A Molecular Phylogenetic Study of the Genus Phedimus for Tracing the Origin of “Tottori Fujita” Cultivars
Abstract
1. Introduction
2. Results
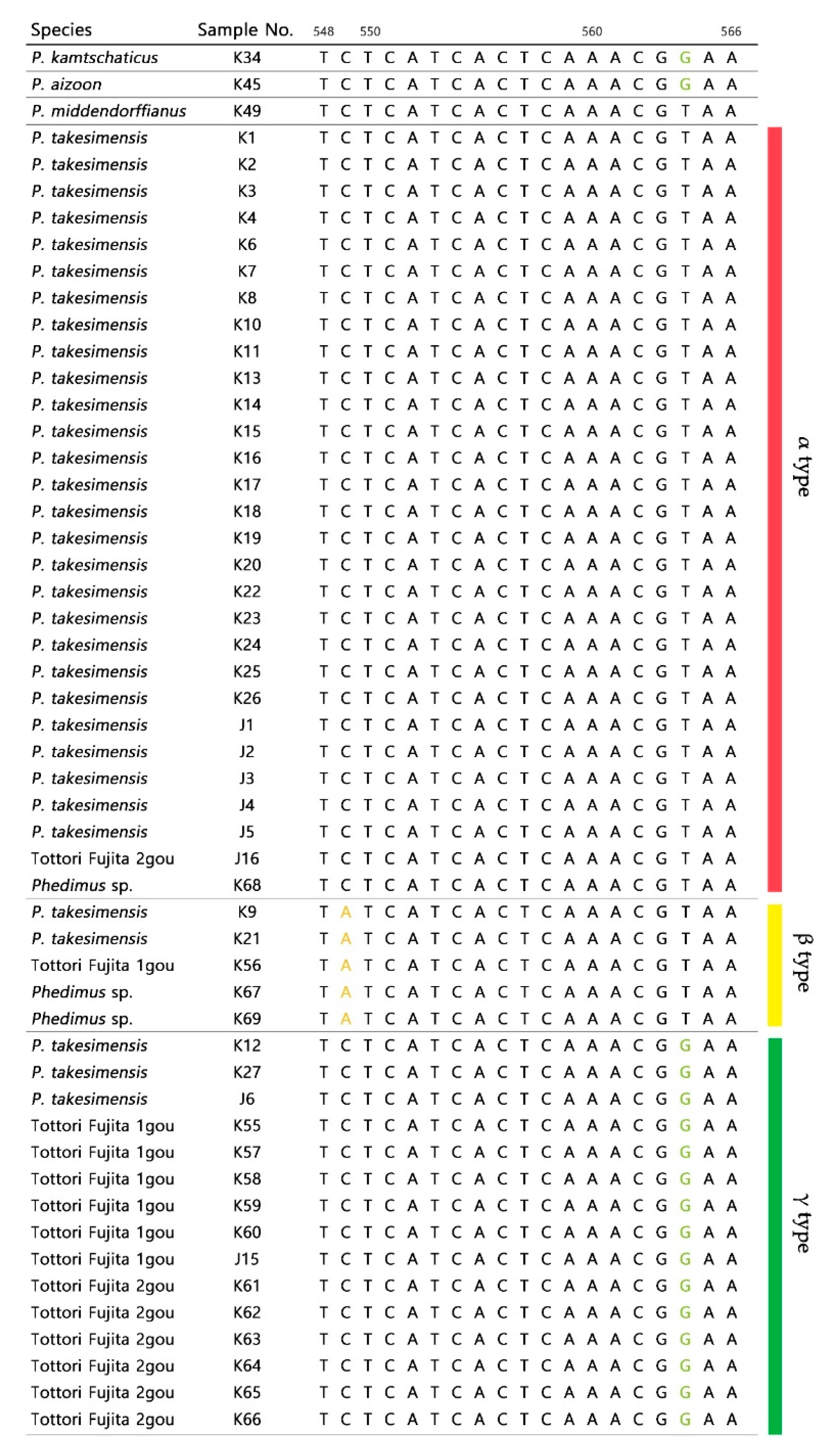
2.1. Sequence Variations within Phedimus
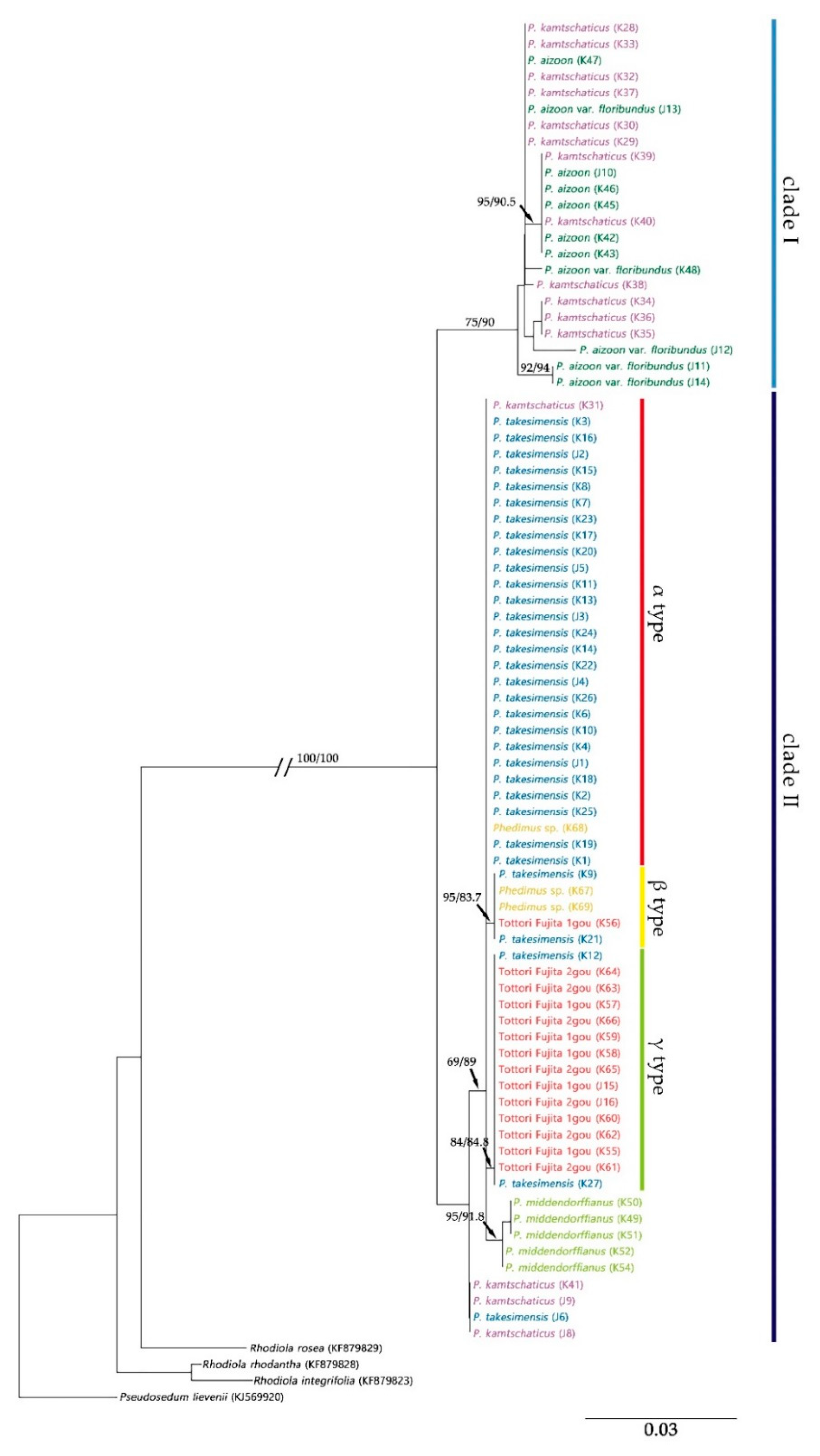
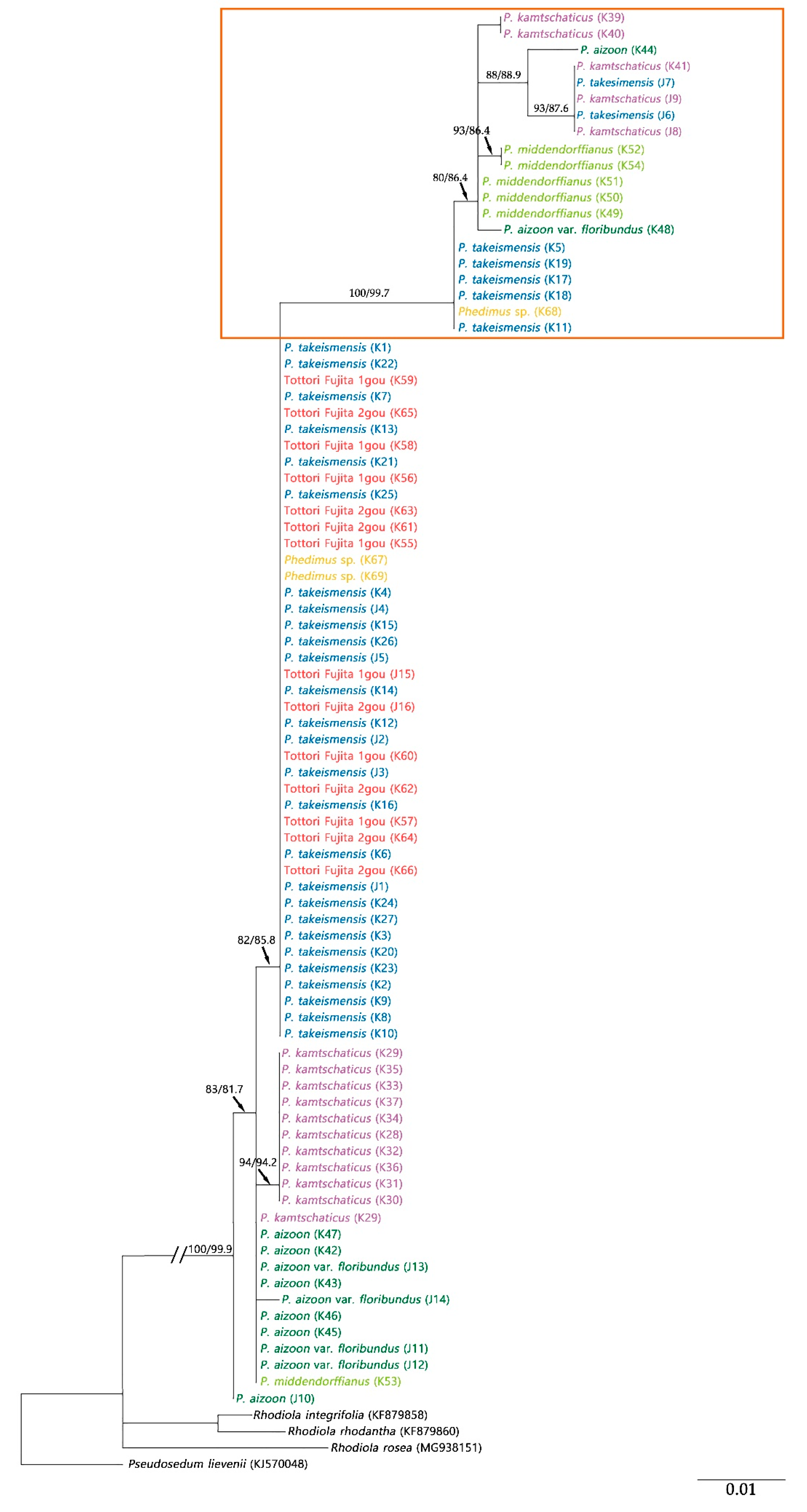
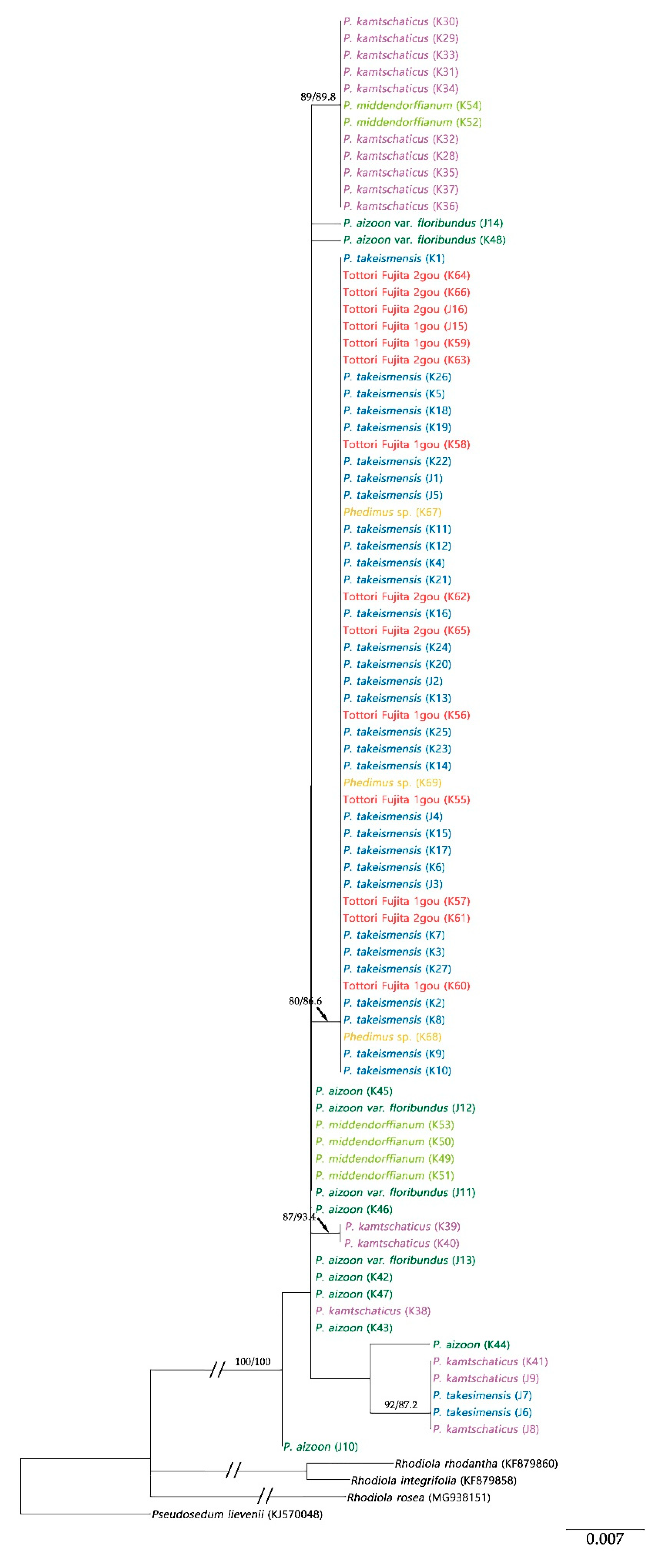
2.2. Origin and Phylogenetic Relationship of Cultivated Phedimus
3. Discussion
3.1. The Origin of Cultivar “Tottori Fujita”
3.2. Application of Molecular Phylogenetic Analysis to Evaluate Misidentified Plants
3.3. Hybridization Leading to Discordance on the Phylogenetic Tree
3.4. Molecular Marker Development for Further Study
4. Materials and Methods
4.1. Plant Materials and Sampling
4.2. DNA Extraction, PCR Amplification, and Sequencing
4.3. Molecular Phylogenetic Analyses of ITS and Plastid DNA Regions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chase, M.W.; Christenhusz, M.; Fay, M.; Byng, J.; Judd, W.S.; Soltis, D.; Mabberley, D.; Sennikov, A.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Berger, A. Crassulaceae. In Die natürlichen Pflanzenfamilien; Engler, A., Prantl, K., Eds.; Verlag Wilhelm Englmann: Leipzig, German, 1930; Volume 2, pp. 352–483. [Google Scholar]
- Rafinesque, C.S. Museum of Natural Sciences. Am. Mon. Ma. Crit. Rev. 1817, 1, 431–442. [Google Scholar]
- Hart, H. Infrafamilial and generic classification of the Crassulaceae. In Evolution and Systematics of the Crassulaceae; Hart, H., Eggli, U., Eds.; Backhuys: Leiden (NL), The Netherlands, 1995; pp. 159–172. [Google Scholar]
- Ohba, H.; Bartholomew, B.M.; Turland, N.J.; Kunjun, F.; Kun-Tsun, F. New combinations in Phedimus (Crassulaceae). Novon 2000, 10, 400–402. [Google Scholar] [CrossRef]
- Mort, M.E.; Soltis, D.E.; Soltis, P.S.; Francisco-Ortega, J.; Santos-Guerra, A. Phylogenetic relationships and evolution of Crassulaceae inferred from matK sequence data. Am. J. Bot. 2001, 88, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Mayuzumi, S.; Ohba, H. The phylogenetic position of eastern Asian Sedoideae (Crassulaceae) inferred from chloroplast and nuclear DNA sequences. Syst. Bot. 2004, 29, 587–598. [Google Scholar] [CrossRef]
- Gontcharova, S.; Gontcharov, A. Molecular phylogeny and systematics of flowering plants of the family Crassulaceae DC. Mol. Biol. 2009, 43, 794. [Google Scholar] [CrossRef]
- Ray, S.; Harris, J.G.S. How Sedum takesimense was interoduced into cultivation. Sedum Soc. Newsl. 1991, 19, 4. [Google Scholar]
- Ray, S.; Harris, J.G.S. Sedum takesimense—Some cultural notes. Sedum Soc. Newsl. 1991, 19, 6–7. [Google Scholar]
- Kentaro, I. Where the naturalized plant attein? Evergreen Sedum (Sedum takesimense NAKAI). J. Jpn. Soc. Reveget. Tech. 2001, 36, 446. (In Japanese) [Google Scholar]
- Arnold, M.L.; Hodges, S.A. Are natural hybrids fit or unfit relative to their parents? Trends Ecol. Evol. 1995, 10, 67–71. [Google Scholar] [CrossRef]
- Rudolf-Pilih, K.; Petkovšek, M.; Jakse, J.; Štajner, N.; Murovec, J.; Bohanec, B. New hybrid breeding method based on genotyping, inter-pollination, phenotyping and paternity testing of selected elite F1 hybrids. Front. Plant Sci. 2019, 10, 1111. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, B.M.; Ryan, M.E.; Johnson, J.R.; Corush, J.; Carter, E.T. Hybridization and the species problem in conservation. Curr. Zool. 2015, 61, 206–216. [Google Scholar] [CrossRef]
- Huamán, Z.; Spooner, D.M. Reclassification of landrace populations of cultivated potatoes (Solanum sect. Petota). Am. J. Bot. 2002, 89, 947–965. [Google Scholar]
- Lema, M.; Ali, M.Y.; Retuerto, R. Domestication influences morphological and physiological responses to salinity in Brassica oleracea seedlings. Aob Plants 2019, 11, plz046. [Google Scholar] [CrossRef]
- Huang, H.; Liu, Y. Natural hybridization, introgression breeding, and cultivar improvement in the genus Actinidia. Tree Genet. Genomes 2014, 10, 1113–1122. [Google Scholar] [CrossRef]
- Chung, Y.H.; Kim, J.H. A taxonomic study of Sedum section Aizoon in Korea. Korean J. Plant Taxon. 1989, 19, 189–227. (In Korean) [Google Scholar] [CrossRef]
- Yoo, Y.; Park, K. A test of the hybrid origin of Korean endemic Sedum latiovalifolium (Crassulaceae). Korean J. Plant Taxon. 2016, 46, 378–391. (In Korean) [Google Scholar] [CrossRef]
- Amano, M. Biosystematic study of Sedum, L. Subgenus Aizoon (Crassulaceae). Bot. Mag. Tokyo 1990, 103, 67–85. [Google Scholar] [CrossRef]
- Zhang, J.; Meng, S.; Allen, G.A.; Wen, J.; Rao, G. Rapid radiation and dispersal out of the Qinghai-Tibetan 1 Plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol. Phylogenet. Evol. 2014, 77, 147–158. [Google Scholar] [CrossRef]
- Guest, H.J.; Allen, G.A. Geographical origins of North American Rhodiola (Crassulaceae) and phylogeography of the western roseroot, Rhodiola integrifolia. J. Biogeogr. 2014, 41, 1070–1080. [Google Scholar] [CrossRef]
- György, Z.; Tóth, E.G.; Incze, N.; Molnár, B.; Höhn, M. Intercontinental migration pattern and genetic differentiation of arctic-alpine Rhodiola rosea L.: A chloroplast DNA survey. Ecol. Evol. 2018, 8, 11508–11521. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR Protocols: A guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- CBOL. Plant Working Group A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.; Von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 2, 461–464. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
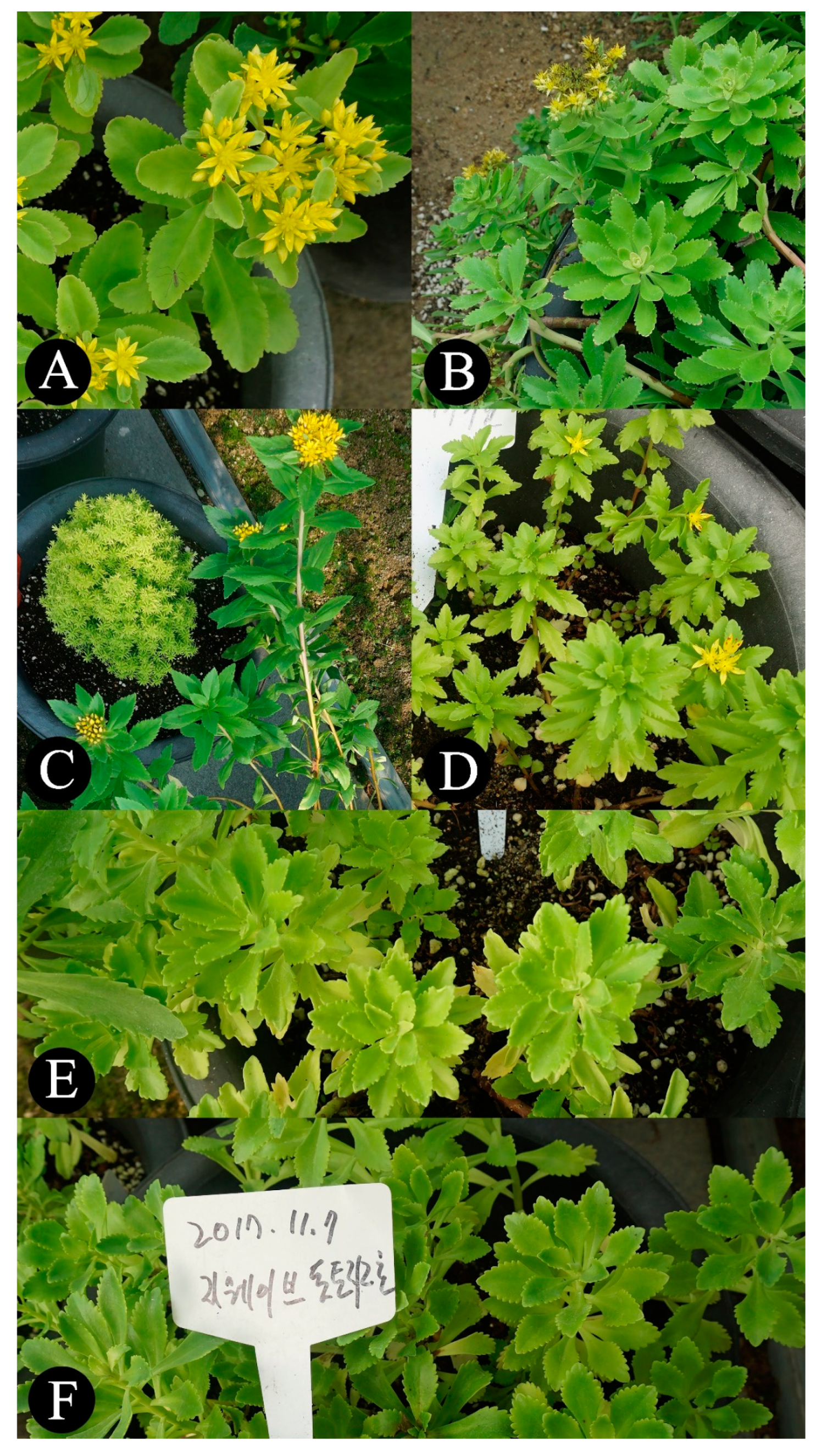
| Sample ID | Species | Collection Information of Locality or Reference | Acc. No. | |
|---|---|---|---|---|
| ITS | psbA-trnH | |||
| K1 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908990 | MN935682 |
| K2 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908991 | MN935683 |
| K3 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908992 | MN935684 |
| K4 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908993 | MN935685 |
| K5 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | - | MN935686 |
| K6 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908994 | MN935687 |
| K7 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908995 | MN935688 |
| K8 | Phedimus takesimensis | collectcollected from the natural locality | MN908996 | MN935689 |
| K9 | Phedimus takesimensis | collected from the natural locality (not specified) | MN908997 | MN935690 |
| K10 | Phedimus takesimensis | collected from the natural locality (not specified) | MN908998 | MN935691 |
| K11 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908999 | MN935692 |
| K12 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN909000 | MN935693 |
| K13 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN909001 | MN935694 |
| K14 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN909002 | MN935695 |
| K15 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN909003 | MN935696 |
| K16 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN909004 | MN935697 |
| K17 | Phedimus takesimensis | Dokdo-ri, Ulleung-gun, Gyeongsangbuk-do, Korea | MN909005 | MN935698 |
| K18 | Phedimus takesimensis | Dokdo-ri, Ulleung-gun, Gyeongsangbuk-do, Korea | MN909006 | MN935699 |
| K19 | Phedimus takesimensis | Dokdo-ri, Ulleung-gun, Gyeongsangbuk-do, Korea | MN909007 | MN935700 |
| K20 | Phedimus takesimensis | collected from the natural locality (not specified) | MN909008 | MN935701 |
| K21 | Phedimus takesimensis | collected from the natural locality (not specified) | MN909009 | MN935702 |
| K22 | Phedimus takesimensis | collected from the natural locality (not specified) | MN909010 | MN935703 |
| K23 | Phedimus takesimensis | Dokdo-ri, Ulleung-gun, Gyeongsangbuk-do, Korea | MN909011 | MN935704 |
| K24 | Phedimus takesimensis | Dokdo-ri, Ulleung-gun, Gyeongsangbuk-do, Korea | MN909012 | MN935705 |
| K25 | Phedimus takesimensis | Dokdo-ri, Ulleung-gun, Gyeongsangbuk-do, Korea | MN909013 | MN935706 |
| K26 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN909014 | MN935707 |
| K27 | Phedimus takesimensis | Ulleung-gun, Gyeongsangbuk-do, Korea | MN909015 | MN935708 |
| K28 | Phedimus kamtschaticus | Pohang-si, Gyeongsangbuk-do, Korea | MN908958 | MN935733 |
| K29 | Phedimus kamtschaticus | Goseong-gun, Gangwon-do, Korea | MN908959 | MN935734 |
| K30 | Phedimus kamtschaticus | Bonghwa-gun, Gyeongsangbuk-do, Korea | MN908960 | MN935735 |
| K31a | Phedimus kamtschaticus | Pocheon-si, Gyeonggi-do, Korea | MN908961 | MN935736 |
| K32 | Phedimus kamtschaticus | Pocheon-si, Gyeonggi-do, Korea | MN908962 | MN935737 |
| K33 | Phedimus kamtschaticus | Pocheon-si, Gyeonggi-do, Korea | MN908963 | MN935738 |
| K34 | Phedimus kamtschaticus | Seocho-gu, Seoul, Korea | MN908964 | MN935739 |
| K35 | Phedimus kamtschaticus | Seocho-gu, Seoul, Korea | MN908965 | MN935740 |
| K36 | Phedimus kamtschaticus | Seocho-gu, Seoul, Korea | MN908966 | MN935741 |
| K37 | Phedimus kamtschaticus | collected from the natural locality (not specified) | MN908967 | MN935742 |
| K38 | Phedimus kamtschaticus | Pohang-si, Gyeongsangbuk-do, Korea | MN908968 | MN935743 |
| K39 | Phedimus kamtschaticus | Pohang-si, Gyeongsangbuk-do, Korea | MN908969 | MN935744 |
| K40 | Phedimus kamtschaticus | Pohang-si, Gyeongsangbuk-do, Korea | MN908970 | MN935745 |
| K41b | Phedimus kamtschaticus | Ulleung-gun, Gyeongsangbuk-do, Korea | MN908971 | MN935748 |
| K42 | Phedimus aizoon | collected from the natural locality (not specified) | MN908974 | MN935749 |
| K43 | Phedimus aizoon | collected from the natural locality (not specified) | MN908975 | MN935750 |
| K44 | Phedimus aizoon | collected from the natural locality (not specified) | - | MN935751 |
| K45 | Phedimus aizoon | farm, Pyeongchang-gun, Gangwon-do, Korea | MN908976 | MN935752 |
| K46 | Phedimus aizoon | farm, Pyeongchang-gun, Gangwon-do, Korea | MN908977 | MN935753 |
| K47 | Phedimus aizoon | farm, Pyeongchang-gun, Gangwon-do, Korea | MN908978 | MN935754 |
| K48 | Phedimus aizoon var. florivundus | Gyeongju-si, Gyeongsangnam-do, Korea | MN908980 | MN935758 |
| K49 | Phedimus middendorffianus | collected from the natural locality (not specified) | MN908985 | MN935761 |
| K50 | Phedimus middendorffianus | collected from the natural locality (not specified) | MN908986 | MN935762 |
| K51 | Phedimus middendorffianus | collected from the natural locality (not specified) | MN908987 | MN935763 |
| K52 | Phedimus middendorffianus | farm, Uiwang-si, Gyeonggi-do, Korea | MN908988 | MN935764 |
| K53 | Phedimus middendorffianus | farm, Uiwang-si, Gyeonggi-do, Korea | - | MN935765 |
| K54 | Phedimus middendorffianus | farm, Uiwang-si, Gyeonggi-do, Korea | MN908989 | MN935766 |
| K55 | “Tottori Fujita 1gou” | generated and submitted by applicant | MN909025 | MN935716 |
| K56 | “Tottori Fujita 1gou” | generated and submitted by applicant | MN909026 | MN935717 |
| K57 | “Tottori Fujita 1gou” | generated and submitted by applicant | MN909027 | MN935718 |
| K58 | “Tottori Fujita 1gou” | generated and submitted by applicant | MN909028 | MN935719 |
| K59 | “Tottori Fujita 1gou” | generated and submitted by applicant | MN909029 | MN935720 |
| K60 | “Tottori Fujita 1gou” | generated and submitted by applicant | MN909030 | MN935721 |
| K61 | “Tottori Fujita 2gou” | generated and submitted by applicant | MN909032 | MN935723 |
| K62 | “Tottori Fujita 2gou” | generated and submitted by applicant | MN909033 | MN935724 |
| K63 | “Tottori Fujita 2gou” | generated and submitted by applicant | MN909034 | MN935725 |
| K64 | “Tottori Fujita 2gou” | generated and submitted by applicant | MN909035 | MN935726 |
| K65 | “Tottori Fujita 2gou” | generated and submitted by applicant | MN909036 | MN935727 |
| K66 | “Tottori Fujita 2gou” | generated and submitted by applicant | MN909037 | MN935728 |
| K67 | Phediums sp. | generated and submitted by applicant | MN909023 | MN935730 |
| K68 | Phediums sp. | generated and submitted by applicant | MN909022 | MN935731 |
| K69 | Phediums sp. | generated and submitted by applicant | MN909024 | MN935732 |
| J1 | Phedimus takesimensis | Seed store, Nagano Pref., Japan | MN909016 | MN935709 |
| J2 | Phedimus takesimensis | Seed store, Nagano Pref., Japan | MN909017 | MN935710 |
| J3 | Phedimus takesimensis | Seed store, Nagano Pref., Japan | MN909018 | MN935711 |
| J4 | Phedimus takesimensis | Seed store, Nagano Pref., Japan | MN909019 | MN935712 |
| J5 | Phedimus takesimensis | Seed store, Nagano Pref., Japan | MN909020 | MN935713 |
| J6 b | Phedimus takesimensis | Seed store, Nagano Pref., Japan | MN909021 | MN935714 |
| J7 b | Phedimus takesimensis | Seed store, Nagano Pref., Japan | - | MN935715 |
| J8 b | Phedimus kamtschaticus | Hokkaido, Obihiro-si, Japan | MN908972 | MN935746 |
| J9 b | Phedimus kamtschaticus | Hokkaido, Memuro-cho, Japan | MN908973 | MN935747 |
| J10 | Phedimus aizoon | Nagano Pref., Japan | MN908979 | MN935755 |
| J11 | Phedimus aizoon var. floribundus | Nigata Pref. Kasiwazaki-si, Japan | MN908981 | MN935756 |
| J12 | Phedimus aizoon var. floribundus | Kagawa Pref. Kankakei, Japan | MN908982 | MN935757 |
| J13 | Phedimus aizoon var. floribundus | Seed store, Osaka Pref., Japan | MN908983 | MN935759 |
| J14 | Phedimus aizoon var. floribundus | Nigata Pref. Sado-si, Japan | MN908984 | MN935760 |
| J15 | “Tottori Fujita 1gou” | Tottori Pref. Iwami-cho, Japan | MN909031 | MN935722 |
| J16 | “Tottori Fujita 2gou” | Tottori Pref. Iwami-cho, Japan | MN909038 | MN935729 |
| Outgroup | ||||
| Pseudosedum lievenii | Zhang, J.Q. et al., 2014 [21] | KJ569920 | KJ570048 | |
| Rhodiola integrifolia | Guest, H.J. et al., 2014 [22] | KF879823 | KF879860 | |
| Rhodiola rhodantha | Guest, H.J. et al., 2014 [22] | KF879828 | KF879860 | |
| Rhodiola rosea | Guest, H.J. et al., 2014 [22], Gyorgy, Z., et al., 2018 [23] | KF879829 | MG938151 | |
| Region | Primer | Sequence (5′→ 3′) | Reference |
|---|---|---|---|
| ITS (rDNA) | ITS1 | TCCGTAGGTGAACCTGCGG | White et al. [25] |
| ITS4 | TCCTCCGCTTATTGATATGC | White et al. [25] | |
| psbA-trnH (cpDNA) | psbA3_f | GTTATGCATGAACGTAATGCTC | CBOL Plant Working Group [26] |
| trnHf_05 | CGCGCATGGTGGATTCACAATCC | CBOL Plant Working Group [26] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.K.; Kim, T.H.; Kim, J.S. A Molecular Phylogenetic Study of the Genus Phedimus for Tracing the Origin of “Tottori Fujita” Cultivars. Plants 2020, 9, 254. https://doi.org/10.3390/plants9020254
Han SK, Kim TH, Kim JS. A Molecular Phylogenetic Study of the Genus Phedimus for Tracing the Origin of “Tottori Fujita” Cultivars. Plants. 2020; 9(2):254. https://doi.org/10.3390/plants9020254
Chicago/Turabian StyleHan, Sung Kyung, Tae Hoon Kim, and Jung Sung Kim. 2020. "A Molecular Phylogenetic Study of the Genus Phedimus for Tracing the Origin of “Tottori Fujita” Cultivars" Plants 9, no. 2: 254. https://doi.org/10.3390/plants9020254
APA StyleHan, S. K., Kim, T. H., & Kim, J. S. (2020). A Molecular Phylogenetic Study of the Genus Phedimus for Tracing the Origin of “Tottori Fujita” Cultivars. Plants, 9(2), 254. https://doi.org/10.3390/plants9020254






