Evolutionary Trends in the Mitochondrial Genome of Archaeplastida: How Does the GC Bias Affect the Transition from Water to Land?
Abstract
1. Introduction
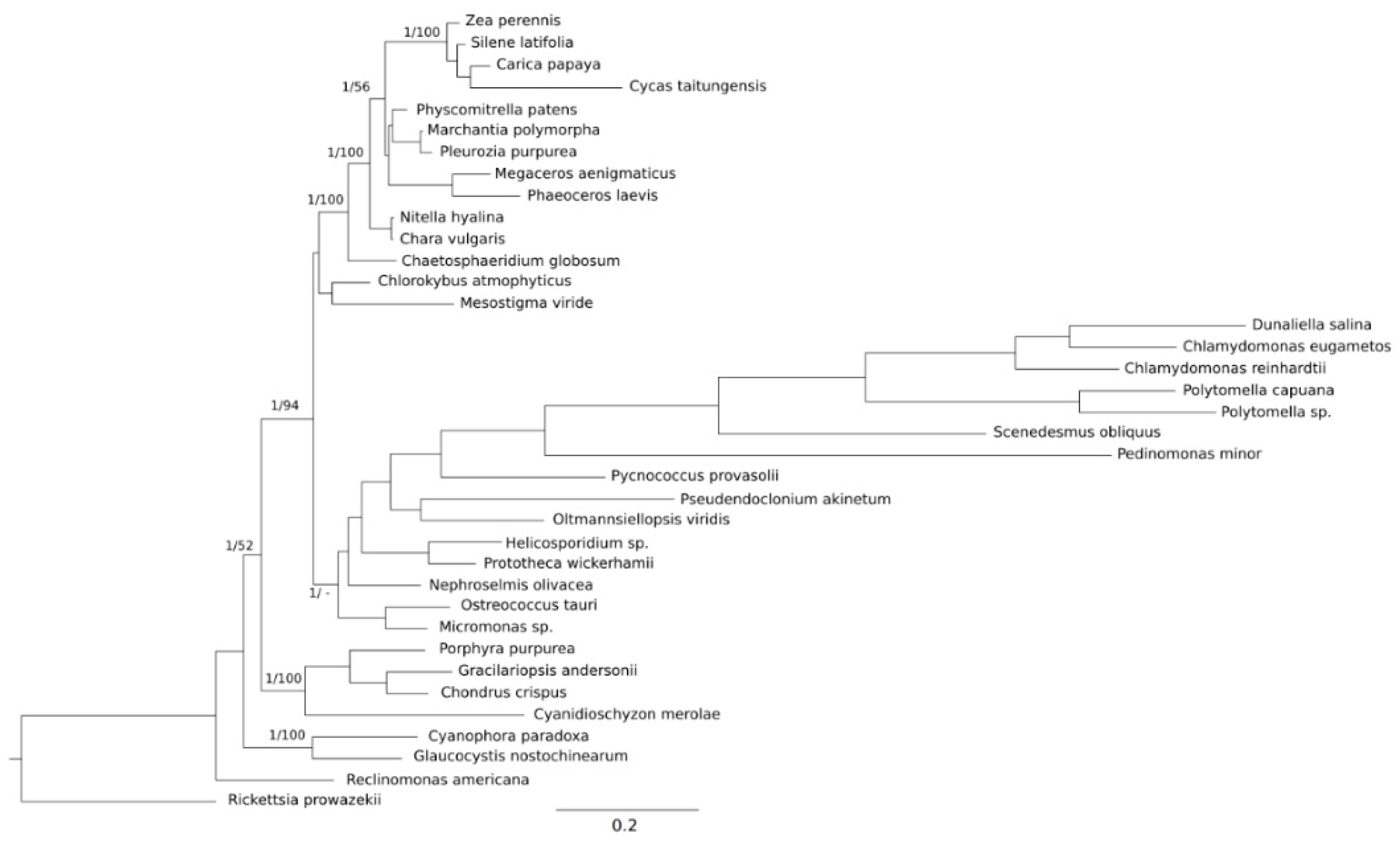
2. Results
2.1. Genome Features
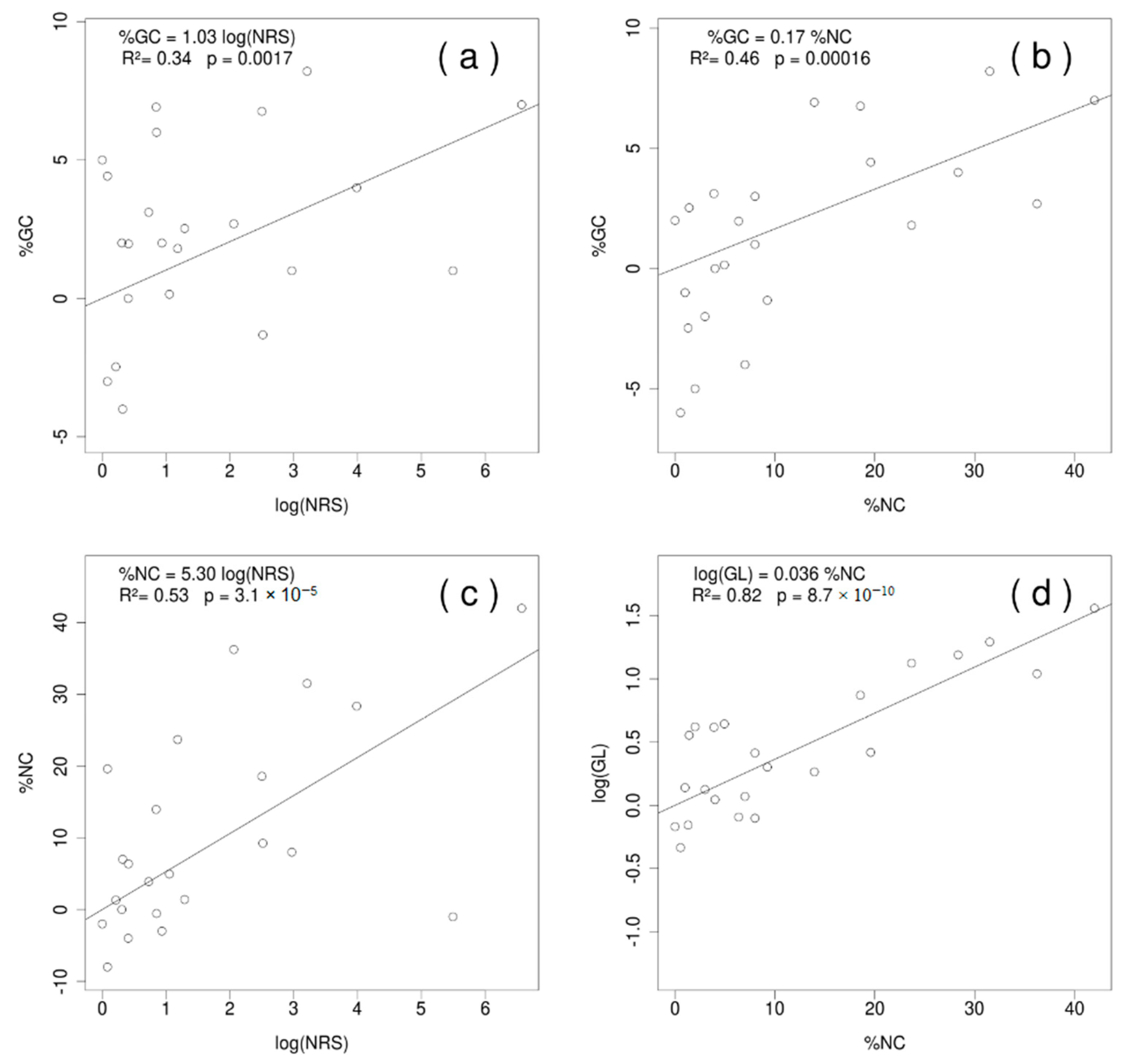
2.2. Heterogeneity in the Base Composition of Ribosomal Subunits in Streptophyta and the Reconstruction of the Ancestral %GC and GC* Content
3. Discussion
3.1. Archaeplastida Mitochondrial Trends
3.2. The Distribution of GC Content and GC* in the Three Genetic Compartments for Streptophyta
4. Materials and Methods
4.1. Archaeplastida Mitochondrial Genome-Wide Characteristics
4.2. Analyses of Heterogeneity in Base Composition
4.3. Streptophyta Ribosomal GC Content and GC*
4.4. Concomitant Evolution between Genetic Compartments and Clades
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| %NC | non-coding DNA |
| cpLSU | chloroplast ribosomal large subunits |
| cpSSU | chloroplast ribosomal small subunits |
| gBGC | GC-biased gene conversion |
| GC* | GC frequency |
| GL | genome lengths |
| GN | gene number |
| mtDNA | mitochondrial genome |
| mtLSU | mitochondrial ribosomal large subunits |
| mtSSU | mitochondrial ribosomal small subunits |
| NPG | number of protein-coding genes |
| NRS | number of repeated sequences |
| nSSU | nuclear ribosomal small subunit |
| nt | nucleotides |
| PGLS | Phylogenetic Generalized Least Squares |
| rDNA | ribosomal DNA |
| RSL | repeated sequences total length |
References
- Melton, J.T., III; Leliaert, F.; Tronholm, A.; Lopez-Bautista, J.M. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta). PLoS ONE 2015, 10, e0121020. [Google Scholar] [CrossRef] [PubMed]
- Green, B.R. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011, 66, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, L.; Li, H.; Sahu, S.K.; Wang, H.; Xu, Y.; Xian, W.; Song, B.; Liang, H.; Cheng, S.; et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 2020, 6, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Graham, L.E. EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata. PLoS ONE 2008, 3, e2621. [Google Scholar] [CrossRef]
- Chan, C.X.; Gross, J.; Yoon, H.S.; Bhattacharya, D. Plastid origin and evolution: New models provide insights into old problems. Plant Physiol. 2011, 155, 1552–1560. [Google Scholar] [CrossRef]
- Adams, K.L.; Palmer, J.D. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenetics Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef]
- Smith, D.R.; Crosby, K.; Lee, R.W. Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol. Evol. 2011, 3, 365–371. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef]
- Kern, R.; Facchinelli, F.; Delwiche, C.; Weber, A.P.; Bauwe, H.; Hagemann, M. Evolution of photorespiratory glycolate oxidase among Archaeplastida. Plants 2020, 9, 106. [Google Scholar] [CrossRef]
- Wang, B.; Liu, J.; Jin, L.; Feng, X.Y.; Chen, J.Q. Complex mutation and weak selection together determined the codon usage bias in bryophyte mitochondrial genomes. J. Integr. Plant. Biol. 2010, 52, 1100–1108. [Google Scholar] [CrossRef]
- Wang, B.; Yuan, J.; Liu, J.; Jin, L.; Chen, J.Q. Codon usage bias and determining forces in green plant mitochondrial genomes. J. Integr. Plant. Biol. 2011, 53, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Sloan, D.B.; Alverson, A.J. Plant Mitochondrial Genome Diversity: The Genomics Revolution; Plant Genome Diversity; Springer: Vienna, Austria, 2012; Volume 1, pp. 123–144. [Google Scholar]
- Galtier, N.; Duret, L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007, 23, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Duret, L.; Glémin, S.; Ranwez, V. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 2009, 25, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Escobar, J.S.; Glémin, S.; Galtier, N. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Mol. Biol. Evol. 2011, 28, 2561–2575. [Google Scholar] [CrossRef]
- Lassalle, F.; Périan, S.; Bataillon, T.; Nesme, X.; Duret, L.; Daubin, V. GC-content evolution in bacterial genomes: The biased gene conversion hypothesis expands. PLoS Genet. 2015, 11, e1004941. [Google Scholar] [CrossRef]
- Sueoka, N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. USA 1962, 48, 582–592. [Google Scholar] [CrossRef]
- Li, L.; Wang, B.; Liu, Y.; Qiu, Y.-L. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J. Mol. Evol. 2009, 68, 665–678. [Google Scholar] [CrossRef]
- Xue, J.Y.; Liu, Y.; Li, L.; Wang, B.; Qiu, Y.L. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: Retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr. Genet. 2010, 56, 53–61. [Google Scholar] [CrossRef]
- Hecht, J.; Grewe, F.; Knoop, V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol. 2011, 3, 344–358. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. The Origins of Genome Architecture; Sinauer Associates: Sunderland, UK, 2007; Volume 98. [Google Scholar]
- Vogel, C.; Chothia, C. Protein family expansions and biological complexity. PLoS Comput. Biol. 2006, 2, e48. [Google Scholar] [CrossRef]
- Turmel, M.; Lemieux, C. Evolution of the plastid genome in green algae. In Advances in Botanical Research; Academic Press: New York, NY, USA, 2018; Volume 85, pp. 157–193. [Google Scholar]
- Wallace, D.C. Bioenergetics, the origins of complexity, and the ascent of man. Proc. Natl. Acad. Sci. USA 2010, 107, 8947–8953. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Sakayama, H.; De Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell 2018, 174, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Chaitanya, K.V. Orgenellar Genome Analysis. In Genome and Genomics; Springer: Singapore, 2019; pp. 89–119. [Google Scholar]
- Gitzendanner, M.A.; Soltis, P.S.; Wong, G.K.S.; Ruhfel, B.R.; Soltis, D.E. Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. Am. J. Bot. 2018, 105, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Wang, B.; Shen, Z.; Wang, Z.; Wang, W.; Liu, H.; Wang, C.; Xin, M. The chloroplast genome sequence of the green macroalga Caulerpa okamurae (Ulvophyceae, Chlorophyta): Its structural features, organization and phylogenetic analysis. Mar. Genom. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Harholt, J.; Moestrup, Ø.; Ulvskov, P. Why plants were terrestrial from the beginning. Trends Plant Sci. 2016, 21, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Kapraun, D.F. Nuclear DNA content estimates in green algal lineages: Chlorophyta and Streptophyta. Ann. Bot. 2007, 99, 677–701. [Google Scholar] [CrossRef]
- Lang, B.F.; Burger, G.; O’Kelly, C.J.; Cedergren, R.; Golding, G.B.; Lemieux, C.; Sankoff, D.; Turmel, M.; Gray, M.W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 1997, 387, 493–497. [Google Scholar] [CrossRef]
- Yang, D.; Oyaizu, Y.; Oyaizu, H.; Olsen, G.J.; Woese, C.R. Mitochondrial origins. Proc. Natl. Acad. Sci. USA 1985, 82, 4443–4447. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Orme, D.; Freckleton, R.; Thomas, G.; Petzoldt, T.; Fritz, S.; Isaac, N.; Thomas, G.; Petzoldt, T.; Pearse, W.; Fritz, S.; et al. Caper: Comparative Analyses of Phylogenetics and Evolution in R. R Package Version 0.5. Available online: http://CRAN.R-project.org/package=caperURL (accessed on 20 February 2020).
- Butler, M.A.; King, A.A. Phylogenetic comparative analysis: A modelling approach for adaptive evolution. Am. Nat. 2004, 164, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Purvis, A.; Rambaut, A. Comparative analysis by independent contrasts (CAIC): An Apple Macintosh application for analysing comparative data. Comput. Appl. Biosci. 1995, 11, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Agapow, P.; Isaac, N.J.B. MacroCAIC: Revealing correlates of species richness by comparative analysis. Divers. Distrib. 2002, 8, 41–43. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, 7–13. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef]
- Schmidt, H.A.; Strimmer, K.; Vingron, M.; von Haeseler, A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 2002, 18, 502–504. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Adachi, J.; Waddell, P.J.; Martin, W.; Hasegawa, M. Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA. J. Mol. Evol. 2000, 50, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Thomson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position- specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Arndt, P.F.; Petrov, D.A.; Hwa, T. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol. Biol. Evol. 2003, 20, 1887–1896. [Google Scholar] [CrossRef]
- Torres, M.; Oliveira da Silva, J. Parallel solution based on collective communication operations for phylogenetic bootstrapping in PhyML 3.0. In Brazilian Symposium on Bioinformatics; Springer: Cham, Switzerland, 2018; pp. 133–145. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Dutheil, J.; Galtier, N. BAOBAB: A java editor for large phylogenetic trees. Bioinformatics 2002, 18, 892–893. [Google Scholar] [CrossRef]
- Timme, R.E.; Bachvaroff, T.R.; Delwiche, C.F. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 2012, 7, 1–8. [Google Scholar] [CrossRef]
- Durtheil, J.; Boussau, B. Non-homogeneous models of sequence evolution in the Bio++ suite of libraries and programs. BMC Evol. Biol. 2008, 8, 255. [Google Scholar]
- Galtier, N.; Gouy, M. Inferring pattern and process: Maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol. Biol. Evol. 1998, 15, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Arndt, P.F. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008, 4, e1000071. [Google Scholar] [CrossRef] [PubMed]

| Hierarchical Models | Mitochondrion | |||||||
|---|---|---|---|---|---|---|---|---|
| Large Ribosomal Subunit (LSU) | Small Ribosomal Subunit (SSU) | |||||||
| −InL | Dev. | Df. | P-Value | −InL | Dev. | Df. | P-Value | |
| Homogeneous | 22486.50 | 14603.13 | ||||||
| NH-Model_M1(1) | 22438.91 | 95.18 | 17 | 6.88 × 10−13 | 14556.56 | 93.14 | 15 | 2.57 × 10−13 |
| NH-Model_M2(2) | 22436.54 | 4.73 | 1 | 0.0296 | 14556.50 | 0.12 | 1 | 0.7234 |
| NH-Terminal clades (as Figure 3) | 22425.82 | 21.43 | 3 | 8.56 × 10−5 | 14548.21 | 16.58 | 3 | 0.0009 |
| NH-One GC* per branch | 22327.36 | 196.92 | 176 | 0.1337 | 14415.88 | 264.66 | 178 | 2.66 × 10−5 |
| Nucleous | ||||||||
| Small Ribosomal Subunit (SSU) | ||||||||
| −InL | Dev. | Df. | P-Value | |||||
| Homogeneous | 14023.18 | |||||||
| NH-Model_M1(1) | 13984.94 | 76.47 | 19 | 7.47 × 10−9 | ||||
| NH-Model_M2(2) | 13970.40 | 29.08 | 1 | 6.94 × 10−8 | ||||
| NH-Terminal clades (as Figure 3) | 13967.78 | 5.23 | 3 | 0.1555 | ||||
| NH-One GC* per branch | 13878.57 | 178.42 | 174 | 0.3933 | ||||
| Chloroplast | ||||||||
| Large Ribosomal Subunit (LSU) | Small Ribosomal Subunit (SSU) | |||||||
| −InL | Dev. | Df. | P-Value | −InL | Dev. | Df. | P-Value | |
| Homogeneous | 23520.96 | 9895.45 | ||||||
| NH-Model_M1(1) | 23324.90 | 392.13 | 19 | 0 | 9816.46 | 157.97 | 19 | 0 |
| NH-Model_M2(2) | 23323.04 | 3.71 | 1 | 0.0540 | 9816.37 | 0.17 | 1 | 0.6795 |
| NH-Terminal clades (as Figure 3) | 23287.39 | 71.31 | 3 | 2.22 × 10−15 | 9808.42 | 15.90 | 3 | 0.0012 |
| NH-One GC* per branch | 23083.32 | 408.13 | 174 | 0 | 9695.43 | 225.98 | 174 | 0.0049 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedrola-Monfort, J.; Lázaro-Gimeno, D.; Boluda, C.G.; Pedrola, L.; Garmendia, A.; Soler, C.; Soriano, J.M. Evolutionary Trends in the Mitochondrial Genome of Archaeplastida: How Does the GC Bias Affect the Transition from Water to Land? Plants 2020, 9, 358. https://doi.org/10.3390/plants9030358
Pedrola-Monfort J, Lázaro-Gimeno D, Boluda CG, Pedrola L, Garmendia A, Soler C, Soriano JM. Evolutionary Trends in the Mitochondrial Genome of Archaeplastida: How Does the GC Bias Affect the Transition from Water to Land? Plants. 2020; 9(3):358. https://doi.org/10.3390/plants9030358
Chicago/Turabian StylePedrola-Monfort, Joan, David Lázaro-Gimeno, Carlos G. Boluda, Laia Pedrola, Alfonso Garmendia, Carla Soler, and Jose M. Soriano. 2020. "Evolutionary Trends in the Mitochondrial Genome of Archaeplastida: How Does the GC Bias Affect the Transition from Water to Land?" Plants 9, no. 3: 358. https://doi.org/10.3390/plants9030358
APA StylePedrola-Monfort, J., Lázaro-Gimeno, D., Boluda, C. G., Pedrola, L., Garmendia, A., Soler, C., & Soriano, J. M. (2020). Evolutionary Trends in the Mitochondrial Genome of Archaeplastida: How Does the GC Bias Affect the Transition from Water to Land? Plants, 9(3), 358. https://doi.org/10.3390/plants9030358







