Structure, Function, Diversity, and Composition of Fungal Communities in Rhizospheric Soil of Coptis chinensis Franch under a Successive Cropping System
Abstract
1. Introduction
2. Method and Materials
2.1. Study Site and Soil Sampling
2.2. Analysis of Physicochemical Properties
2.3. DNA Extraction and PCR Amplification
2.4. Illumina MiSeq Sequencing
2.5. Processing of Sequencing Data
2.6. Statistic Analysis
3. Result
3.1. Soil Physicochemical Properties
3.2. Biodiversity
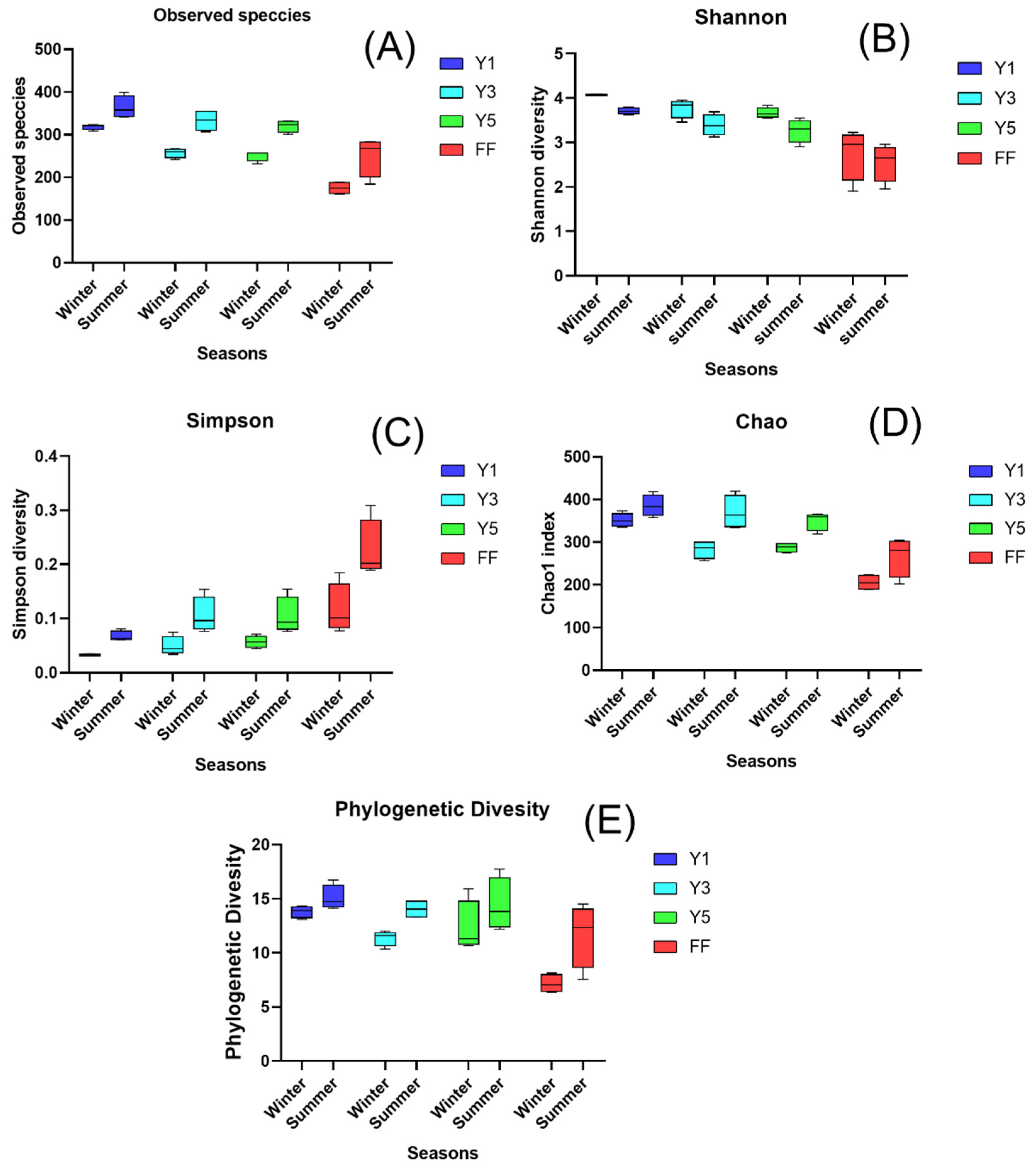
3.2.1. Alpha Diversity
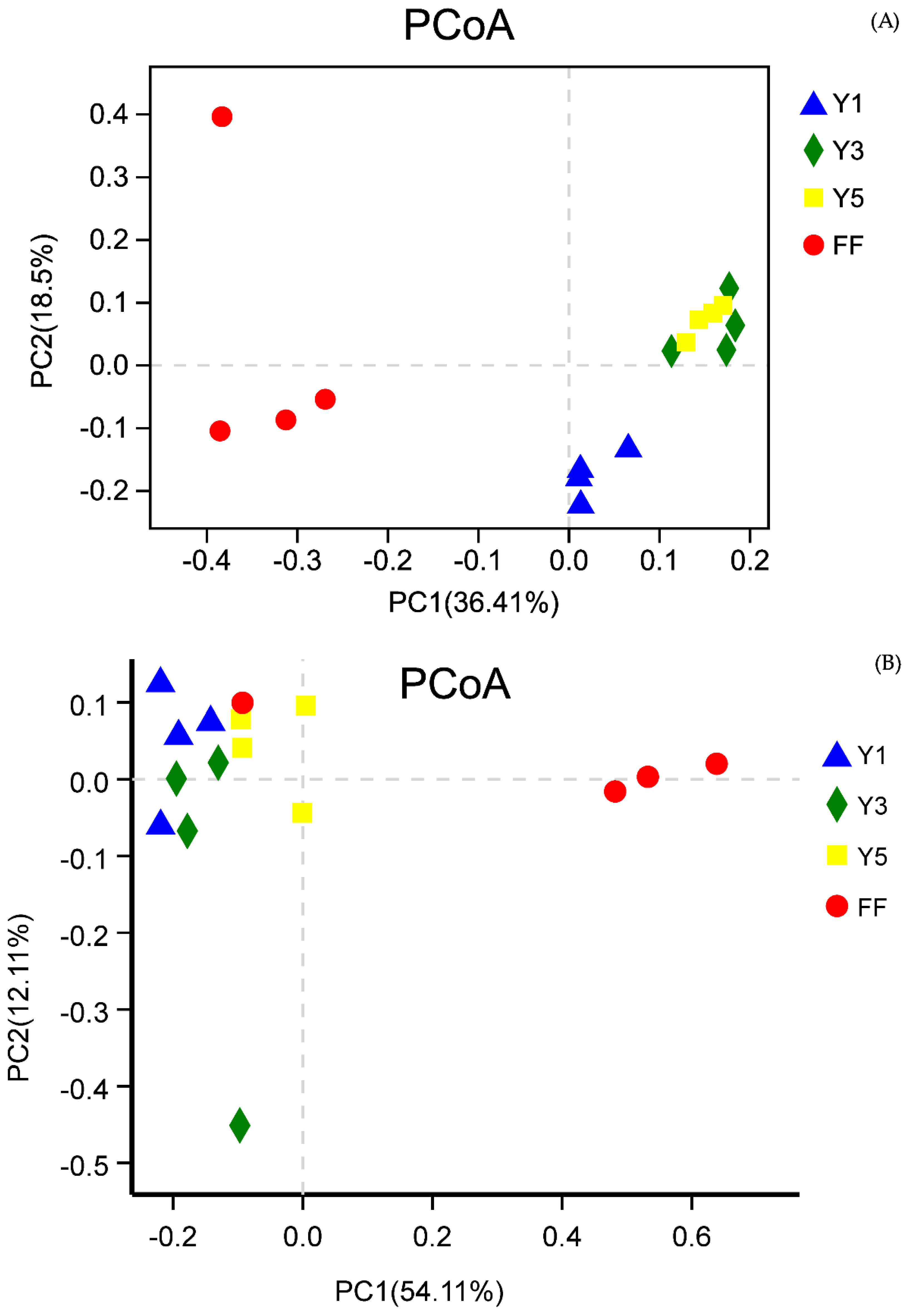
3.2.2. Beta Diversity
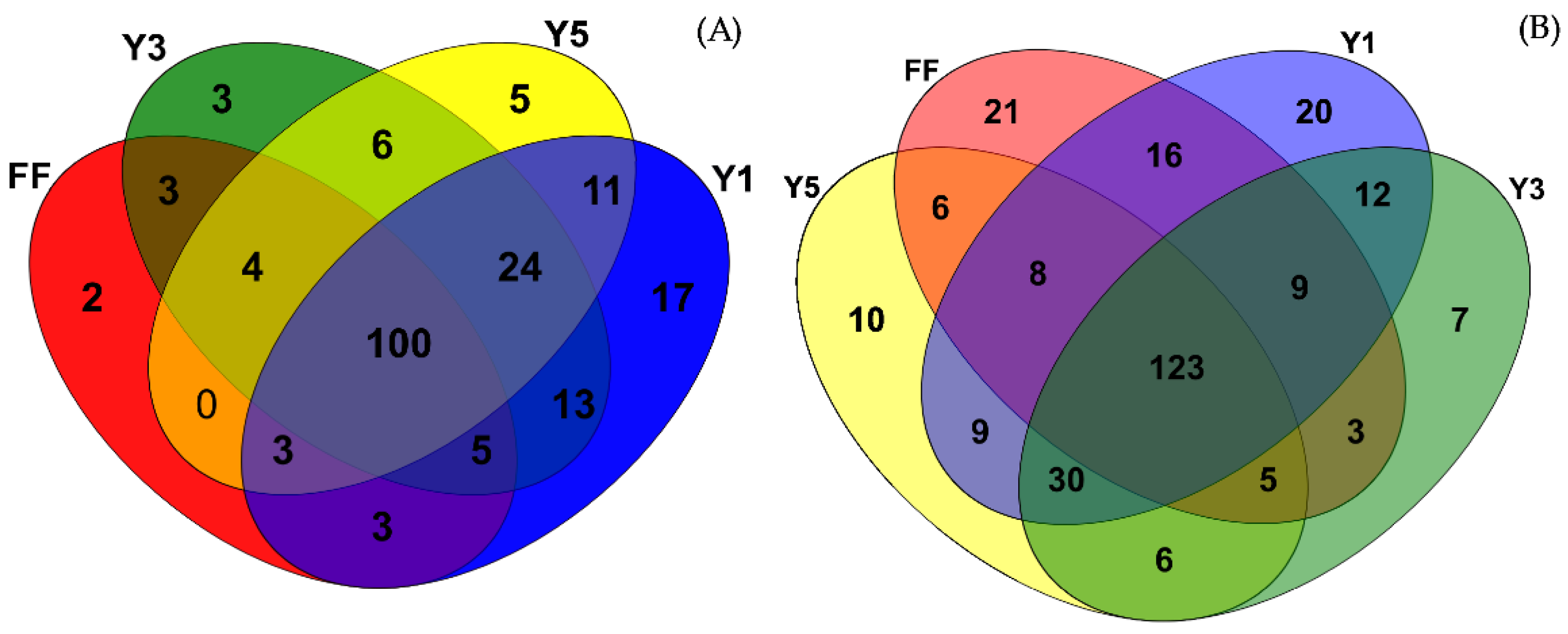
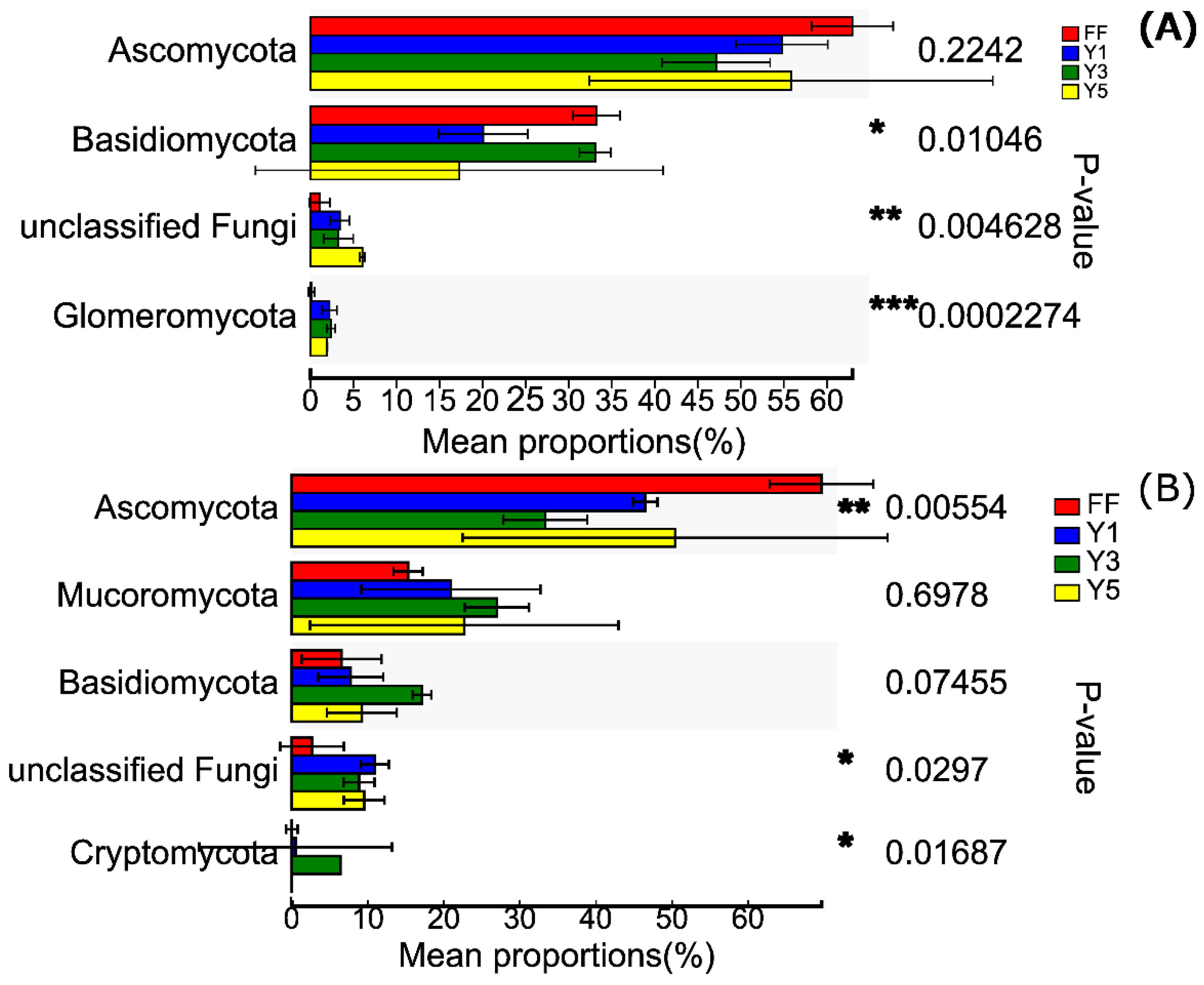
3.3. Description of Fungal Communities
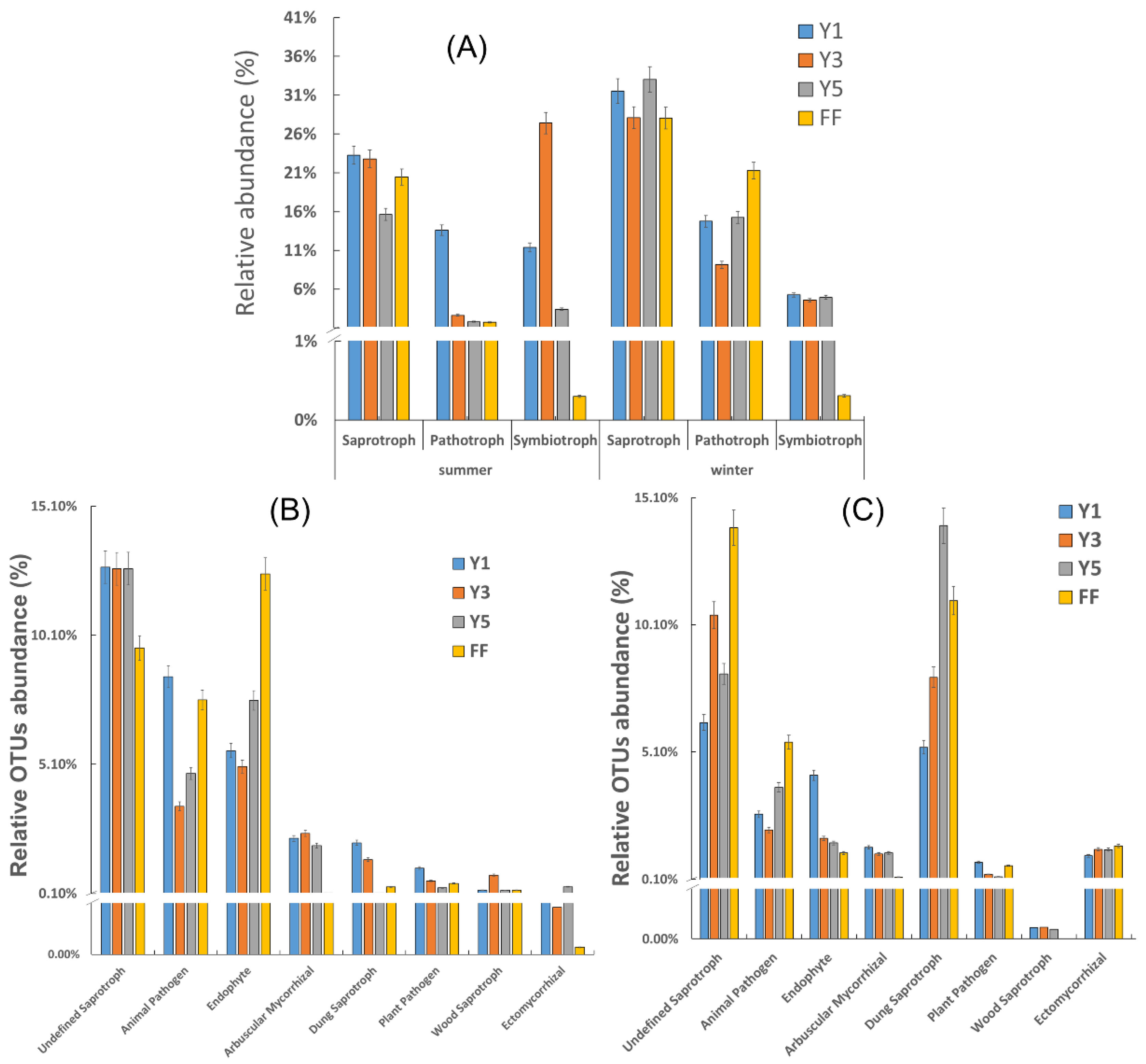
3.4. Functional Prediction of Fungal Communities
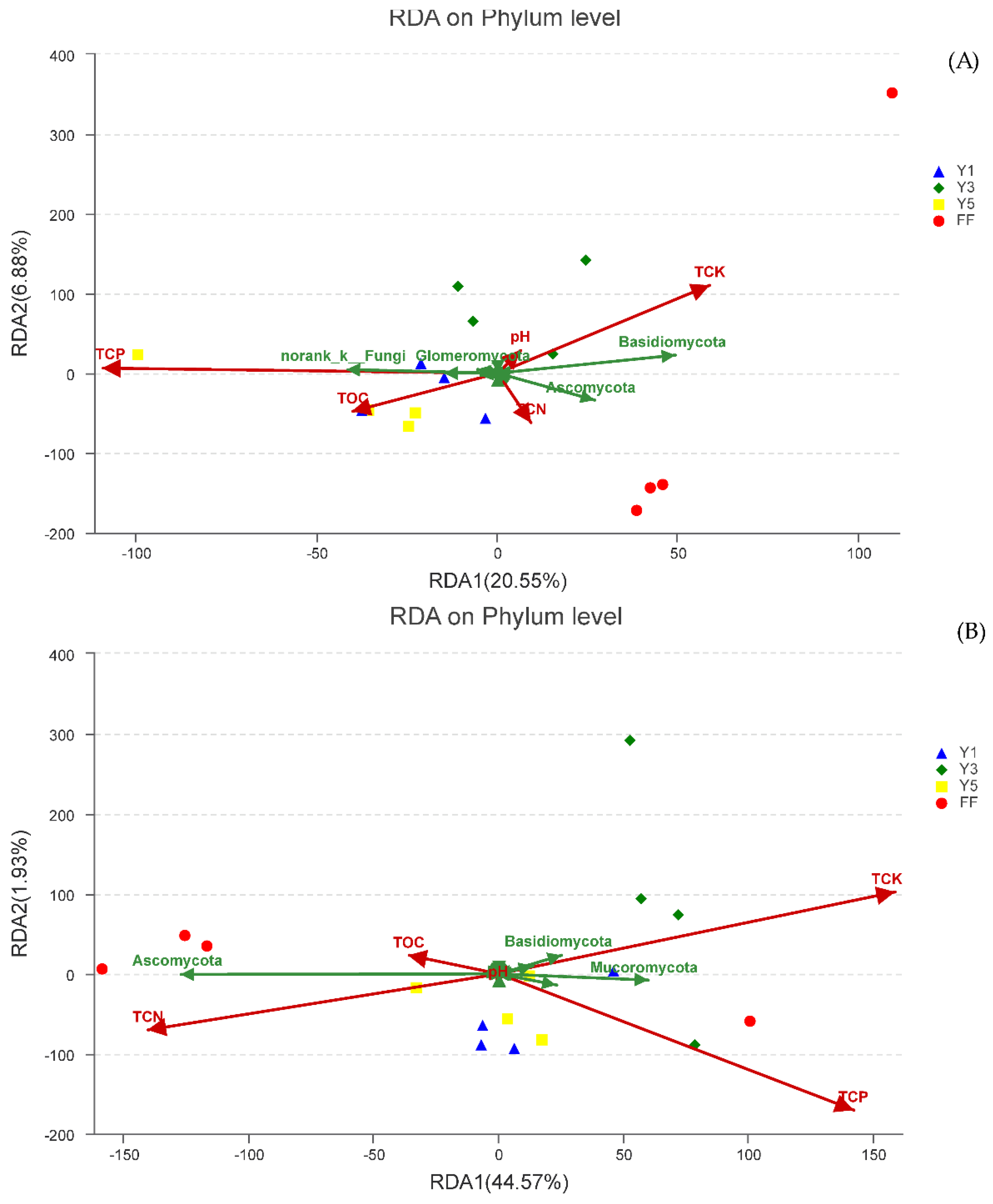
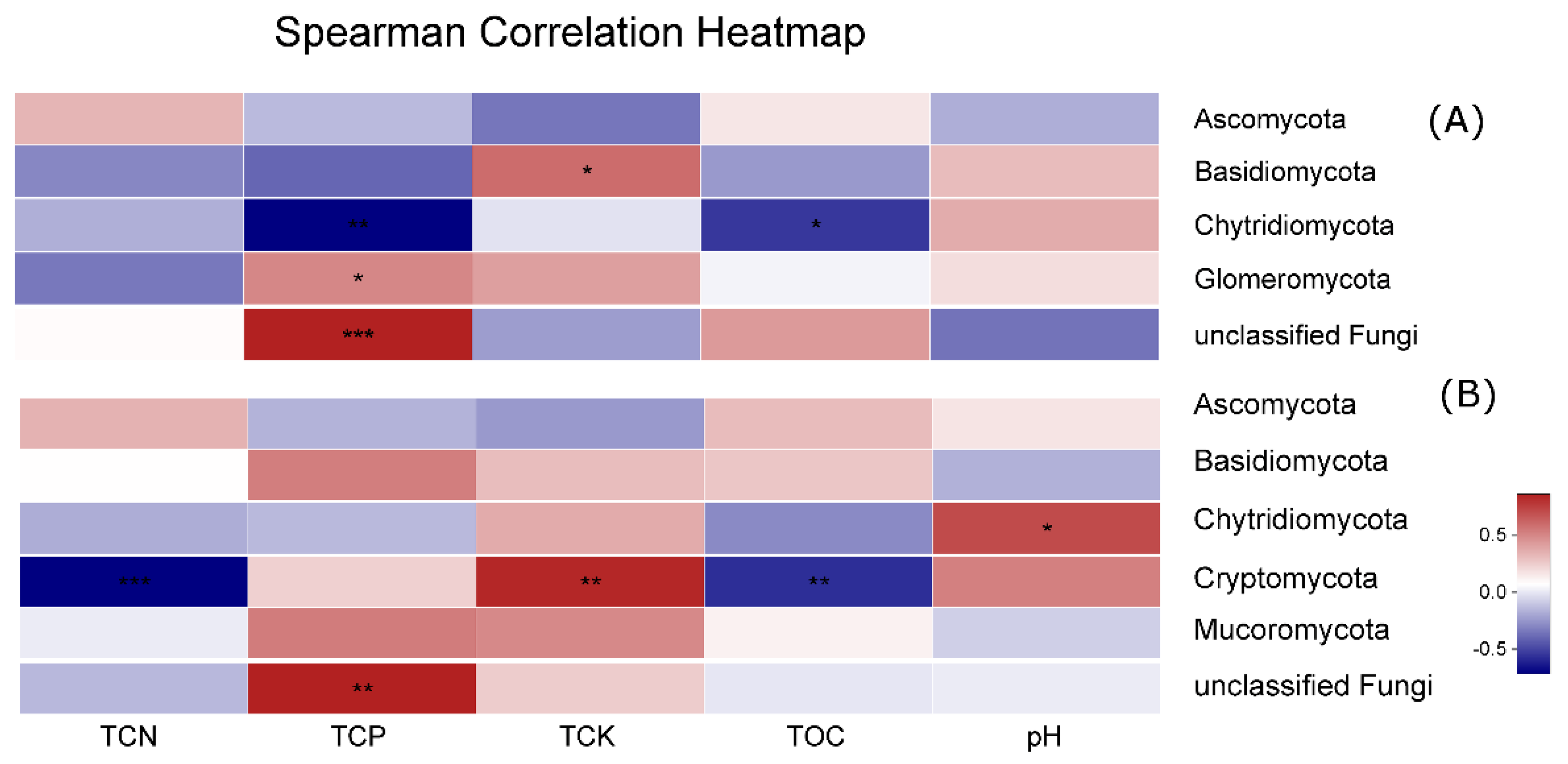
3.5. Effect of Environmental Factors on Fungal Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NGS | Next-Generation Sequencing |
| RDA | Redundancy Analysis |
| PcoA | Principle Coordinate Analysis |
| OTUs | Operational Taxonomic Units |
References
- Teng, H.; Choi, Y.H. Optimization of ultrasonic-assisted extraction of bioactive alkaloid compounds from Rhizoma coptidis (Coptis chinensis Franch.) using response surface methodology. Food Chem. 2014, 142, 299–305. [Google Scholar] [CrossRef]
- Song, X.; Pan, Y.; Li, L.; Wu, X.; Wang, Y. Composition and diversity of rhizosphere fungal community in Coptis chinensis Franch. continuous cropping fields. PLoS ONE 2018, 13, e0193811. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.-Y.; Chen, L.; Huang, X.-J.; Zhang, Q.-W.; Jiang, R.-W.; Yao, F.; Ye, W.-C. New enantiomeric isoquinoline alkaloids from Coptis chinensis. Phytochem. Lett. 2014, 7, 89–92. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Gao, J.; Huang, T.; Kendall, J.R.A.; Shen, Q.; Zhang, R. Parental material and cultivation determine soil bacterial community structure and fertility. Fems Microbiol. Ecol. 2015, 91, 1–10. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Beckers, B.; Op De Beeck, M.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef]
- Bron, P.A.; van Baarlen, P.; Kleerebezem, M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 2012, 10, 66–78. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant. Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Huang, L.-F.; Song, L.-X.; Xia, X.-J.; Mao, W.-H.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q. Plant-Soil Feedbacks and Soil Sickness: From Mechanisms to Application in Agriculture. J. Chem. Ecol. 2013, 39, 232–242. [Google Scholar] [CrossRef]
- Niu, J.; Chao, J.; Xiao, Y.; Chen, W.; Zhang, C.; Liu, X.; Rang, Z.; Yin, H.; Dai, L. Insight into the effects of different cropping systems on soil bacterial community and tobacco bacterial wilt rate. J. Basic Microbiol. 2017, 57, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, Q.; Zhou, J.; Wei, Q. Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ni, T.; Li, Y.; Xiong, W.; Ran, W.; Shen, B.; Shen, Q.; Zhang, R. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS ONE 2014, 9, e85301. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, C.; Zhang, T.; Wang, X. Soil Biology & Biochemistry Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar]
- Zhao, J.; Zhang, R.; Xue, C.; Xun, W.; Sun, L.; Xu, Y.; Shen, Q. Pyrosequencing reveals contrasting soil bacterial diversity and community structure of two main winter wheat cropping systems in China. Microb. Ecol. 2014, 67, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Manici, L.M.; Caputo, F. Fungal community diversity and soil health in intensive potato cropping systems of the east Po valley, northern Italy. Ann. Appl. Biol. 2009, 155, 245–258. [Google Scholar] [CrossRef]
- Xu, L.; Ravnskov, S.; Larsen, J.; Nilsson, R.H.; Nicolaisen, M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol. Biochem. 2012, 46, 26–32. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Wu, H.; Wang, J.; Wu, Y.; Lin, S.; Khan, M.U.; Zhang, Z.; Lin, W. Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 2016, 6, 26601. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, L.; Chu, L.; Yang, Y.; Li, Z.; Azeem, S.; Zhang, Z.; Fang, C.; Lin, W. Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f.sp. heterophylla mediated by its root exudates in a consecutive monoculture system. Sci. Rep. 2015, 5, 8197. [Google Scholar] [CrossRef]
- Song, X.; Wang, Y.; Li, L.; Tan, J. Research on bacteria microecology in root rot rhizosphere soil of Coptis chinensis produced in Shizhu city. China J. Chin. Mater. Med. 2017, 42, 1–8. [Google Scholar]
- Xiao, D.; Huang, Y.; Feng, S.; Ge, Y.; Zhang, W.; He, X.; Wang, K. Soil organic carbon mineralization with fresh organic substrate and inorganic carbon additions in a red soil is controlled by fungal diversity along a pH gradient. Geoderma 2018, 321, 79–89. [Google Scholar] [CrossRef]
- Yuan, C.; Zhang, L.; Hu, H.; Wang, J.; Shen, J.; He, J. The biogeography of fungal communities in paddy soils is mainly driven by geographic distance. J. Soils Sediments 2018, 18, 1795–1805. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open source, platform independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Gao, Z.; Han, M.; Hu, Y.; Li, Z.; Liu, C.; Wang, X.; Tian, Q.; Jiao, W.; Hu, J.; Liu, L.; et al. Effects of continuous cropping of sweet potato on the fungal community structure in rhizospheric soil. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Lin, W.-X. Continuous cropping obstacle and allelopathic autotoxicity of medicinal plants. Chin. J. Eco-Agric. 2009, 17, 189–196. [Google Scholar] [CrossRef]
- Xiong, W.; Li, Z.; Liu, H.; Xue, C.; Zhang, R.; Wu, H.; Li, R.; Shen, Q. The effect of long-term continuous cropping of black pepper on soil bacterial communities as determined by 454 pyrosequencing. PLoS ONE 2015, 10, e0136946. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Gu, T.; Zhang, W.; Shen, Q.; Yin, S.; Qiu, H. Microbial community diversities and taxa abundances in soils along a seven-year gradient of potato monoculture using high throughput pyrosequencing approach. PLoS ONE 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Tan, Y.; Cui, Y.; Li, H.; Kuang, A.; Li, X.; Wei, Y.; Ji, X. Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol. Res. 2017, 194, 10–19. [Google Scholar] [CrossRef]
- Zu, C.; Li, Z.; Yang, J.; Yu, H.; Sun, Y.; Tang, H.; Yost, R.; Wu, H. Acid soil is associated with reduced yield, root growth and nutrient uptake in black pepper. Agric. Sci. 2014, 05, 466–473. [Google Scholar]
- Ehlers, B.K. Soil microorganisms alleviate the allelochemical effects of a thyme monoterpene on the performance of an associated grass species. PLoS ONE 2011, 6, e26321. [Google Scholar] [CrossRef] [PubMed]
- Arafat, Y.; Tayyab, M.; Khan, M.U.; Chen, T.; Amjad, H.; Awais, S.; Lin, X.; Lin, W.; Lin, S. Long-term monoculture negatively regulates fungal community composition and abundance of tea orchards. Agronomy 2019, 9, 466. [Google Scholar] [CrossRef]
- Dong, L.; Xu, J.; Feng, G.; Li, X.; Chen, S. Soil bacterial and fungal community dynamics in relation to Panax notoginseng death rate in a continuous cropping system. Sci. Rep. 2016, 6, 31802. [Google Scholar] [CrossRef]
- Zhao, Q.; Xiong, W.; Xing, Y.; Sun, Y.; Lin, X.; Dong, Y. Long-term coffee monoculture alters soil chemical properties and microbial communities. Sci. Rep. 2018, 8, 6116. [Google Scholar] [CrossRef]
- Bai, L.; Sun, H.; Zhang, X.; Cai, B. Next-generation sequencing of root fungal communities in continuous cropping soybean. Chil. J. Agric. Res. 2018, 78, 528–538. [Google Scholar] [CrossRef]
- Maron, P.-A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, 1–13. [Google Scholar] [CrossRef]
- Singh, B.K.; Quince, C.; Macdonald, C.A.; Khachane, A.; Thomas, N.; Al-Soud, W.A.; Sørensen, S.J.; He, Z.; White, D.; Sinclair, A.; et al. Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ. Microbiol. 2014, 16, 2408–2420. [Google Scholar] [CrossRef]
- Bai, L.; Cui, J.; Jie, W.; Cai, B. Analysis of the community compositions of rhizosphere fungi in soybeans continuous cropping fields. Microbiol. Res. 2015, 180, 49–56. [Google Scholar] [CrossRef]
- Lu, L.; Yin, S.; Liu, X.; Zhang, W.; Gu, T.; Shen, Q.; Qiu, H. Fungal networks in yield-invigorating and -debilitating soils induced by prolonged potato monoculture. Soil Biol. Biochem. 2013, 65, 186–194. [Google Scholar] [CrossRef]
- Liu, J.; Wu, F.; Yang, Y. Effects of cinnamic acid on bacterial community diversity in rhizosphere soil of cucumber seedlings under salt stress. Agric. Sci. China 2010, 9, 266–274. [Google Scholar] [CrossRef]
- Buée, M.; Reich, M.; Murat, C.; Morin, E.; Nilsson, R.H.; Uroz, S.; Martin, F. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 2009, 184, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- de Scally, S.Z.; Makhalanyane, T.P.; Frossard, A.; Hogg, I.D.; Cowan, D.A. Antarctic microbial communities are functionally redundant, adapted and resistant to short term temperature perturbations. Soil Biol. Biochem. 2016, 103, 160–170. [Google Scholar] [CrossRef]
- Treseder, K.K.; Lennon, J.T. Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. 2015, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Kennedy, P.G.; Liew, F.J.; Schilling, J.S. Fungal endophytes as priority colonizers initiating wood decomposition. Funct. Ecol. 2017, 31, 407–418. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, Y.; Feng, K.; Li, S.; Wang, S.; Jin, D.; Zhang, Y.; Chen, H.; Yin, H.; Xu, M.; et al. The divergence between fungal and bacterial communities in seasonal and spatial variations of wastewater treatment plants. Sci. Total Environ. 2018, 628, 969–978. [Google Scholar] [CrossRef]
- Gladieux, P.; Byrnes, E.J.; Aguileta, G.; Fisher, M.; Billmyre, R.B.; Heitman, J.; Giraud, T. Epidemiology and Evolution of Fungal Pathogens in Plants and Animals. In Genetics and Evolution of Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2017; pp. 71–98. ISBN 9780127999425. [Google Scholar]
- Tayyab, M.; Islam, W.; Lee, C.G.; Pang, Z.; Khalil, F.; Lin, S.; Lin, W.; Zhang, H. Short-term effects of different organic amendments on soil fungal composition. Sustainability 2019, 11, 198. [Google Scholar] [CrossRef]
- Freedman, Z.; Zak, D.R. Soil bacterial communities are shaped by temporal and environmental filtering: Evidence from a long-term chronosequence. Environ. Microbiol. 2015, 17, 3208–3218. [Google Scholar] [CrossRef]
- Xu, H.J.; Wang, X.H.; Li, H.; Yao, H.Y.; Su, J.Q.; Zhu, Y.G. Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environ. Sci. Technol. 2014, 48, 9391–9399. [Google Scholar] [CrossRef]
- Xu, J.M.; Tang, C.; Chen, Z.L. The role of plant residues in pH change of acid soils differing in initial pH. Soil Biol. Biochem. 2006, 38, 709–719. [Google Scholar] [CrossRef]

| Seasons | Samples | TCN (g/kg) | TCP (g/kg) | TCK (g/kg) | TOM (g/kg) | pH |
|---|---|---|---|---|---|---|
| Winter | FF | 2.66 ± 0.33ab | 1.17 ± 0.04d | 21.95 ± 0.58c | 21.70 ± 0.64b | 4.88 ± 0.08b |
| Y1 | 0.99 ± 0.16c | 1.66 ± 0.03c | 27.88 ± 1.13b | 11.16 ± 1.45c | 5.84 ± 0.33a | |
| Y3 | 2.09 ± 0.26b | 2.82 ± 0.44b | 30.17 ± 1.03a | 21.62 ± 1.57b | 5.11 ± 0.32b | |
| Y5 | 3.27 ± 0.68a | 4.18 ± 0.23a | 9.27 ± 0.22d | 36.04 ± 3.33a | 4.49 ± 0.01c | |
| Summer | FF | 3.13 ± 0.91a | 1.47 ± 0.35c | 21.86 ± 2.10b | 21.44 ± 1.23b | 6.20 ± 0.21b |
| Y1 | 1.02 ± 0.28b | 1.97 ± 0.27bc | 31.21 ± 2.97a | 8.35 ± 1.12c | 6.48 ± 0.25a | |
| Y3 | 1.52 ± 0.78b | 2.35 ± 0.71b | 31.86 ± 1.71a | 19.23 ± 0.68b | 6.09 ± 0.13b | |
| Y5 | 2.68 ± 0.48a | 4.18 ± 0.38a | 9.30 ± 1.17c | 33.90 ± 4.49a | 5.59 ± 0.10c |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alami, M.M.; Xue, J.; Ma, Y.; Zhu, D.; Abbas, A.; Gong, Z.; Wang, X. Structure, Function, Diversity, and Composition of Fungal Communities in Rhizospheric Soil of Coptis chinensis Franch under a Successive Cropping System. Plants 2020, 9, 244. https://doi.org/10.3390/plants9020244
Alami MM, Xue J, Ma Y, Zhu D, Abbas A, Gong Z, Wang X. Structure, Function, Diversity, and Composition of Fungal Communities in Rhizospheric Soil of Coptis chinensis Franch under a Successive Cropping System. Plants. 2020; 9(2):244. https://doi.org/10.3390/plants9020244
Chicago/Turabian StyleAlami, Mohammad Murtaza, Jinqi Xue, Yutao Ma, Dengyan Zhu, Aqleem Abbas, Zedan Gong, and Xuekui Wang. 2020. "Structure, Function, Diversity, and Composition of Fungal Communities in Rhizospheric Soil of Coptis chinensis Franch under a Successive Cropping System" Plants 9, no. 2: 244. https://doi.org/10.3390/plants9020244
APA StyleAlami, M. M., Xue, J., Ma, Y., Zhu, D., Abbas, A., Gong, Z., & Wang, X. (2020). Structure, Function, Diversity, and Composition of Fungal Communities in Rhizospheric Soil of Coptis chinensis Franch under a Successive Cropping System. Plants, 9(2), 244. https://doi.org/10.3390/plants9020244







