In Arabidopsis thaliana Heterosis Level Varies among Individuals in an F1 Hybrid Population
Abstract
1. Introduction
2. Results
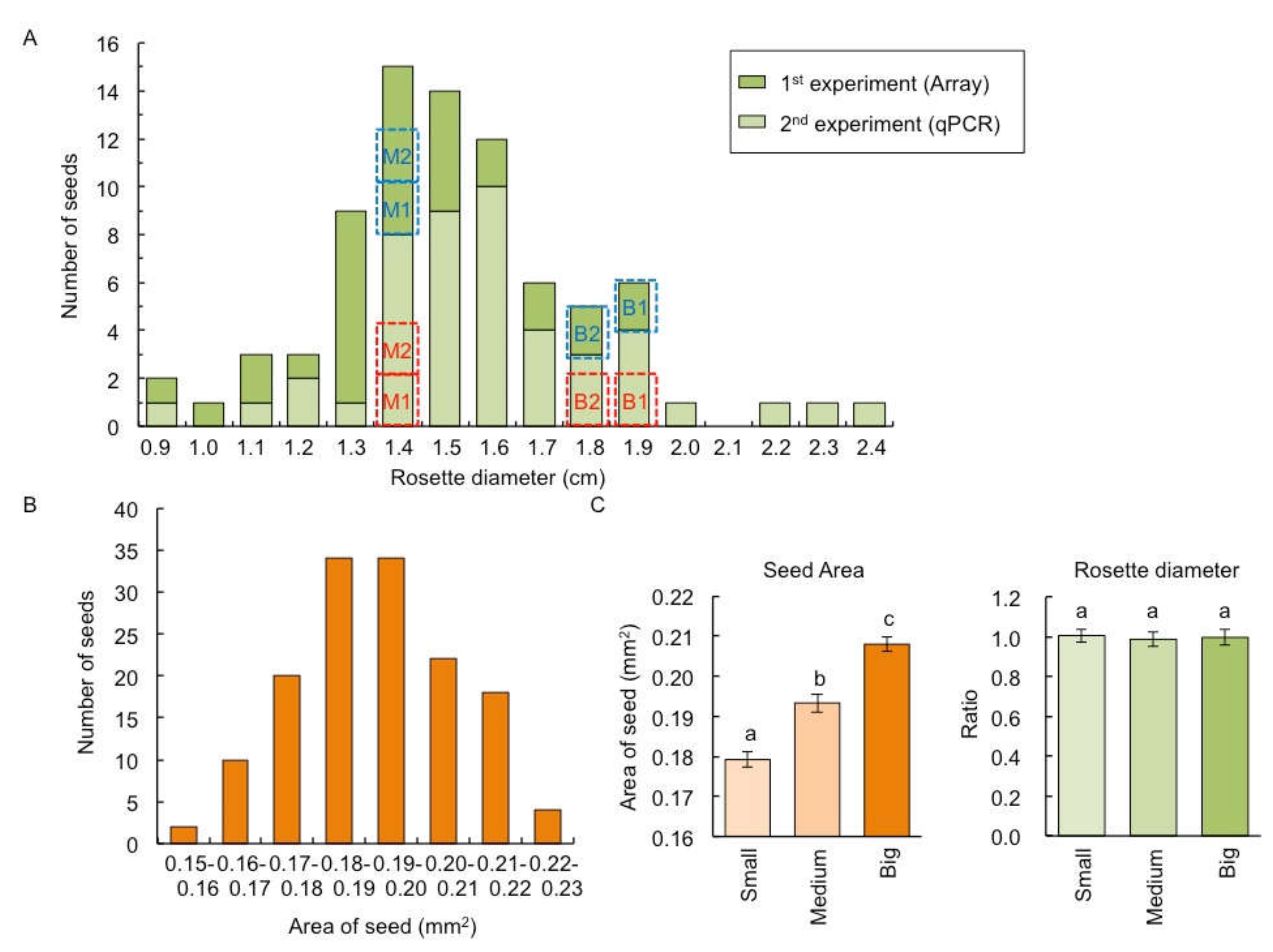
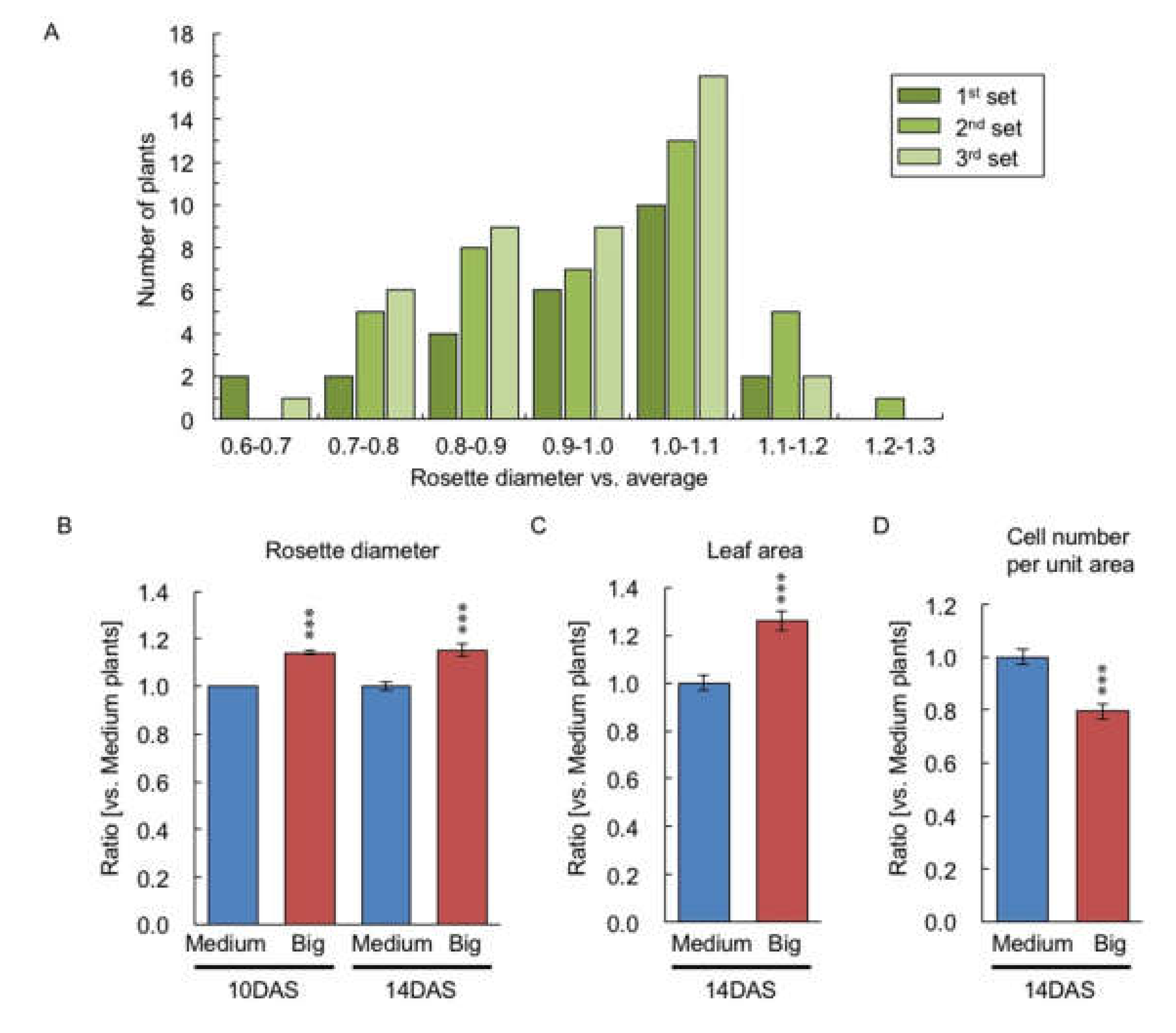
2.1. Variation of Shoot Size in the F1 between C24 and Col
2.2. The Transcriptome Divergence between Big- and Medium-Sized F1 Plants
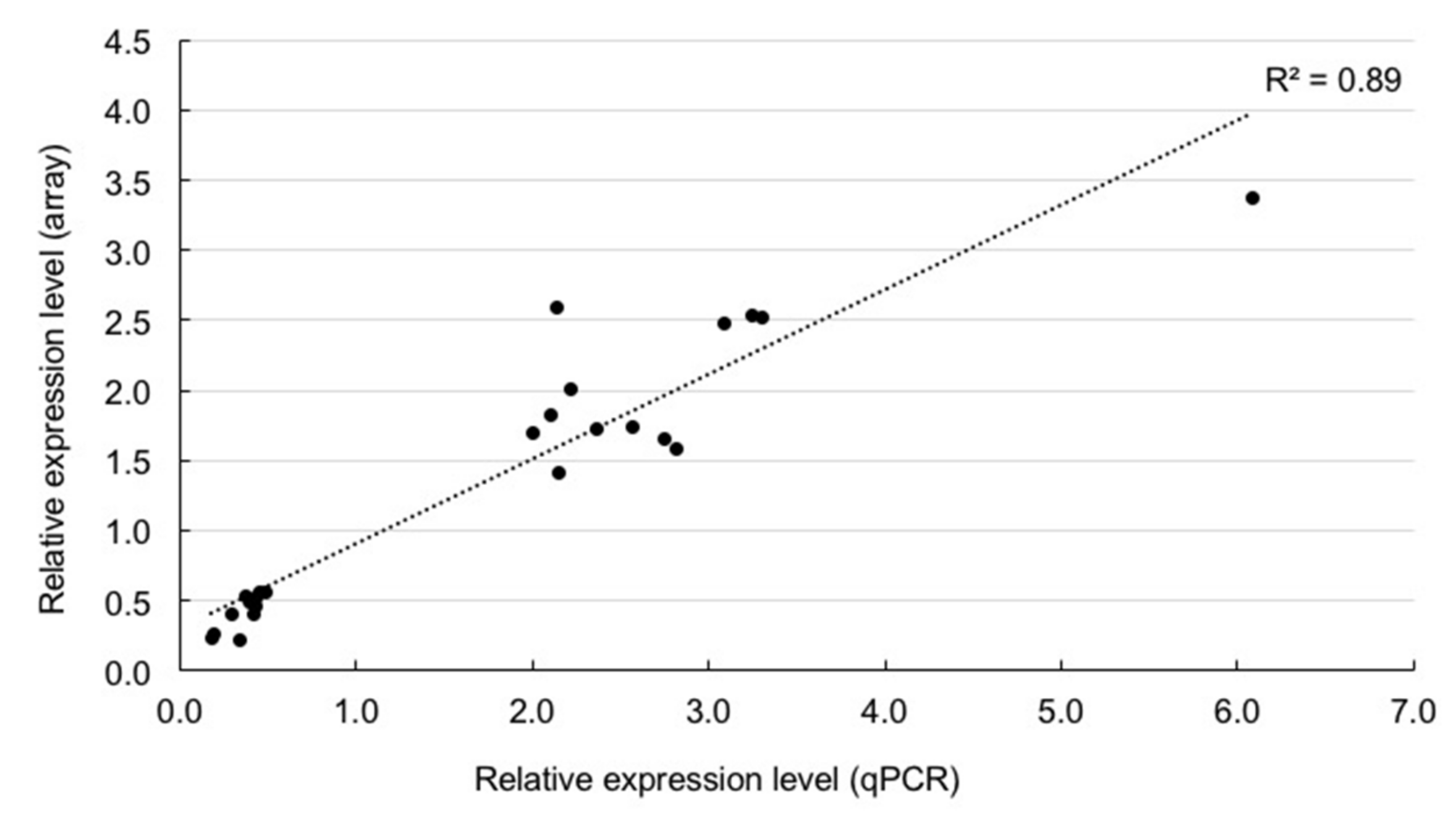
2.3. Confirmation of Differential Gene Expression by Quantitative RT-PCR
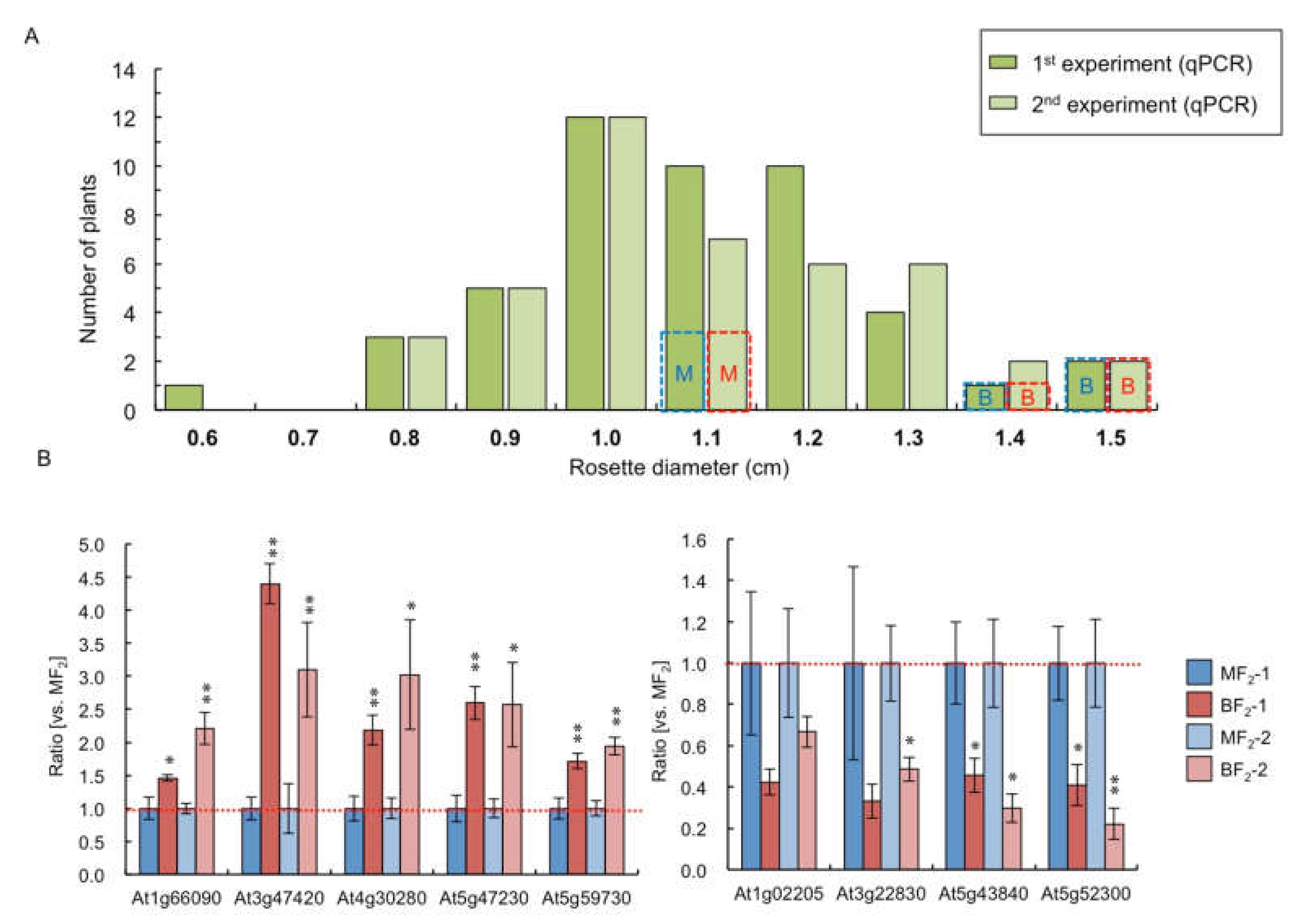
2.4. Comparison of Gene Expression between Big- and Medium-Sized F2 Plants
2.5. Classification of the Differentially Expressed Genes between Big- and Medium-Sized F1 Plants
2.6. Overexpression Resulted in Plant Size Difference
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Measuring Seed Size, Rosette Diameter, Leaf Area, and Cell Size
4.3. Expression Analysis
4.4. Microarray Analysis
4.5. Gene Ontology Analysis
4.6. Constructs and Plant Transformation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fujimoto, R.; Uezono, K.; Ishikura, S.; Osabe, K.; Peacock, W.J.; Dennis, E.S. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed. Sci. 2018, 68, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.F. 90 years ago: The beginning of hybrid maize. Genetics 1998, 148, 923–928. [Google Scholar]
- Lippman, Z.B.; Zamir, D. Heterosis: Revisiting the magic. Trends Genet. 2007, 23, 60–66. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yao, H.; Chudalayandi, S.; Vaiman, D.; Veitia, R.A. Heterosis. Plant Cell 2010, 22, 2105–2112. [Google Scholar] [CrossRef]
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Springer, N.M. Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 2013, 64, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Groszmann, M.; Greaves, I.K.; Fujimoto, R.; Peacock, W.J.; Dennis, E.S. The role of epigenetics in hybrid vigour. Trends Genet. 2013, 29, 684–690. [Google Scholar] [CrossRef]
- Miyaji, N.; Fujimoto, R. Hybrid vigor: Importance of epigenetic processes and consequences for breeding. Adv. Bot. Res. 2018, 88, 247–275. [Google Scholar]
- Birchler, J.A.; Auger, D.L.; Riddle, N.C. In Search of the molecular basis of heterosis. Plant Cell 2003, 15, 2236–2239. [Google Scholar] [CrossRef]
- Swanson-Wagner, R.A.; Jia, Y.; DeCook, R.; Borsuk, L.A.; Nettleton, D.; Schnable, P.S. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 2006, 103, 6805–6810. [Google Scholar] [CrossRef]
- Wei, G.; Tao, Y.; Liu, G.; Chen, C.; Luo, R.; Xia, H.; Gan, Q.; Zeng, H.; Lu, Z.; Han, Y.; et al. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc. Natl. Acad. Sci. USA 2009, 106, 7695–7701. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Zhu, X.; Elling, A.A.; Chen, L.; Wang, X.; Guo, L.; Liang, M.; He, H.; Zhang, H.; Chen, F.; et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 2010, 22, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Song, G.S.; Zhai, H.L.; Peng, Y.G.; Zhang, L.; Wei, G.; Chen, X.Y.; Xiao, Y.G.; Wang, L.; Chen, Y.J.; Wu, B.; et al. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol. Plant. 2010, 3, 1012–1025. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, R.; Taylor, J.M.; Shirasawa, S.; Peacock, W.J.; Dennis, E.S. Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc. Natl. Acad. Sci. USA 2012, 109, 7109–7114. [Google Scholar] [CrossRef]
- Meyer, R.C.; Witucka-Wall, H.; Becher, M.; Blacha, A.; Boudichevskaia, A.; Dörmann, P.; Fiehn, O.; Friedel, S.; von Korff, M.; Lisec, J.; et al. Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 2012, 71, 669–683. [Google Scholar] [CrossRef]
- Shen, H.; He, H.; Li, J.; Chen, W.; Wang, X.; Guo, L.; Peng, Z.; He, G.; Zhong, S.; Qi, Y.; et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 2012, 24, 875–892. [Google Scholar] [CrossRef]
- Groszmann, M.; Gonzalez-Bayon, R.; Lyons, R.L.; Greaves, I.K.; Kazan, K.; Peacock, W.J.; Dennis, E.S. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc. Natl. Acad. Sci. USA 2015, 112, E6397–E6406. [Google Scholar] [CrossRef]
- Zhu, A.; Greaves, I.K.; Liu, P.C.; Wu, L.; Dennis, E.S.; Peacock, W.J. Early changes of gene activity in developing seedlings of Arabidopsis hybrids relative to parents may contribute to hybrid vigour. Plant J. 2016, 88, 597–607. [Google Scholar] [CrossRef]
- Saeki, N.; Kawanabe, T.; Ying, H.; Shimizu, M.; Kojima, M.; Abe, H.; Okazaki, K.; Kaji, M.; Taylor, J.M.; Sakakibara, H.; et al. Molecular and cellular characteristics of hybrid vigour in a commercial hybrid of Chinese cabbage. BMC Plant. Biol. 2016, 16, 45. [Google Scholar] [CrossRef]
- Barth, S.; Busimi, A.K.; Utz, H.F.; Melchinger, A.E. Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity 2003, 91, 36–42. [Google Scholar] [CrossRef]
- Meyer, R.C.; Törjék, O.; Becher, M.; Altmann, T. Heterosis of biomass production in Arabidopsis: Establishment during early development. Plant Physiol. 2004, 134, 1813–1823. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.; Lukens, L. An evaluation of Arabidopsis thaliana hybrid traits and their genetic control. G3 2011, 1, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Seymour, D.K.; Chae, E.; Grimm, D.G.; Pizarro, C.M.; Habring-Müller, A.; Vasseur, F.; Rakitsch, B.; Borgwardt, K.M.; Koenig, D.; Weigel, D. Genetic architecture of nonadditive inheritance in Arabidopsis thaliana hybrids. Proc. Natl. Acad. Sci. USA 2016, 113, E7317–E7326. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wang, X.; Ren, D.; Huang, H.; Xu, M.; He, G.; Deng, X.W. Genomic architecture of biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 8101–8106. [Google Scholar] [CrossRef]
- Vasseur, F.; Fouqueau, L.; de Vienne, D.; Nidelet, T.; Violle, C.; Weigel, D. Nonlinear phenotypic variation uncovers the emergence of heterosis in Arabidopsis thaliana. PLoS Biol. 2019, 17, e3000214. [Google Scholar] [CrossRef]
- Groszmann, M.; Gonzalez-Bayon, R.; Greaves, I.K.; Wang, L.; Huen, A.K.; Peacock, W.J.; Dennis, E.S. Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol. 2014, 166, 265–280. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Xu, T.; Srivastava, A.K.; Wang, D.; Zeng, L.; Yang, L.; He, L.; Zhang, H.; Zheng, Z.; et al. The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov. 2016, 2, 16027. [Google Scholar] [CrossRef]
- Berlan, J.P. Hybrid corn and the unsettled question of heterosis. J. Genet. 2018, 97, 1075–1082. [Google Scholar] [CrossRef]
- Bomblies, K.; Yant, L.; Laitinen, R.A.; Kim, S.T.; Hollister, J.D.; Warthmann, N.; Fitz, J.; Weigel, D. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 2010, 6, e1000890. [Google Scholar] [CrossRef]
- Greaves, I.K.; Groszmann, M.; Ying, H.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. Trans chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. USA 2012, 109, 3570–3575. [Google Scholar] [CrossRef]
- Greaves, I.K.; Groszmann, M.; Dennis, E.S.; Peacock, W.J. Trans-chromosomal methylation. Epigenetics 2012, 7, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Morota, G.; Rosa, G.J.M.; Gianola, D. Prediction of plant height in Arabidopsis thaliana using DNA methylation data. Genetics 2015, 201, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Song, Q.; Shi, X.; Juenger, E.T.; Chen, Z.J. Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat. Commun. 2015, 6, 7453. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Ando, A.; Xu, D.; Fang, L.; Zhang, T.; Huq, E.; Qiao, H.; Deng, X.W.; Chen, Z.J. Diurnal down-regulation of ethylene biosynthesis mediates biomass heterosis. Proc. Natl. Acad. Sci. USA 2018, 115, 5606–5611. [Google Scholar] [CrossRef]
- Gonzalez-Bayon, R.; Shen, Y.; Groszmann, M.; Zhu, A.; Wang, A.; Allu, A.D.; Dennis, E.S.; Peacock, W.J.; Greaves, I.K. Senescence and defense pathways contribute to heterosis. Plant Physiol. 2019, 180, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Kawanabe, T.; Ishikura, S.; Miyaji, N.; Sasaki, T.; Wu, L.M.; Itabashi, E.; Takada, S.; Shimizu, M.; Takasaki-Yasuda, T.; Osabe, K.; et al. Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, E6704–E6711. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Wang, L.; Wu, L.M.; Greaves, I.K.; Dennis, E.S.; William James Peacock, W.J. In Arabidopsis hybrids and Hybrid Mimics, up-regulation of cell wall biogenesis is associated with the increased plant size. Plant Direct 2019, 3, 1–15. [Google Scholar] [CrossRef]
- Pitaksaringkarn, W.; Matsuoka, K.; Asahina, M.; Miura, K.; Sage-Ono, K.; Ono, M.; Yokoyama, R.; Nishitani, K.; Ishii, T.; Iwai, H.; et al. XTH20 and XTH19 regulated by ANAC071 under auxin flow are involved in cell proliferation in incised Arabidopsis inflorescence stems. Plant J. 2014, 80, 604–614. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
- Tonosaki, K.; Osabe, K.; Kawanabe, T.; Fujimoto, R. The importance of reproductive barriers and the effect of allopolyploidization on crop breeding. Breed. Sci. 2016, 66, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Maekawa, S.; Yasuda, S.; Sonoda, Y.; Katoh, E.; Ichikawa, T.; Nakazawa, M.; Seki, M.; Shinozaki, K.; Matsui, M.; et al. CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 2009, 60, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Sato, T.; Asada, Y.; Yasuda, S.; Yoshida, M.; Chiba, Y.; Yamaguchi, J. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol. Biol. 2012, 79, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, S.; Reyes, T.H.; Guglielminetti, L.; Lu, Y.; Morita, Y.; Sato, T.; Yamaguchi, J. Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol. 2014, 55, 293–305. [Google Scholar] [CrossRef]
- Hiruma, K.; Nishiuchi, T.; Kato, T.; Bednarek, P.; Okuno, T.; Schulze-Lefert, S.; Takano, Y. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 2011, 67, 980–992. [Google Scholar] [CrossRef]
- Wang, J.; Tian, L.; Lee, H.S.; Wei, N.E.; Jiang, H.; Watson, B.; Madlung, A.; Osborn, T.C.; Doerge, R.W.; Comai, L.; et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 2006, 172, 507–517. [Google Scholar] [CrossRef]
- Fujimoto, R.; Taylor, J.M.; Sasaki, T.; Kawanabe, T.; Dennis, E.S. Genome wide gene expression in artificially synthesized amphidiploids of Arabidopsis. Plant Mol. Biol. 2011, 77, 419–431. [Google Scholar] [CrossRef]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
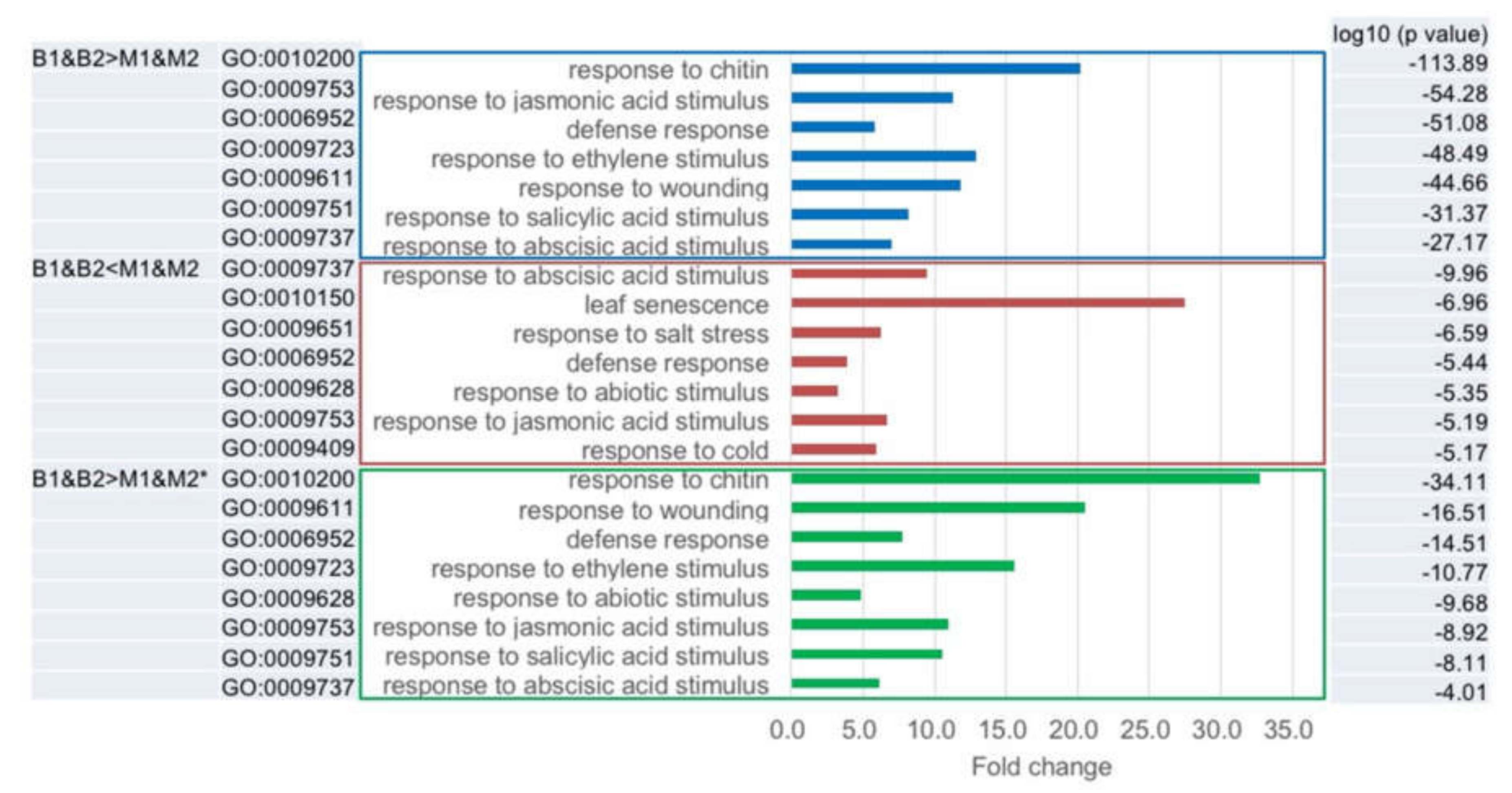

| Transcriptome Data | B1&B2 > M1&M2 (361) | B1&B2 < M1&M2 (80) | |||
|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | ||
| C24/Col DEG | F1 > MPV (863) | 102 | 28.3% | 8 | 10.0% |
| F1 < MPV (1,234) | 53 | 14.7% | 25 | 31.3% | |
| C24/Ler DEG | F1 > MPV (669) | 114 | 31.6% | 10 | 12.5% |
| F1 < MPV (1,050) | 47 | 13.0% | 18 | 22.5% | |
| Col/Ler DEG | F1 > MPV (464) | 21 | 5.8% | 7 | 8.8% |
| F1 < MPV (907) | 63 | 17.5% | 11 | 13.8% | |
| C24xCol vs. ddm1C24xddm1Col | WT > ddm1 (73) | 3 | 0.8% | 0 | 0.0% |
| WT < ddm1 (1,128) | 25 | 6.9% | 0 | 0.0% | |
| ColxC24 vs. ddm1Colxddm1C24 | WT > ddm1 (69) | 0 | 0.0% | 4 | 5.0% |
| WT < ddm1 (1,208) | 36 | 10.0% | 0 | 0.0% | |
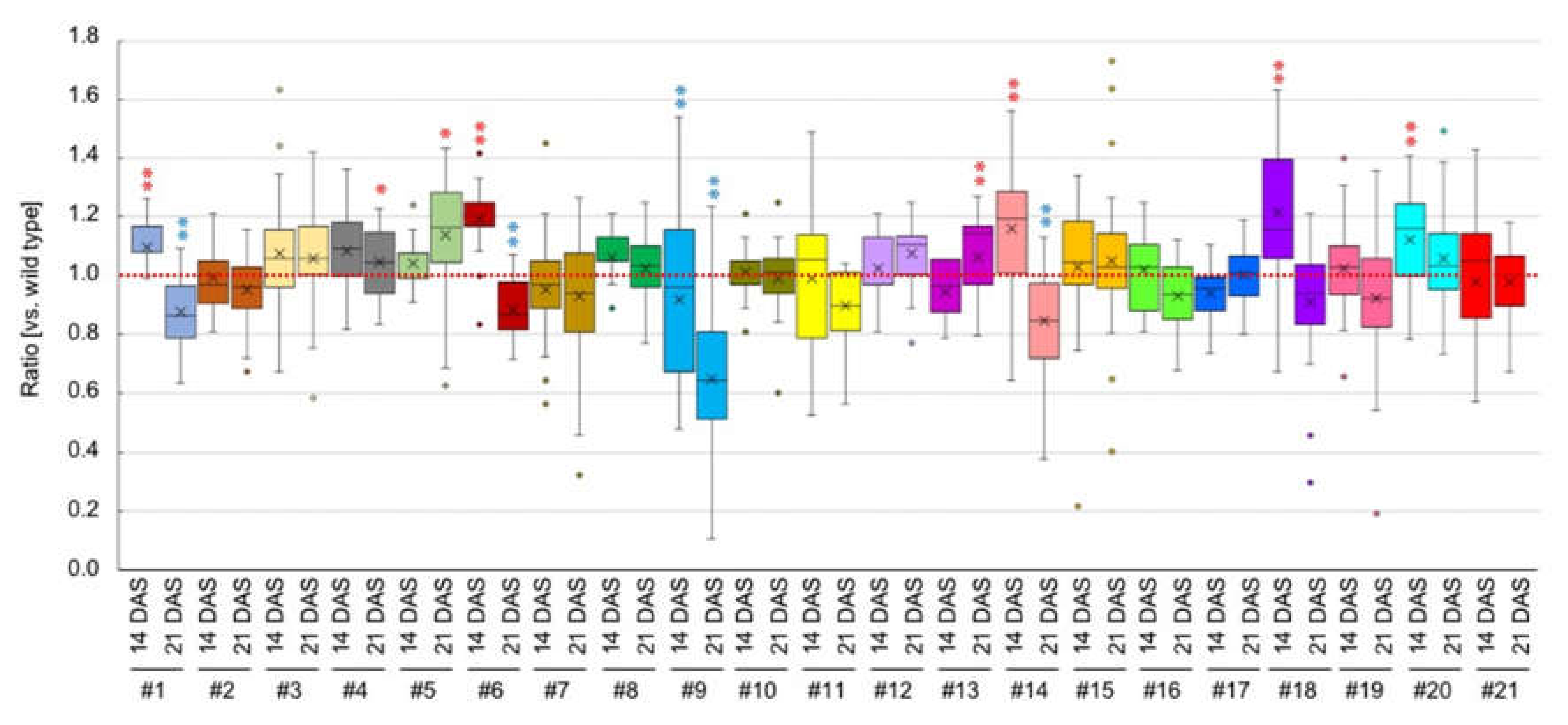
| Number | Gene Model | Name | Description | 14 DAS # | 21 DAS # |
|---|---|---|---|---|---|
| #1 | At1g05575 | Unknown protein | 1.10** | 0.88** | |
| #2 | At1g19210 | ERF17 | Encodes a member of the DREB subfamily A-5 of ERF/AP2 transcription factor family | 1.00 | 0.95 |
| #3 | At1g22810 | ERF19 | Encodes a member of the DREB subfamily A-5 of ERF/AP2 transcription factor family | 1.07 | 1.06 |
| #4 | At1g33760 | ERF22 | Encodes a member of the DREB subfamily A-5 of ERF/AP2 transcription factor family | 1.08 | 1.04 * |
| #5 | At2g26020 | PDF1.2b | Predicted to encode a PR (pathogenesis-related) protein | 1.04 | 1.14 * |
| #6 | At2g35290 | SAUR79 | SMALL AUXIN UPREGULATED RNA 79 | 1.19 ** | 0.88 ** |
| #7 | At2g44840 | ERF13 | ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13 | 0.95 | 0.93 |
| #8 | At4g17490 | ERF6 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6 | 1.06 | 1.02 |
| #9 | At4g30290 | XTH19 | XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19 | 0.92 ** | 0.65 ** |
| #10 | At4g38840 | SAUR14 | SMALL AUXIN UPREGULATED RNA 14 | 1.01 | 0.99 |
| #11 | At5g07100 | WRKY26 | Encode WRKY DNA-binding protein 26 | 0.99 | 0.90 |
| #12 | At5g09570 | Cox-19-like CHCH family protein | 1.02 | 1.08 | |
| #13 | At5g27420 | CNI1 | CARBON/NITROGEN INSENSITIVE 1 | 0.95 | 1.06 ** |
| #14 | At5g42380 | CML37 | Calmodulin like 37 | 1.16 ** | 0.85 ** |
| #15 | At5g47230 | ERF5 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5 | 1.03 | 1.05 |
| #16 | At5g51190 | ERF105 | Encodes a member of the ERF subfamily B-3 of ERF/AP2 transcription factor family | 1.02 | 0.93 |
| #17 | At3g02040 | GDPD1 | GLYCEROPHOSPHODIESTER PHOSPHODIESTERASE 1 | 0.94 | 1.00 |
| #18 | At3g23250 | MYB15 | MYB DOMAIN PROTEIN 15 | 1.22 ** | 0.91 |
| #19 | At4g34410 | RRTF1/ERF109 | REDOX RESPONSIVE TRANSCRIPTION FACTOR 1 | 1.03 | 0.92 |
| #20 | At2g20750 | EXPB1 | EXPANSIN B1 | 1.24 ** | 1.06 |
| #21 | At4g28250 | EXPB3 | EXPANSIN B3 | 0.98 | 0.98 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehraj, H.; Kawanabe, T.; Shimizu, M.; Miyaji, N.; Akter, A.; Dennis, E.S.; Fujimoto, R. In Arabidopsis thaliana Heterosis Level Varies among Individuals in an F1 Hybrid Population. Plants 2020, 9, 414. https://doi.org/10.3390/plants9040414
Mehraj H, Kawanabe T, Shimizu M, Miyaji N, Akter A, Dennis ES, Fujimoto R. In Arabidopsis thaliana Heterosis Level Varies among Individuals in an F1 Hybrid Population. Plants. 2020; 9(4):414. https://doi.org/10.3390/plants9040414
Chicago/Turabian StyleMehraj, Hasan, Takahiro Kawanabe, Motoki Shimizu, Naomi Miyaji, Ayasha Akter, Elizabeth S. Dennis, and Ryo Fujimoto. 2020. "In Arabidopsis thaliana Heterosis Level Varies among Individuals in an F1 Hybrid Population" Plants 9, no. 4: 414. https://doi.org/10.3390/plants9040414
APA StyleMehraj, H., Kawanabe, T., Shimizu, M., Miyaji, N., Akter, A., Dennis, E. S., & Fujimoto, R. (2020). In Arabidopsis thaliana Heterosis Level Varies among Individuals in an F1 Hybrid Population. Plants, 9(4), 414. https://doi.org/10.3390/plants9040414







