Scholar] [CrossRef]
- Petit, R.J.; El Mousadik, A.; Pons, O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 1998, 12, 844–855. [Google Scholar] [CrossRef]
- Zeng, Z.B.; Cockerham, C.C. Long-term response to artificial selection with multiple alleles—study by simulations. Theor. Pop. Biol. 1990, 37, 254–272. [Google Scholar] [CrossRef]
- Wagner, A. Robustness and evolvability: A paradox resolved. Proc. R. Soc. B Biol. Sci. 2008, 275, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, G.; Templeton, A.R.; Zarmi, Y.; Bar-David, S. Allelic richness following population founding events–A stochastic modeling framework incorporating gene flow and genetic drift. PLoS ONE 2014, 9, e115203. [Google Scholar] [CrossRef]
- Norman, P.E.; Asfaw, A.; Tongoona, P.B.; Danquah, A.; Danquah, E.Y.; Koeyer, D.D.; Asiedu, R. Can parentage analysis facilitate breeding activities in root and tuber crops? Agriculture 2018, 8, 95. [Google Scholar] [CrossRef]
- Snow, A.A.; Spira, T.P. Pollen-tube competition and male fitness in Hibiscus moscheutos. Evolution 1996, 50, 1866–1870. [Google Scholar] [CrossRef] [PubMed]
- Pasonen, H.L.; Pulkkinen, P.; Käpylä, M.; Blom, A. Pollen-tube growth rate and seed-siring success among Betula pendula clones. New Phytol. 1999, 143, 243–251. [Google Scholar] [CrossRef]
- Aronen, T.; Nikkanen, T.; Harju, A.; Tiimonen, H.; Häggman, H. Pollen competition and seed-siring success in Picea abies. Theor. Appl. Genet. 2002, 104, 638–642. [Google Scholar] [CrossRef]
- Spira, T.; Snow, A.A.; Puterbaugh, M.N. The timing and effectiveness of sequential pollinations in Hibiscus moscheutos. Oecologia 1996, 105, 230–235. [Google Scholar] [CrossRef]
- Parantainen, A.; Pasonen, H.L. Pollen-pollen interactions in Pinus sylvestris. Scand. J. For. Res. 2004, 19, 199–205. [Google Scholar] [CrossRef]
- Nakamura, R.R.; Wheeler, N.C. Pollen competition and paternal success in Douglas-fir. Evolution 1992, 46, 846–851. [Google Scholar] [CrossRef]
- Moriguchi, Y.; Ishiduka, D.; Kaneko, T.; Itoo, S.; Taira, H.; Tsumura, Y. The Contribution of pollen germination rates to uneven paternity among polycrosses of Cryptomeria japonica. Silvae Genet. 2009, 58, 139–144. [Google Scholar] [CrossRef]
- Mignouna, H.D.; Abang, M.M.; Asiedu, R.; Geeta, R. Producing yam (Dioscorea) seeds through controlled crosses. Cold Spring Harbor Protoc. 2009, prot5327. [Google Scholar] [CrossRef]
- Segnou, M.; Fatokun, C.A.; Akoroda, M.O.; Hahn, S. Studies on the reproductive biology of white yam (Dioscorea rotundata Poir.). Euphytica 1992, 64, 197–203. [Google Scholar] [CrossRef]
- Ardlie, K.G.; Lunetta, K.L.; Seielstad, M. Testing for population subdivision and association in four case-control studies. Amer. J. Hum. Genet. 2002, 71, 304–311. [Google Scholar] [CrossRef]
- Vos, P.G.; Uitdewilligen, J.G.; Voorrips, R.E.; Visser, R.G.; van Eck, H.J. Development and analysis of a 20K SNP array for potato (Solanum tuberosum): An insight into the breeding history. Theor. Appl. Genet. 2015, 128, 2387–2401. [Google Scholar] [CrossRef]
- Tamiru, M.; Natsume, S.; Takagi, H.; White, B.; Yaegashi, H.; Shimizu, M.; Yoshida, K.; Uemura, A.; Oikawa, K.; Abe, A.; et al. Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination. BMC Biol. 2017, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Vincent, K.; Robert, K.; Morag, F.; Tadeo, K.; Yona, B.; Peter, V. Identification of F1 cassava (Manihot esculenta Crantz) progeny using microsatellite markers and capillary electrophoresis. Am. J. Plant Sci. 2014, 5, 119–125. [Google Scholar] [CrossRef]
- Rabbi, I.Y.; Kulakow, P.A.; Manu-Aduening, J.A.; Dankyi, A.A.; Asibuo, J.Y.; Parkes, E.Y.; Abdoulaye, T.; Girma, G.; Gedil, M.A.; Ramu, P.; et al. Tracking crop varieties using genotyping by-sequencing markers: A case study using cassava (Manihot esculenta Crantz). BMC Genet. 2015, 16, 115. [Google Scholar] [CrossRef]
- Da Silva, D.C.S.; Martins, M.L.L.; Santos, A.S.; da Silva Santos, V.; Alves, A.A.C.; da Silva Ledo, C.A. Obtaining hybrids of cultivars and wild subspecies of cassava. Pesqui. Agropecu. Bras. 2018, 53, 182–188. [Google Scholar] [CrossRef]
- Smyda-Dajmund, P.; Sliwka, J.; Wasilewicz-Flis, I.; Jakuczun, H.; Zimnoch-Guzowska, E. Genetic composition of interspecific potato somatic hybrids and autofused 4x plants evaluated by DArT and cytoplasmic DNA markers. Plant Cell Rep. 2016, 35, 1345–1358. [Google Scholar] [CrossRef]
- Endelman, J.B.; Carley, C.A.S.; Douches, D.S.; Coombs, J.J.; Bizimungu, B.; De Jong, W.S.; Holm, K.G.; Holm, D.G.; Creighton Miller, J., Jr.; Novy, R.G.; et al. Pedigree reconstruction with genome-wide markers in potato. Am. J. Potato Res. 2017, 94, 184–190. [Google Scholar] [CrossRef]
- Buteler, M.I.; LaBonte, R.L.; Jarret, D.R.; Macchiavelli, R.E. Microsatellite-based paternity analysis in polyploid sweetpotato. J. Am. Soc. Hortic. Sci. 2002, 127, 392–396. [Google Scholar] [CrossRef]
- Jones, A.G.; Ardren, W.R. Methods of parentage analysis in natural populations. Mol. Ecol. 2003, 12, 2511–2523. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, P.R.; Resende, M.F.R., Jr.; Huber, D.A.; Quesada, T.; Resende, M.D.V.; Neale, D.B.; Wegrzyn, J.L.; Kirst, M.; Peter, G.F. Genomic relationship matrix for correcting pedigree errors in breeding populations: Impact on genetic parameters and genomic selection accuracy. Crop Sci. 2014, 54, 1115–1123. [Google Scholar] [CrossRef]
- Cao, J.; Jiang, D.; Zhao, Z.; Yuan, S.; Zhang, Y.; Zhang, T.; Zhong, W.; Yuan, Q.; Huang, L. Development of Chloroplast Genomic Resources in Chinese Yam (Dioscorea polystachya). BioMed Res. Int. 2018, 2018, 6293847. [Google Scholar] [CrossRef] [PubMed]
- Exposito-Alonso, M.; Becker, C.; Schuenemann, V.J.; Reiter, E.; Setzer, C.; Slovak, R.; Brachi, B.; Hagmann, J.; Grimm, D.G.; Chen, J.; et al. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 2018, 14, e1007155. [Google Scholar] [CrossRef]
- International Soil Reference and Information Centre and the Food and Agriculture Organization (ISRIC/FAO). Procedures for Soil Analysis, 6th ed.; Technical Paper 9; FAO: Roma, Italy, 2002; p. 120. [Google Scholar]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA mini preparation Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR- based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- McIlhagga, W. Penalized: A MATLAB Toolbox for fitting generalized linear models with penalties. J. Stat. Soft 2016, 72, 1–21. [Google Scholar] [CrossRef]
- Shaw, P.D.; Graham, M.; Kennedy, J.; Milne, I.; Marshall, D.F. Helium: Visualization of large scale plant pedigrees. BMC Bioinform. 2014, 15, 259. [Google Scholar] [CrossRef]
- Shaw, P.D. Visualizing Genetic Transmission Patterns in Plant Pedigrees. Ph.D. Thesis, Edinburgh Napier University, Edinburgh, UK, 2016; p. 221. [Google Scholar]

| Family | Sample | Sample after QC | Hybrid | Proportion of Heterozygous | Minor Allele Frequency | Major Allele Frequency | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | ||||||||
| TDr1685 | 50 | 50 | 50 | 0.276 | 0.193 | 0.486 | 0.002 | 0.043 | 0.205 | 0.794 |
| TDr1686 | 50 | 50 | 37 | 0.276 | 0.116 | 0.531 | 0.004 | 0.065 | 0.211 | 0.788 |
| TDr1687 | 50 | 46 | 46 | 0.291 | 0.229 | 0.397 | 0.001 | 0.030 | 0.209 | 0.791 |
| TDr1688 | 50 | 50 | 50 | 0.327 | 0.294 | 0.407 | 0.000 | 0.020 | 0.233 | 0.766 |
| TDr1689 | 50 | 28 | 28 | 0.328 | 0.161 | 0.517 | 0.002 | 0.048 | 0.238 | 0.762 |
| TDr1690 | 45 | 44 | 44 | 0.272 | 0.255 | 0.437 | 0.001 | 0.029 | 0.186 | 0.813 |
| TDr1691 | 49 | 48 | 48 | 0.281 | 0.216 | 0.360 | 0.001 | 0.029 | 0.204 | 0.795 |
| TDr1692 | 50 | 50 | 49 | 0.293 | 0.251 | 0.460 | 0.001 | 0.038 | 0.209 | 0.791 |
| Female | Family | Male | Progeny with Fully Recovered Pedigree | Percent of Progenies with Fully Recovered Pedigree | ||
|---|---|---|---|---|---|---|
| TDr9501932 | TDr9902789 | TDr9902607 | ||||
| TDr9700793 | TDr1685 | 26 (52%) | 1 (2.0%) | 23 (46%) | 50 | 100 |
| TDr8902157 | TDr1686 | 12 (24%) | 0 (0%) | 25 (50%) | 37 | 74 |
| TDr8902475 | TDr1687 | 46 (92%) | 0 (0%) | 0 (0%) | 46 | 92 |
| TDr9700632 | TDr1688 | 23 (46%) | 1 (2%) | 26 (52%) | 50 | 100 |
| TDr9700205 | TDr1689 | 18 (36%) | 6 (12%) | 4 (8%) | 28 | 56 |
| TDr9519158 | TDr1690 | 39 (86.7%) | 0 (0%) | 5 (11.1%) | 44 | 97.8 |
| TDr9518988 | TDr1691 | 45 (91.8%) | 0 (0%) | 3 (6.1%) | 48 | 97.9 |
| Ojuiyawo | TDr1692 | 22 (44.0%) | 27 (54%) | 0 (0%) | 49 | 98 |
| Total progenies | 231 | 35 | 86 | 352 | ||
| % contribution | 65.63 | 9.94 | 24.43 | 100 | 96.2 | |
| Family | Number of Nonmissing Gametes | Proportion of Nonmissing Gametes | Number of Missing Gametes | Proportion of Missing Gametes |
|---|---|---|---|---|
| TDr1685 | 669930 | 0.940 | 42870 | 0.060 |
| TDr1686 | 660336 | 0.926 | 52680 | 0.074 |
| TDr1687 | 672606 | 0.943 | 40410 | 0.057 |
| TDr1688 | 676128 | 0.948 | 36888 | 0.052 |
| TDr1689 | 681738 | 0.939 | 44482 | 0.061 |
| TDr1690 | 601600 | 0.949 | 32192 | 0.051 |
| TDr1691 | 643928 | 0.938 | 42680 | 0.062 |
| TDr1692 | 674222 | 0.946 | 38794 | 0.054 |
| Family | SNP Type | Tv to Ts Ratio | |||||
|---|---|---|---|---|---|---|---|
| Transitions (Ts) | Transversions (Tv) | ||||||
| AG | CT | AT | AC | GT | GC | ||
| TDr1685 | 805 (29.10) | 854 (30.87) | 394 (14.24) | 257 (9.29) | 261 (9.44) | 195 (7.05) | 1.67 |
| TDr1686 | 822 (28.20) | 938 (32.18) | 413 (14.17) | 251 (8.61) | 280 (9.61) | 211 (7.24) | 1.66 |
| TDr1687 | 785 (30.05) | 852 (32.62) | 344 (13.17) | 227 (8.69) | 230 (8.81) | 174 (6.66) | 1.60 |
| TDr1688 | 829 (28.57) | 924 (31.84) | 416 (14.33) | 259 (8.92) | 266 (9.17) | 208 (7.17) | 1.66 |
| TDr1689 | 869 (27.86) | 1005 (32.22) | 419 (13.43) | 292 (9.36) | 301 (9.65) | 233 (7.47) | 1.66 |
| TDr1690 | 756 (29.62) | 820 (32.13) | 349 (13.68) | 223 (8.74) | 228 (8.93) | 176 (6.90) | 1.62 |
| TDr1691 | 755 (29.13) | 841 (32.45) | 353 (13.62) | 237 (9.14) | 238 (9.18) | 168 (6.48) | 1.62 |
| TDr1692 | 831 (29.37) | 888 (31.39) | 403 (14.25) | 256 (9.05) | 256 (9.05) | 195 (6.89) | 1.65 |
| Mean | 806.5 (28.99) | 890.25 (31.96) | 386.38 (13.86) | 250.25 (8.98) | 257.5 (9.23) | 195.0 (6.98) | 1.64 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norman, P.E.; Paterne, A.A.; Danquah, A.; Tongoona, P.B.; Danquah, E.Y.; De Koeyer, D.; Ikeogu, U.N.; Asiedu, R.; Asfaw, A. Paternity Assignment in White Guinea Yam (Dioscorea Rotundata) Half-Sib Progenies from Polycross Mating Design Using SNP Markers. Plants 2020, 9, 527. https://doi.org/10.3390/plants9040527
Norman PE, Paterne AA, Danquah A, Tongoona PB, Danquah EY, De Koeyer D, Ikeogu UN, Asiedu R, Asfaw A. Paternity Assignment in White Guinea Yam (Dioscorea Rotundata) Half-Sib Progenies from Polycross Mating Design Using SNP Markers. Plants. 2020; 9(4):527. https://doi.org/10.3390/plants9040527
Chicago/Turabian StyleNorman, Prince E., Agre A. Paterne, Agyemang Danquah, Pangirayi B. Tongoona, Eric Y. Danquah, David De Koeyer, Ugochukwu N. Ikeogu, Robert Asiedu, and Asrat Asfaw. 2020. "Paternity Assignment in White Guinea Yam (Dioscorea Rotundata) Half-Sib Progenies from Polycross Mating Design Using SNP Markers" Plants 9, no. 4: 527. https://doi.org/10.3390/plants9040527
APA StyleNorman, P. E., Paterne, A. A., Danquah, A., Tongoona, P. B., Danquah, E. Y., De Koeyer, D., Ikeogu, U. N., Asiedu, R., & Asfaw, A. (2020). Paternity Assignment in White Guinea Yam (Dioscorea Rotundata) Half-Sib Progenies from Polycross Mating Design Using SNP Markers. Plants, 9(4), 527. https://doi.org/10.3390/plants9040527








