Co-Suppression of NbClpC1 and NbClpC2, Encoding Clp Protease Chaperons, Elicits Significant Changes in the Metabolic Profile of Nicotiana benthamiana
Abstract
1. Introduction
2. Results
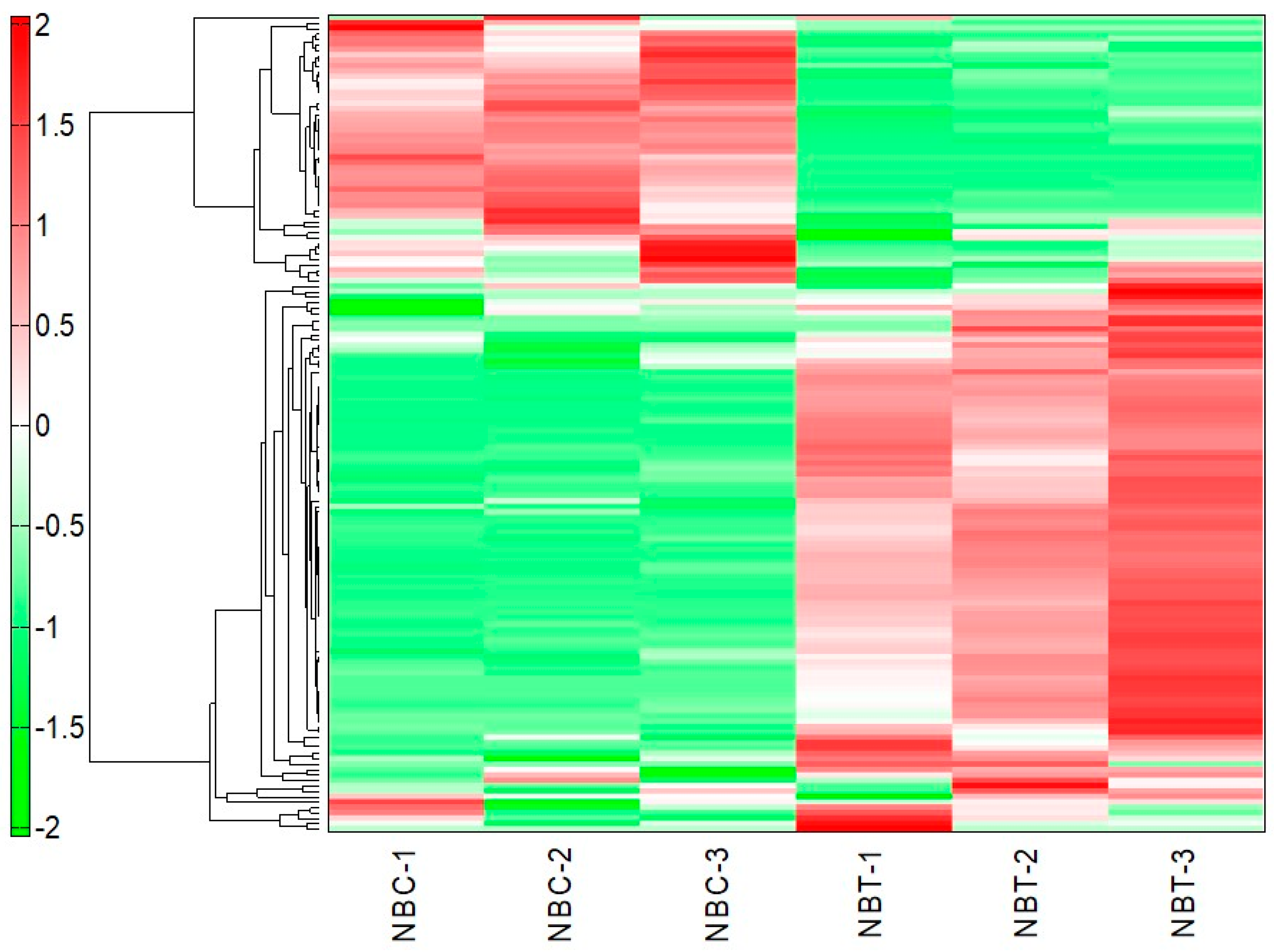
2.1. Metabolites of the NbClpC1/C2 Co-Suppressed Leaves
2.2. Effects of NbClpC1/C2 Co-Suppression on the Content of Intermediate Metabolites of Glycolysis
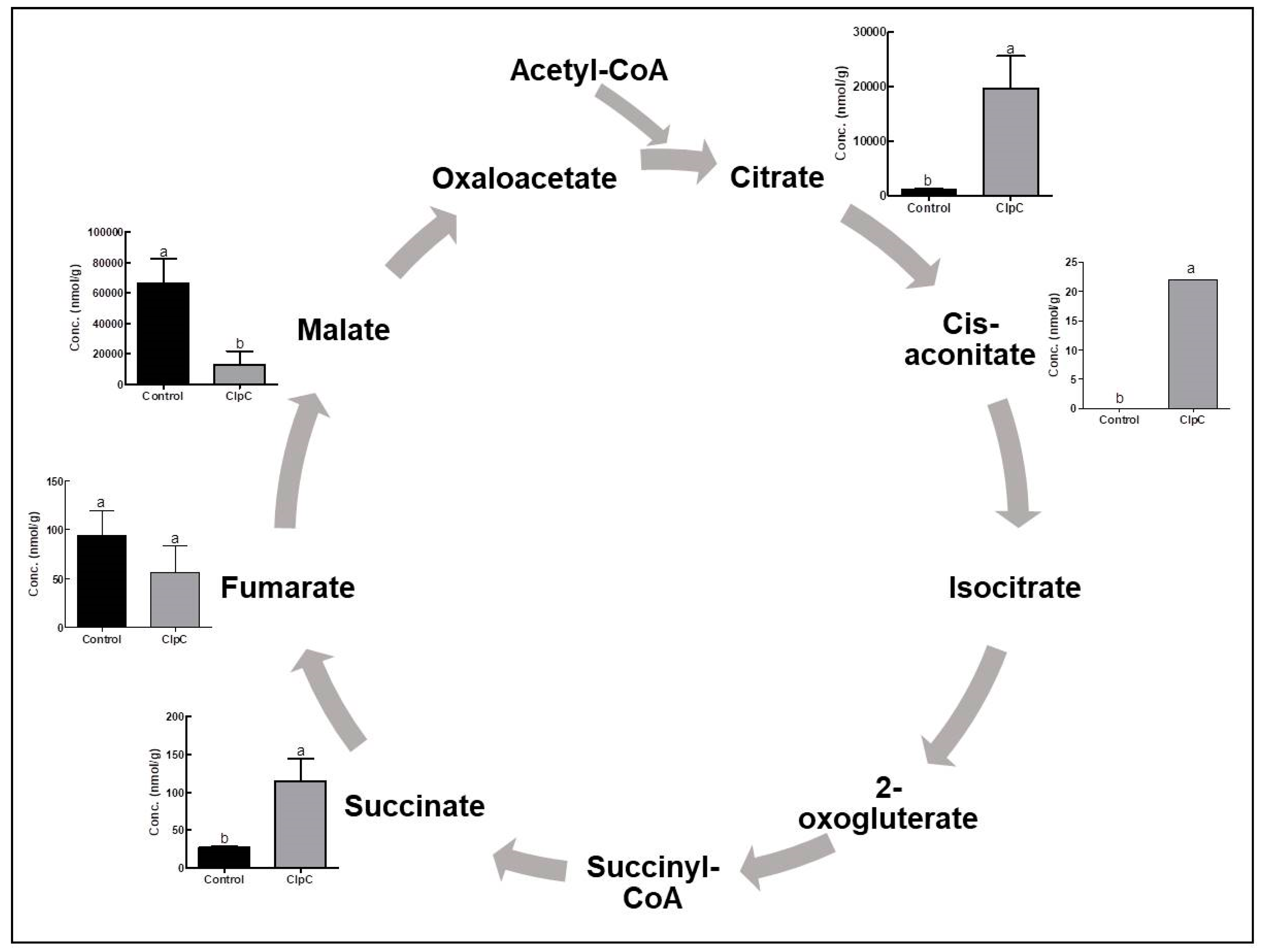
2.3. Effects of NbClpC1/C2 Co-Suppression on the Levels of Intermediate Metabolites of the TCA Cycle
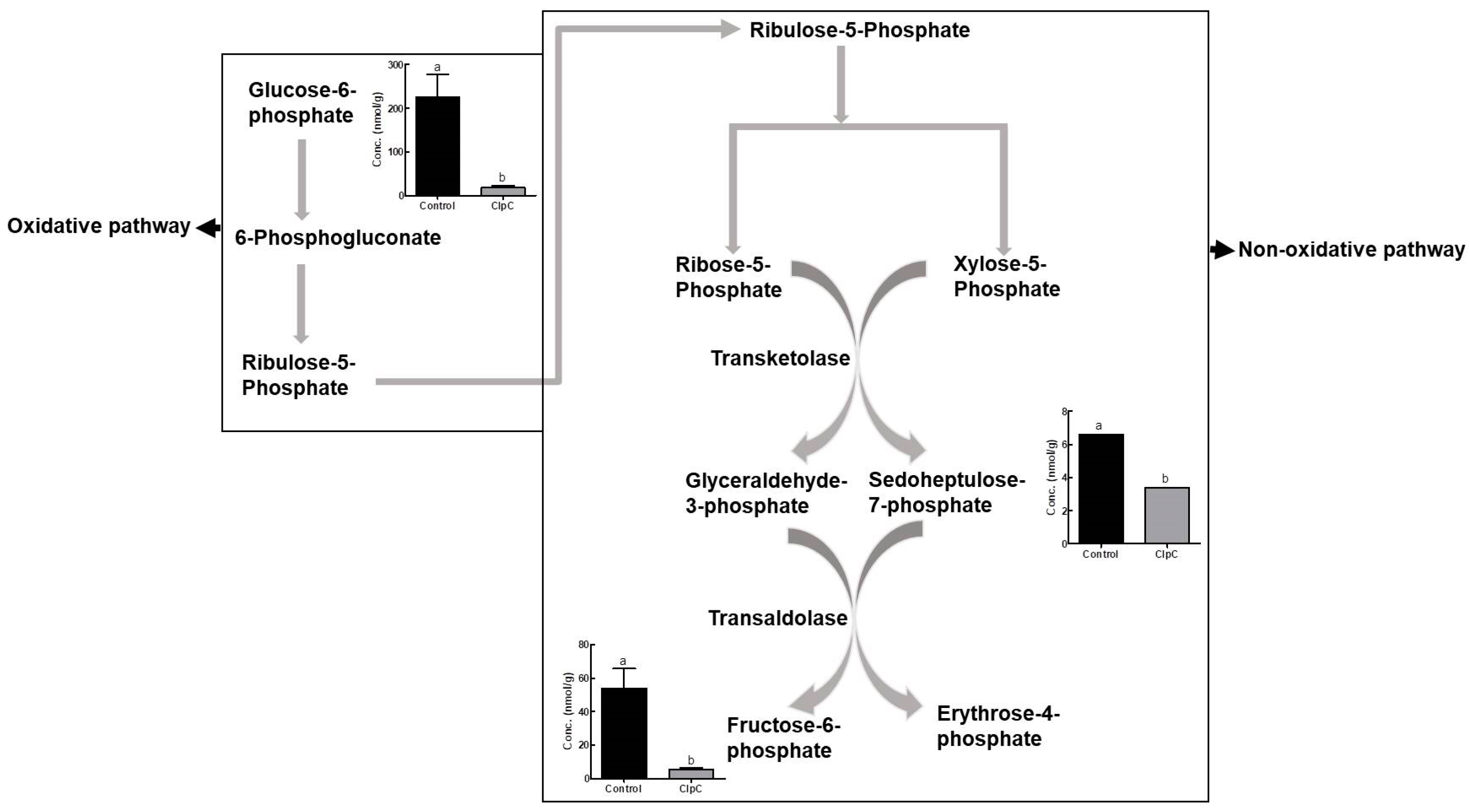
2.4. Effects of NbClpC1/C2 Co-Suppression on the Levels of Intermediate Metabolites of the Pentose Phosphate Pathway
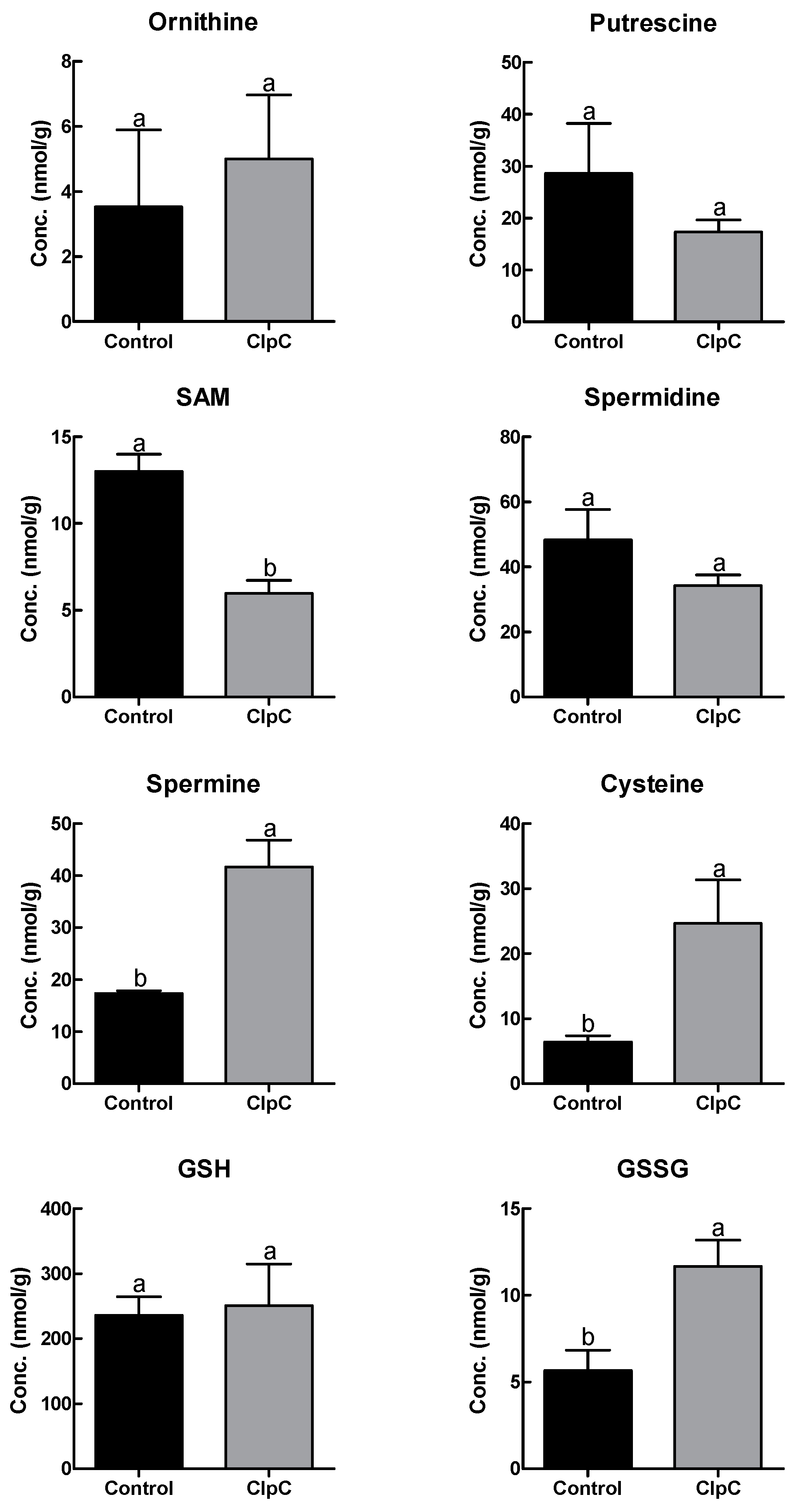
2.5. Effects of NbClpC1/C2 Co-Suppression on Polyamine and Antioxidant Contents
2.6. Effects of NbClpC1/C2 Co-Suppression on Purine Nucleotide Levels
3. Discussion
4. Materials and Methods
4.1. Plant Materials, VIGS, and Semi-Quantitative RT-PCR
4.2. Preparation of Samples for Metabolic Profiling
4.3. CE-TOF-MS Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Livneh, I.; Cohen-Kaplan, V.; Cohen-Rosenzweig, C.; Avni, N.; Ciechanover, A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016, 26, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Adam, Z.; Rudella, A.; van Wijk, K.J. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 2006, 9, 234–240. [Google Scholar] [CrossRef]
- Nishimura, K.; Kato, Y.; Sakamoto, W. Essentials of proteolytic machineries in chloroplasts. Mol. Plant 2017, 10, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Olinares, P.D.B.; Kim, J.; van Wijk, K.J. The Clp protease system; A central component of the chloroplast protease network. Biochim. Biophys. Acta 2011, 1807, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Schuhmann, H.; Adamska, I. Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol. Plant. 2012, 145, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.; Aigner, H.; Funk, C. FtsH proteases located in the plant chloroplast. Physiol. Plant. 2012, 145, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, L.L.E.; Tanabe, N.; Lymperopoulos, P.; Khan, N.Z.; Rodermel, S.R.; Aronsson, H.; Clarke, A.K. Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J. Biol. Chem. 2014, 289, 11318–11330. [Google Scholar] [CrossRef]
- Nishimura, K.; van Wijk, K.J. Organization, function and substrates of the essential Clp protease system in plastids. Biochim. Biophys. Acta 2015, 1847, 915–930. [Google Scholar] [CrossRef]
- Nishimura, K.; Kato, Y.; Sakamoto, W. Chloroplast proteases: Updates on proteolysis within and across suborganellar compartments. Plant Physiol. 2016, 171, 2280–2293. [Google Scholar] [CrossRef]
- Sjogren, L.L.E.; Stanne, T.M.; Zheng, B.; Sutinen, S.; Clarke, A.K. Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 2006, 18, 2635–2649. [Google Scholar] [CrossRef]
- Zheng, B.; MacDonald, T.M.; Sutinen, S.; Hurry, V.; Clarke, A.K. A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 2006, 224, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Koussevitzky, S.; Stanne, T.M.; Peto, C.A.; Giap, T.; Sjo¨gren, L.L.E.; Zhao, Y.; Clarke, A.K.; Chory, J. An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 2007, 63, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Rudella, A.; Rodriguez, V.R.; Zybailov, B.; Olinares, P.D.B.; van Wijk, K.J. Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 2009, 21, 1669–1692. [Google Scholar] [CrossRef] [PubMed]
- Nakagawara, E.; Sakuraba, Y.; Yamasato, A.; Tanaka, R.; Tanaka, A.; Corporation, T. Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J. 2007, 49, 800–809. [Google Scholar] [CrossRef]
- Zybailov, B.; Friso, G.; Kim, J.; Rudella, A.; Rodríguez, V.R.; Asakura, Y.; Sun, Q.; van Wijk, K.J. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteom. 2009, 8, 1789–1810. [Google Scholar] [CrossRef]
- Welsch, R.; Zhou, X.; Yuan, H.; Álvarez, D.; Sun, T.; Schlossarek, D.; Yang, Y.; Shen, G.; Zhang, H.; Rodriguez-Concepcion, M.; et al. Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol. Plant 2018, 11, 149–162. [Google Scholar] [CrossRef]
- Adam, Z.; Clarke, A.K. Cutting edge of chloroplast proteolysis. Trends Plant Sci. 2002, 7, 451–456. [Google Scholar] [CrossRef]
- Nishimura, K.; Asakura, Y.; Giulia Friso, J.K.; Oh, S.; Rutschow, H.; Ponnala, L.; van Wijk, K.J. ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 2013, 25, 2276–2301. [Google Scholar] [CrossRef]
- Moreno, J.C.; Tiller, N.; Diez, M.; Karcher, D.; Tillich, M.; Schöttler, M.A.; Bock, R. Generation and characterization of a collection of knock-down lines for the chloroplast Clp protease complex in tobacco. J. Exp. Bot. 2017, 68, 2199–2218. [Google Scholar] [CrossRef]
- Peltier, J.-B.; Ripoll, D.R.; Friso, G.; Rudella, A.; Cai, Y.; Ytterberg, J.; Giacomelli, L.; Pillardy, J.; van Wijk, K.J. Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 2004, 279, 4768–4781. [Google Scholar] [CrossRef]
- Kovacheva, S.; Bedard, J.; Wardle, A.; Patel, R.; Jarvis, P. Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 2007, 50, 364–379. [Google Scholar] [CrossRef] [PubMed]
- Flores-Pérez, Ú.; Bédard, J.; Tanabe, N.; Lymperopoulos, P.; Clarke, A.K.; Jarvis, P. Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol. 2016, 170, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Akita, M.; Nielsen, E.; Keegstra, K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 1997, 136, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, E.; Akita, M.; Davila-aponte, J.; Keegstra, K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997, 16, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Paila, Y.D.; Richardson, L.G.L.; Schnell, D.J. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 2015, 427, 1038–1060. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.G.L.; Singhal, R.; Schnell, D.J. The integration of chloroplast protein targeting with plant developmental and stress responses. BMC Biol. 2017, 15, 118. [Google Scholar] [CrossRef]
- Shanklin, J.; DeWitt, N.D.; Flanagan, J.M. The stroma of higher plant plastids contain ClpP and CIpC, functional homobgs of Escheríchia colí ClpP and CIpA: An archetypal two-component ATP-dependent protease. Plant Cell 1995, 7, 1713–1722. [Google Scholar]
- Constan, D.; Froehlich, J.E.; Rangarajan, S.; Keegstra, K. A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 2004, 136, 3605–3615. [Google Scholar] [CrossRef]
- Sjogren, L.L.E.; MacDonald, T.M.; Sutinen, S.; Clarke, A.K. Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 2004, 136, 4114–4126. [Google Scholar] [CrossRef]
- Pulido, P.; Llamas, E.; Llorente, B.; Ventura, S.; Wright, L.P.; Rodríguez-Concepción, M. Specific Hsp100 chaperones determine the fate of the first enzyme of the plastidial isoprenoid pathway for either refolding or degradation by the stromal Clp protease in Arabidopsis. PLoS Genet. 2016, 12, 1–19. [Google Scholar] [CrossRef]
- Wu, G.-Z.; Chalvin, C.; Hoelscher, M.P.; Meyer, E.H.; Wu, X.N.; Bock, R. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol. 2018, 176, 2472–2495. [Google Scholar] [CrossRef] [PubMed]
- Remmers, I.M.; Adamo, S.D.; Martens, D.E.; De Vos, R.C.H.; Mumm, R.; America, A.H.P.; Cordewener, J.H.G.; Bakker, L.V.; Peters, S.A.; Wij, R.H.; et al. Orchestration of transcriptome, proteome and metabolome in the diatom Phaeodactylum tricornutum during nitrogen limitation. Algal Res. 2018, 35, 33–49. [Google Scholar] [CrossRef]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Krumsiek, J.; Bartel, J.; Theis, F.J. Computational approaches for systems metabolomics. Curr. Opin. Biotechnol. 2016, 39, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Baran, R.; Suematsu, M.; Ueno, Y.; Ikeda, S.; Sakurakawa, T.; Kakazu, Y.; Ishikawa, T.; Robert, M.; Nishioka, T.; et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006, 281, 16768–16776. [Google Scholar] [CrossRef]
- Cho, H.S. DNA Gyrase is involved in chloroplast nucleoid partitioning. Plant Cell 2004, 16, 2665–2682. [Google Scholar] [CrossRef][Green Version]
- Ahn, C.S.; Pai, H.S. Physiological function of IspE, a plastid MEP pathway gene for isoprenoid biosynthesis, in organelle biogenesis and cell morphogenesis in Nicotiana benthamiana. Plant Mol. Biol. 2008, 66, 503–517. [Google Scholar] [CrossRef]
- Velásquez, A.C.; Chakravarthy, S.; Martin, G.B. Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J. Vis. Exp. 2009, 10. [Google Scholar] [CrossRef]
- Kang, Y.W.; Lee, J.Y.; Jeon, Y.; Cheong, G.W.; Kim, M.; Pai, H.S. In vivo effects of NbSiR silencing on chloroplast development in Nicotiana Benthamiana. Plant Mol. Biol. 2010, 72, 569–583. [Google Scholar] [CrossRef]
- Cui, H.; Wang, A. An efficient viral vector for functional genomic studies of Prunus fruit trees and its induced resistance to Plum pox virus via silencing of a host factor gene. Plant Biotechnol. J. 2017, 15, 344–356. [Google Scholar] [CrossRef]
- Gottesman, S.; Squirest, C.; Pichersky, E.; Carrington, M.; Hobbsii, M.; Mattickii, J.S.; Dalrymple, B.; Kuramitsuttft, H.; Shirozatt, T.; Foster, T.; et al. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Biochemistry 1990, 87, 3513–3517. [Google Scholar] [CrossRef] [PubMed]
- Adam, Z.; Adamska, I.; Nakabayashi, K.; Ostersetzer, O.; Haussuhl, K.; Zheng, B.; Vallon, O.; Rodermel, S.R.; Shinozaki, K.; Clarke, A.K. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 2001, 125, 1912–1918. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Kim, K.W.; Dhakal, R.; Choi, D.; Baek, K.-H. Accumulation of high contents of free amino acids in the leaves of Nicotiana benthamiana by the co-suppression of NbClpC1 and NbClpC2 genes. Plant Cell Rep. 2015, 34, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Yu, Y.; Oh, W.; Cho, J.Y.; Choi, J.; Dhakal, R.; Park, Y.I.; Baek, K.H. Co-suppression of NbClpCl and NbClpC2 in Nicotiana benthamiana lowers photosynthetic capacity via altered leaf structure. Plant Omics 2015, 8, 508–516. [Google Scholar]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.; Hartl, D.; Knoll, A.; Lue, R.; Michael, M. Biology: How Life Works, 3rd ed.; W. H. Freeman and Company: New York, NY, USA, 2017. [Google Scholar]
- Taiz, L.; Zeiger, E.; Møller, I.M.; Murphy, A. Plant Physiology and Development, 6th ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2015. [Google Scholar]
- Majumdar, R.; Shao, L.; Minocha, R.; Long, S.; Minocha, S.C. Ornithine: The overlooked molecule in the regulation of polyamine metabolism. Plant Cell Physiol. 2013, 54, 990–1004. [Google Scholar] [CrossRef]
- Bürstenbinder, K.; Sauter, M. Early Events in the Ethylene Biosynthetic Pathway–Regulation of the Pools of Methionine and S-Adenosylmethionine. In The Plant Hormone Ethylene; Mcmanus, M.T., Ed.; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2012; Volume 44, pp. 19–52. [Google Scholar]
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: Current state. Nutr. J. 2016, 15, 1–22. [Google Scholar] [CrossRef]
- Roxas, V.P.; Smith, R.K., Jr.; Allen, E.R.; Allen, R.D. Overexpression of glutathione S-transferase/glutathioneperoxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997, 15, 988–991. [Google Scholar] [CrossRef]
- Cheng, M.C.; Ko, K.; Chang, W.L.; Kuo, W.C.; Chen, G.H.; Lin, T.P. Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 2015, 83, 926–939. [Google Scholar] [CrossRef]
- Moffatt, B.A.; Ashihara, H. Purine and pyrimidine nucleotide synthesis and metabolism. Arab. Book 2002, e0018. [Google Scholar] [CrossRef]
- Soga, T.; Heiger, D.N. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2000, 72, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Ueno, Y.; Naraoka, H.; Ohashi, Y.; Tomita, M.; Nishioka, T. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2002, 74, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Ohashi, Y.; Ueno, Y.; Naraoka, H.; Tomita, M.; Nishioka, T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003, 2, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Ooga, T.; Sato, H.; Nagashima, A.; Sasaki, K.; Tomita, M.; Ohashi, Y. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol. Biosyst. 2011, 7, 1217–1223. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.S.; Baek, K.-H. Co-Suppression of NbClpC1 and NbClpC2, Encoding Clp Protease Chaperons, Elicits Significant Changes in the Metabolic Profile of Nicotiana benthamiana. Plants 2020, 9, 259. https://doi.org/10.3390/plants9020259
Ali MS, Baek K-H. Co-Suppression of NbClpC1 and NbClpC2, Encoding Clp Protease Chaperons, Elicits Significant Changes in the Metabolic Profile of Nicotiana benthamiana. Plants. 2020; 9(2):259. https://doi.org/10.3390/plants9020259
Chicago/Turabian StyleAli, Md. Sarafat, and Kwang-Hyun Baek. 2020. "Co-Suppression of NbClpC1 and NbClpC2, Encoding Clp Protease Chaperons, Elicits Significant Changes in the Metabolic Profile of Nicotiana benthamiana" Plants 9, no. 2: 259. https://doi.org/10.3390/plants9020259
APA StyleAli, M. S., & Baek, K.-H. (2020). Co-Suppression of NbClpC1 and NbClpC2, Encoding Clp Protease Chaperons, Elicits Significant Changes in the Metabolic Profile of Nicotiana benthamiana. Plants, 9(2), 259. https://doi.org/10.3390/plants9020259





