Transcriptome-Wide Identification, Evolutionary Analysis, and GA Stress Response of the GRAS Gene Family in Panax ginseng C. A. Meyer
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of GRAS Domain Genes in the Whole Transcriptome
2.2. Conserved Motifs Analysis and Evolutionary Analysis of GRAS Genes
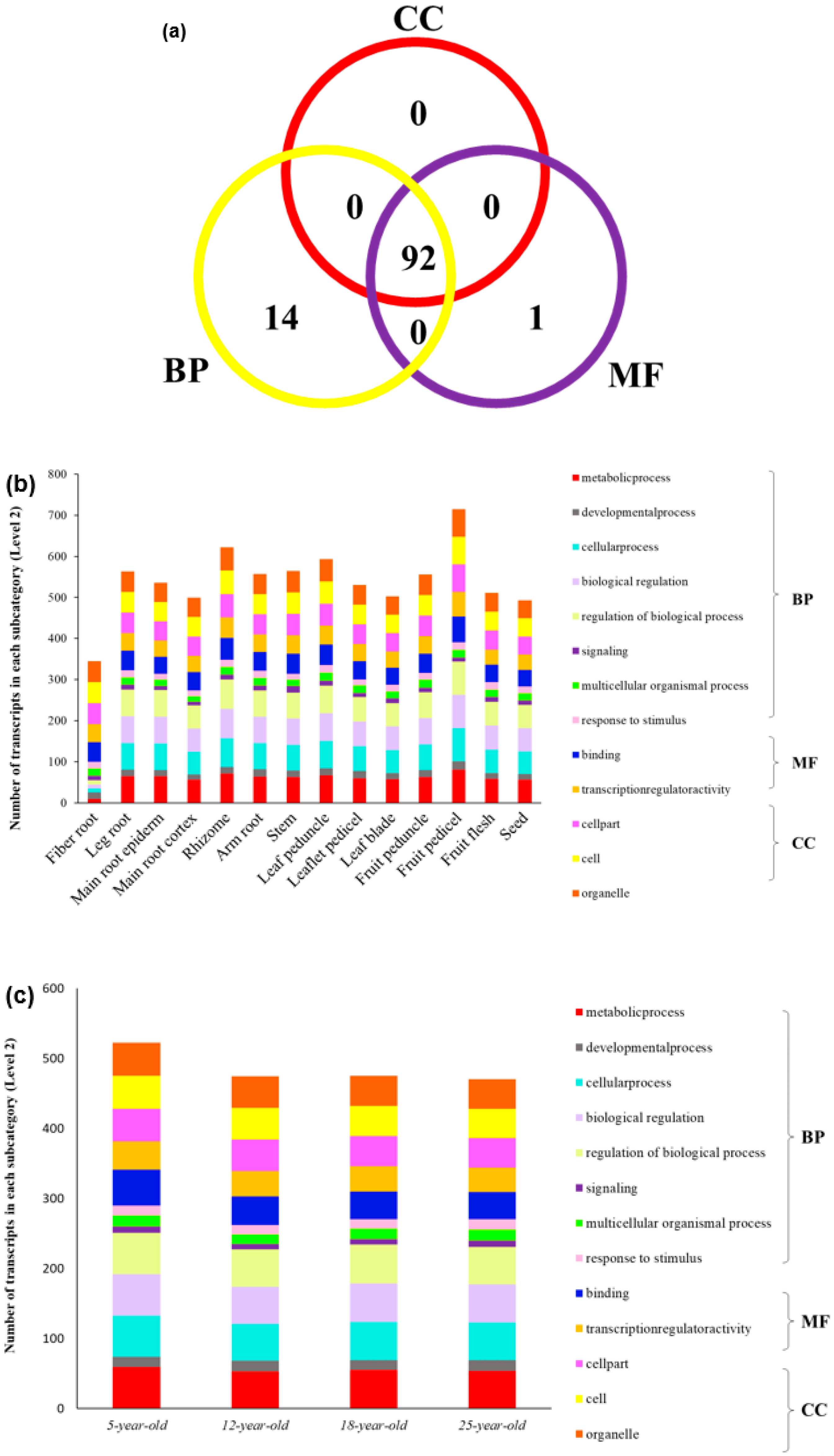
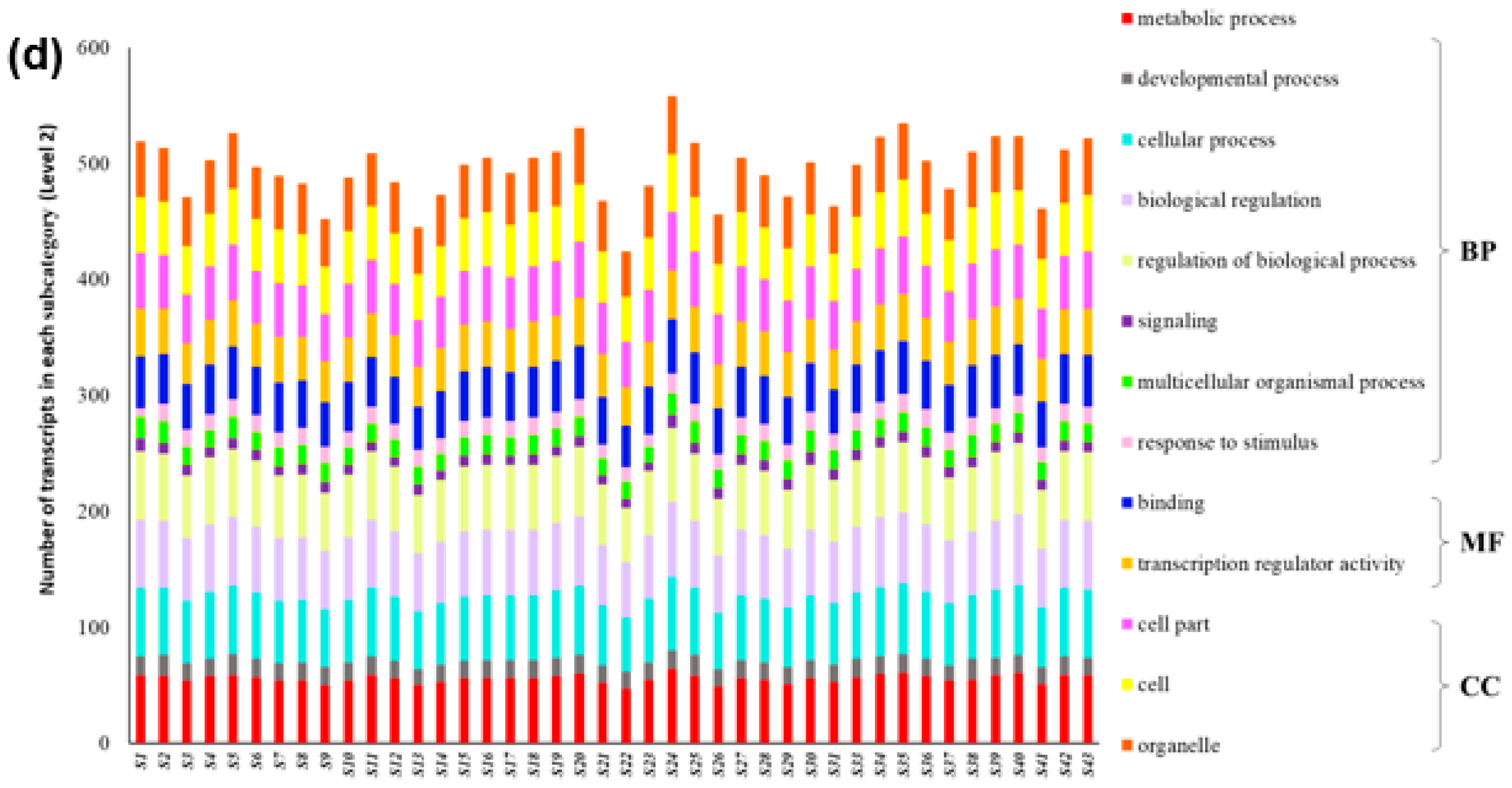
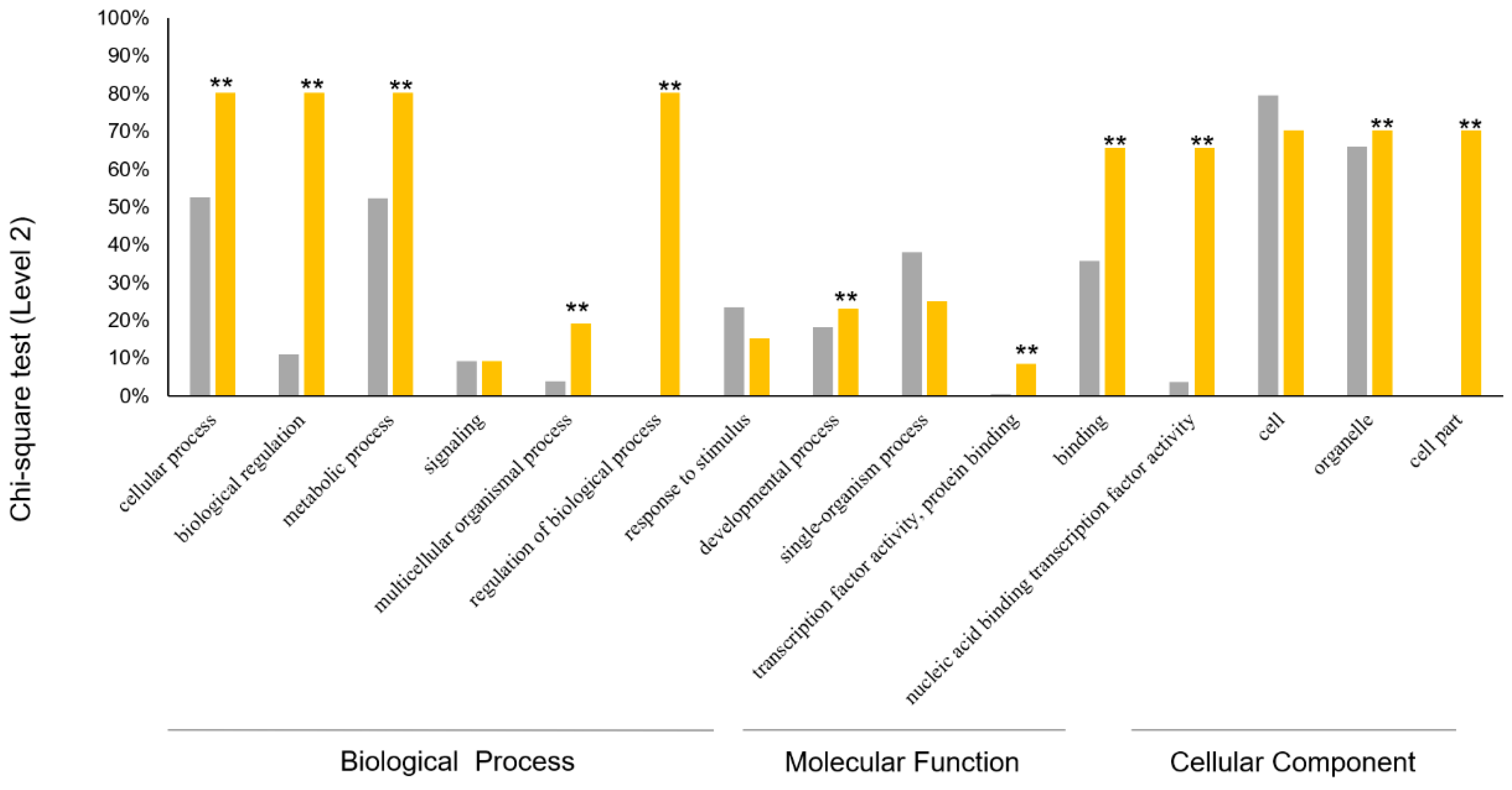
2.3. Gene Expression Pattern, GO Function Classification and Enrichment Analysis
2.4. Network Analysis of PgGRAS Transcripts Genes
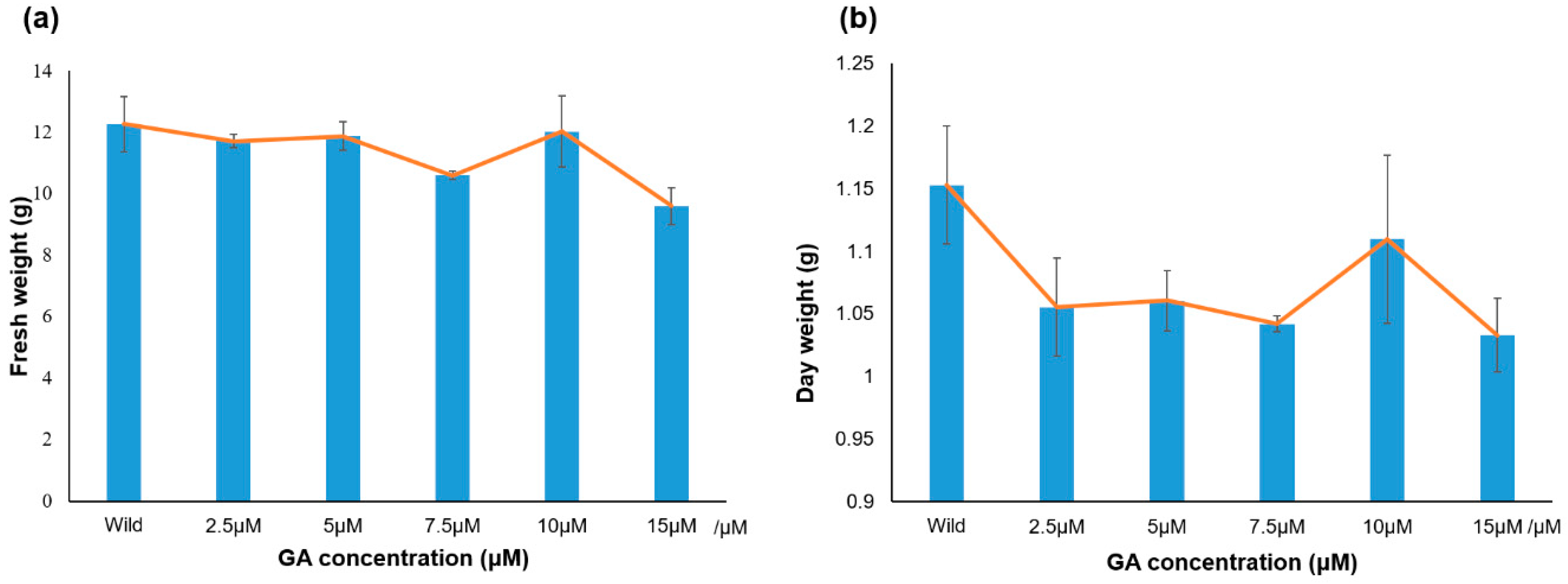
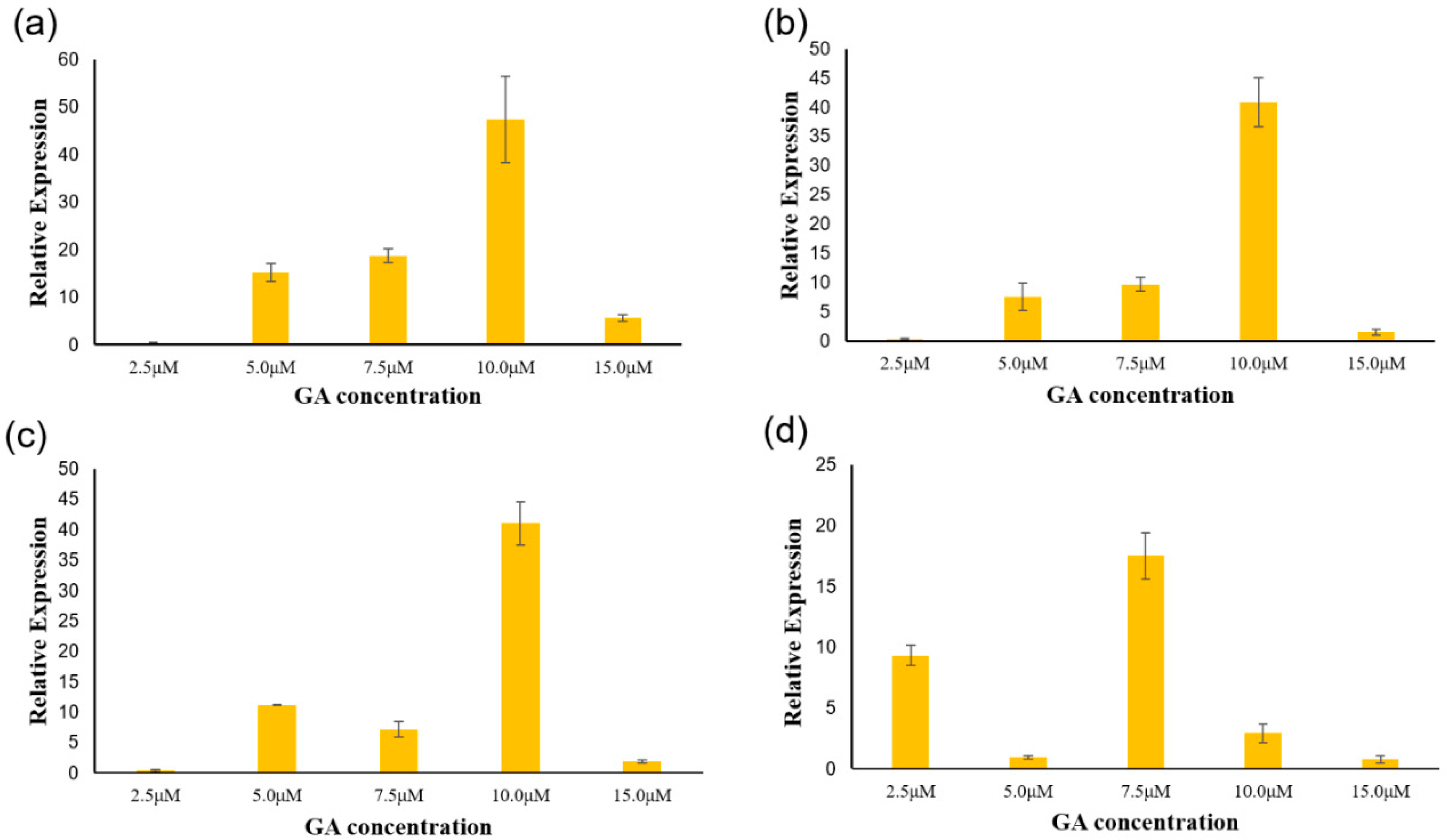
2.5. Response of PgGRAS Genes to Different Concentrations of GA
3. Results
3.1. Transcriptome-Wide Identification GRAS Gene Family in Ginseng
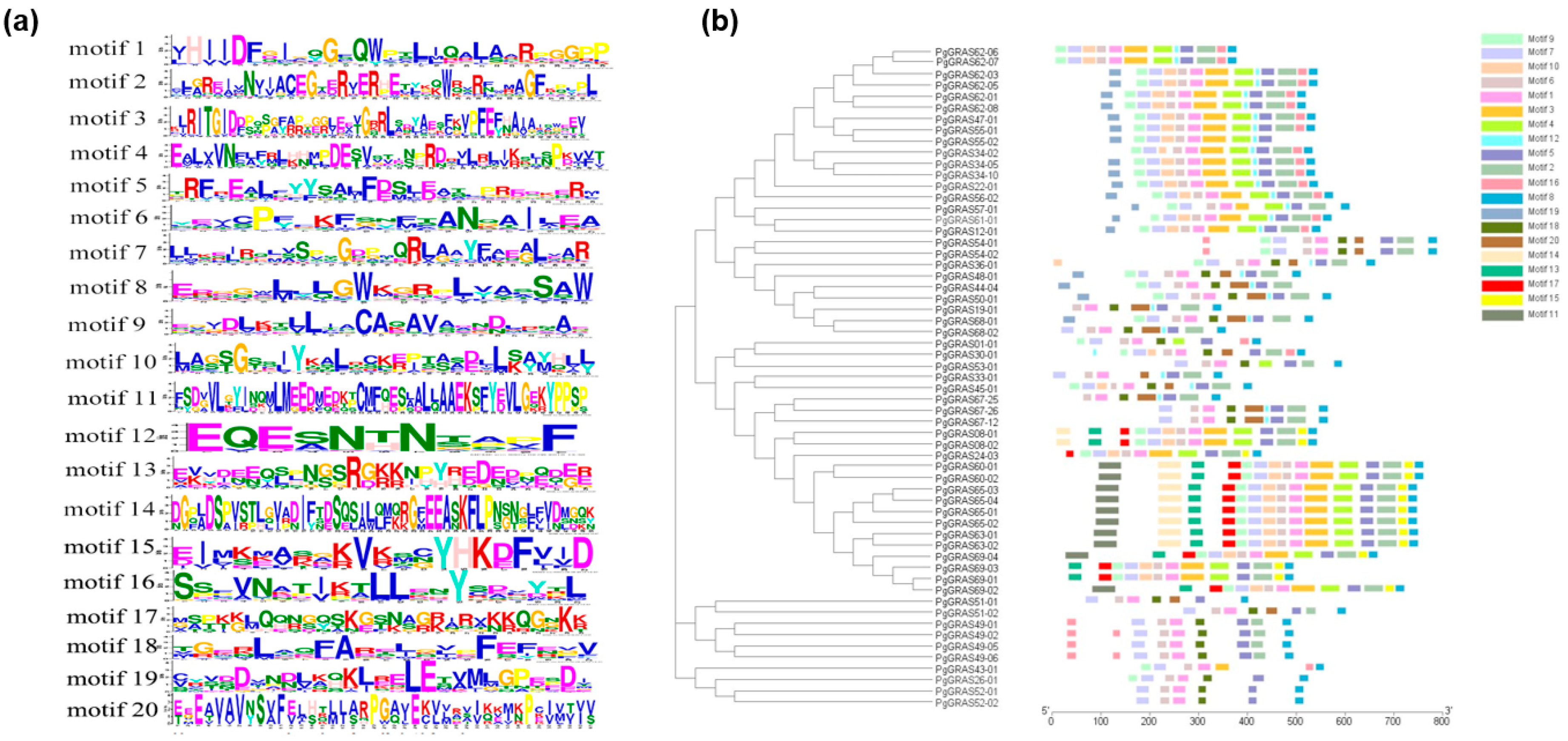
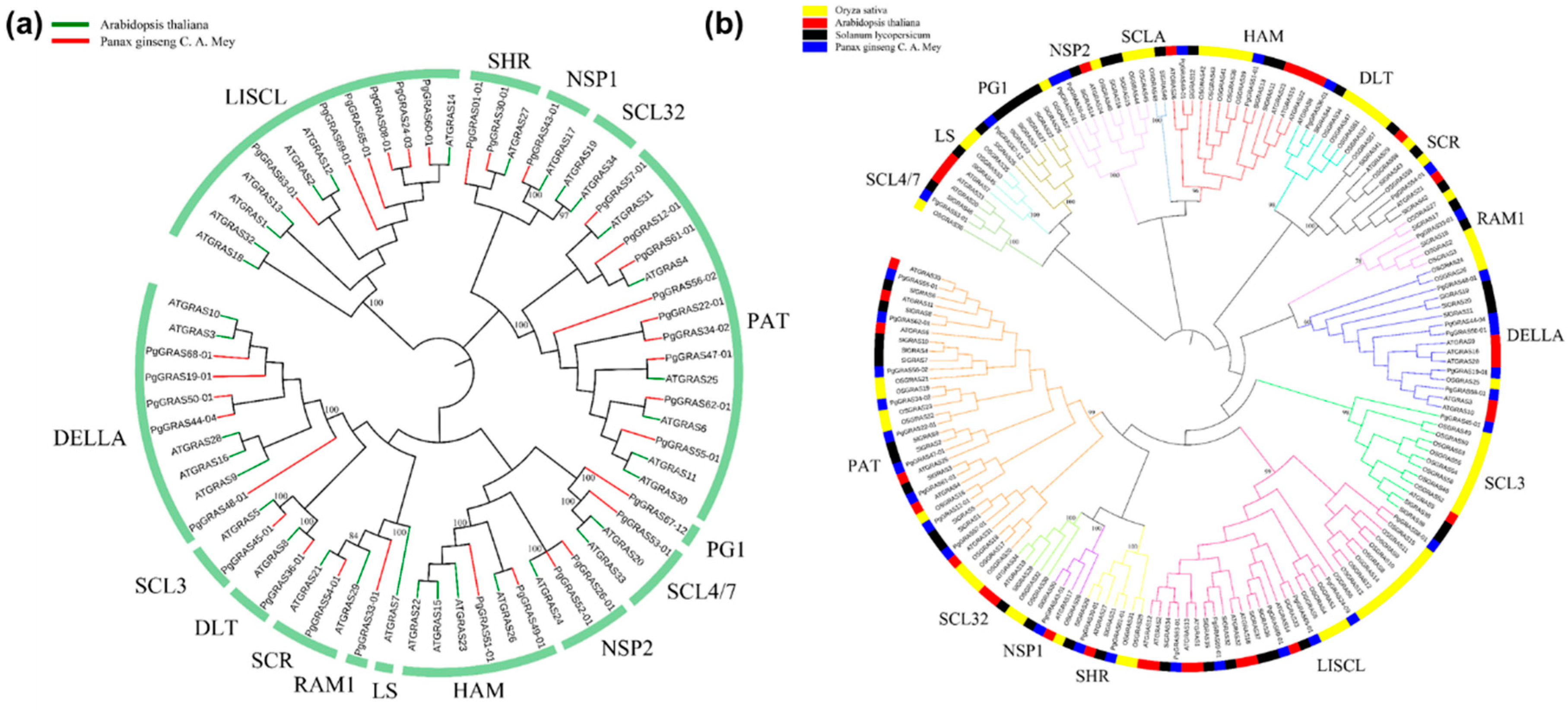
3.2. Conserved Motif Analysis and Systematic Analysis of PgGRAS Gene Family
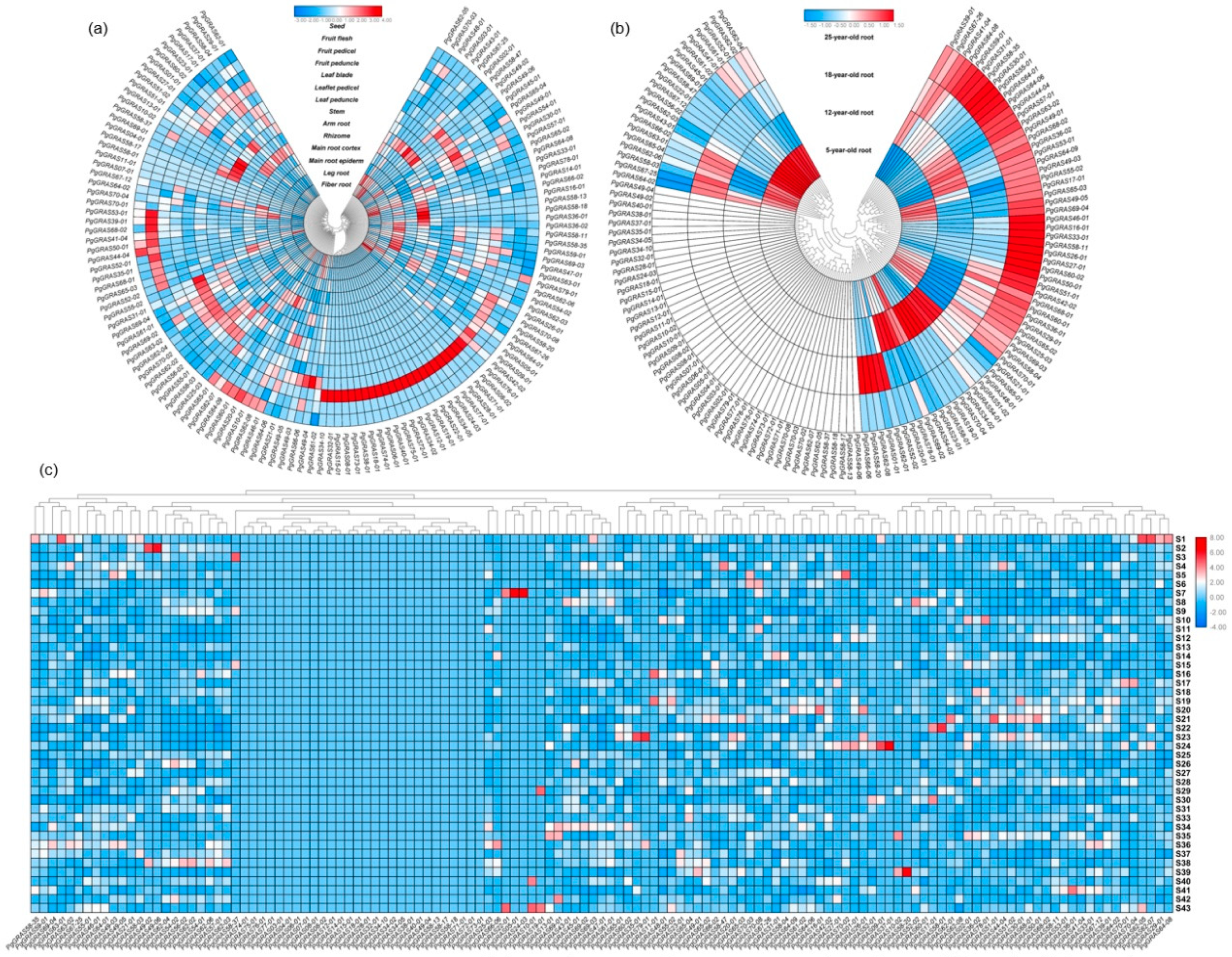
3.3. Network Analysis of Gene Expression in PgGRAS Gene Family
3.4. Expression Analysis and Functional Evolution of PgGRAS Gene Family
3.5. Analysis of PgGRAS Genes Expression under Different Concentrations of GA
4. Discussion
4.1. Function and Analysis of GRAS Transcription Factor Family
4.2. Response of DELLA Sub-Family to Gibberellin and Its Effect on Ginsenosides
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Additional Information
References
- Wang, J.; Gao, W.Y.; Zhang, J.; Zuo, B.M.; Zhang, L.M.; Huang, L.Q. Advances in study of ginsenoside biosynthesis pathway in Panax ginseng C. A. Meyer. Acta Physiol. Plant. 2012, 34, 397–403. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.; Yang, Z.E.; Chen, E.Y.; Zhang, C.J.; Zhang, X.Y.; Li, F.G. Genome-wide analysis of GRAS transcription factor gene family in Gossypium hirsutum L. BMC Genom. 2018, 19, 348. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.T.; Molinero, R.N.; Fariña, F.D.; Villena, D.M.; Garcíae, G.J. Identification and expression analysis of GRAS transcription factor genes involved in the control of Arbuscular mycorrhizal development in tomato. Front. Plant Sci. 2019, 10, 268. [Google Scholar]
- Liu, B.L.; Sun, Y.; Xue, J.A.; Li, R.Z. Genome-wide characterization and expression analysis of GRAS gene family in pepper (Capsicum annuum L.). PeerJ 2018, 6, e4796. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Jiang, J.J.; Ma, S.H.; Ye, N.H.; Jiang, M.; Cao, J.S.; Zhang, J.H. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Li, M.Y.; Liu, J.X.; Hao, J.N.; Feng, K.; Duan, A.Q.; Yang, Q.Q.; Xu, Z.S.; Xiong, A.S. Genomic identification of AP2/ERF transcription factors and functional characterization of two cold resistance-related AP2/ERF genes in celery (Apium graveolens L.). Planta 2019, 250, 1265–1280. [Google Scholar] [CrossRef]
- Cenci, A.; Rouard, M. Evolutionary analyses of GRAS transcription factors in angiosperms. Front. Plant Sci. 2017, 8, 273. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T.P. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Cao, Y.P.; Shang, C.; Li, J.K.; Wang, J.L.; Wu, Z.Y.; Ma, L.C.; Qi, T.X.; Fu, C.X.; Bai, Z.T.; et al. Genome-wide characterization of GRAS family genes in Medicago truncatula reveals their evolutionary dynamics and functional diversification. PLoS ONE 2017, 12, e0185439. [Google Scholar] [CrossRef] [PubMed]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Kim, B.; Song, S.K.; Heo, J.O.; Yu, N.; Lee, S.A.; Kim, M.; Kim, D.G.; Sohn, S.O.; Lim, C.E.; et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 2018, 67, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011, 77, 205–223. [Google Scholar] [CrossRef]
- Liu, X.Y.; Widmer, A. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and rice. Plant Mol. Biol. Report. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Bolle, C.; Koncz, C.; Chua, N.H. PAT1, a new member of the GRAS family, is involved in phytochrome a signal transduction. Genes Dev. 2000, 14, 1269–1278. [Google Scholar]
- Engstrom, E.M. Phylogenetic analysis of GRAS proteins from moss, lycophyte and vascular plant lineages reveals that GRAS genes arose and underwent substantial diversifification in the ancestral lineage common to bryophytes and vascular plants. Plant Signal. Behav. 2011, 6, 850–854. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Shi, S.L.; Zhou, Y.; Yang, J.; Tang, X.Q. Genome-wide identification and characterization of GRAS transcription factors in sacred lotus (Nelumbo nucifera). PeerJ 2016, 4, e2388. [Google Scholar] [CrossRef]
- Miyashima, S.; Hashimoto, T.; Nakajima, K. ARGONAUTE1 acts in Arabidopsis root radial pattern formation independently of the SHR/SCR pathway. Plant Cell Physiol. 2009, 50, 626–634. [Google Scholar] [CrossRef]
- Smit, P.; Raedts, J.; Portyanko, V.; Debellé, F.; Gough, C.; Bisseling, T.; Geurts, R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 2005, 308, 1789–1791. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.S.; Liang, D.; Shuai, P.; Xia, X.L.; Yin, W.L. The salt-and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2010, 61, 4011–4019. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.N.; Jin, Y.; Liu, W.B.; Li, F.; Fang, J.; Yin, Y.H.; Qian, Q.; Zhu, L.H.; Chu, C.C. DWARF ANDLOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Cui, H.T.; Buer, B.; Vijayakumar, V.; Delaux, P.M.; Junkermann, S.; Bucher, M. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 2015, 167, 854–871. [Google Scholar] [CrossRef] [PubMed]
- Heck, C.; Kuhn, H.; Heidt, S.; Walter, S.; Rieger, N.; Requena, N. Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 2016, 26, 2770–2778. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.; Jung, H.S.; Sun, T.P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 2001, 98, 14162–14167. [Google Scholar] [CrossRef]
- Zhang, D.P.; Iyer, L.M.; Aravind, L. Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 2012, 28, 2407–2411. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Human Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA 7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.H.; Lercher, M.J.; Hu, S.N.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Xia, R.; Chen, H.; He, Y.H. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25. [Google Scholar] [CrossRef]
- Theocharidis, A.; Van, D.S.; Enright, A.J.; Freeman, T.C. Network visualization and analysis of gene expression data using BioLayout Express (3D). Nat. Protoc. 2009, 4, 1535. [Google Scholar] [CrossRef]
- Jong, M.D.; Mariani, C.; Vriezen, W.H. The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 2009, 60, 1523–1532. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, X.; Cai, W.; Huang, W.; Zhou, X.; Luo, Q.; Yang, H.; Wang, J.; Huang, J. Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLoS Genet. 2014, 10, e1004519. [Google Scholar] [CrossRef]
- Li, L.; Wang, K.Y.; Zhao, M.Z.; Li, S.K.; Jiang, Y.; Zhu, L.; Chen, J.; Wang, Y.F.; Sun, C.Y.; Chen, P.; et al. Selection and validation of reference genes desirable for gene expression analysis by qRT-PCR in MeJA-treated ginseng hairy roots. PLoS ONE 2019, 14, e0226168. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Gonda, I.; Sun, H.H.; Ma, Q.Y.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.P.; Wu, Y.H.; Lee, M.K.; Liu, Y.H.; Rong, Y.; Santos, T.S.; Wu, C.C.; Xie, F.M.; Nelson, R.L.; Zhang, H.B. Numbers of genes in the NBS and RLK families vary by more than four-fold within a plant species and are regulated by multiple factors. Nucleic Acids Res. 2010, 38, 6513–6525. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chen, Z.X.; Ahmed, N.; Han, B.; Cui, Q.H.; Liu, A.Z. Genome-wide identification, evolutionary analysis, and stress responses of the GRAS gene family in castor beans. Int. J. Mol. Sci. 2016, 17, 1004. [Google Scholar] [CrossRef] [PubMed]
- Grimplet, J.; Agudelo-Romero, P.; Teixeira, R.T.; Martinez-Zapater, J.M.; Fortes, A.M. Structural and functional analysis of the GRAS gene family in grapevine indicates a role of GRAS proteins in the control of development and stress responses. Front. Plant Sci. 2016, 7, 353. [Google Scholar] [CrossRef]
- Gu, X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol. Biol. Evol. 2001, 18, 453–464. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Gao, C.; Zhao, M.; Wu, H.; Li, Y.; Shen, Y.; Han, M. Identification, classification, and expression analysis of GRAS gene family in Malus domestica. Front. Physiol. 2017, 8, 253. [Google Scholar] [CrossRef]
- Morohashi, K.; Minami, M.; Takase, H.; Hotta, Y.; Hiratsuka, K. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 2003, 278, 20865–20873. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome a signal transduction. Plant Physiol. 2013, 161, 291–304. [Google Scholar] [CrossRef]
- Li, W.; Wu, J.; Weng, S.; Zhang, Y.; Zhang, D.; Shi, C. Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta 2010, 232, 1383–1396. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Liu, J.; Lin, S.; Wang, J.; Lin, W.; Xu, W. GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation. Plant Cell Rep. 2017, 36, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Wu, H.Y.; Li, X.; Li, Q.; Zhao, X.Y.; Duan, X.Q.; An, Y.R.; Lv, W.; An, H.L. Identification and expression of GRAS family genes in maize (Zea mays L.). PLoS ONE 2017, 12, e0185418. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Zhao, M.Z.; Wang, K.Y.; Lin, Y.P.; Wang, Y.F.; Sun, C.Y.; Wang, Y.; Zhang, M.P. Functional differentiation and spatial-temporal co-expression networks of the NBS-encoding gene family in Jilin ginseng, Panax ginseng C. A. Meyer. PLoS ONE 2017, 12, e0181596. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.Y.; Jiang, S.C.; Sun, C.Y.; Lin, Y.P.; Yin, R.; Wang, Y.; Zhang, M.P. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C. A. Meyer. Sci. Rep. 2015, 5, 18283. [Google Scholar] [CrossRef]
- Lin, Y.P.; Wang, K.Y.; Li, X.Y.; Sun, C.Y.; Yin, R.; Wang, Y.F.; Wang, Y.; Zhang, M.P. Evolution, functional differentiation, and co-expression of the RLK gene family revealed in Jilin ginseng, Panax ginseng C. A. Meyer. Mol. Genet. Genom. 2018, 293, 845–859. [Google Scholar] [CrossRef]
- Itoh, H.; Ueguchi-Tanaka, M.; Sato, Y.; Ashikari, M.; Matsuoka, M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 2002, 14, 57–70. [Google Scholar] [CrossRef]
- Sun, T.P.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef]
- Zeng, X.; Ling, H.; Chen, X.M.; Guo, S.X. Genome-wide identification, phylogeny and function analysis of GRAS gene family in Dendrobium catenatum (Orchidaceae). Gene 2019, 705, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Nguyen, K.T.; Park, E.; Jeon, J.S.; Choi, G. DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 2013, 25, 927–943. [Google Scholar] [CrossRef]
- Tyler, L.; Thomas, S.G.; Hu, J.H.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.P. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]

| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| PgGRAS44-04 | GGCAATAGAAATGGGAATAAGCGAA | CTCTTGAGAATCGACAAGTACCAAC |
| PgGRAS48-01 | TCGGACCTCAGTTACGTTGAC | TTCCTCCATCGCGGTTACAAC |
| PgGRAS50-01 | CTGCACAAATATAGCACCACCG | ATTACCCCTTCCGACCAGAATG |
| PgGRAS68-01 | GTGGAGGAACGGGTGATTGA | TCCGGCAGAGTCGTCTACTA |
| GADPH | GAGAAGGAATACACACCTGACC | CAGTAGTCATAAGCCCCTCAAC |
| Name | Transcriptome ID | Nucleic Acid Length (bp) | Amino Acid Length | Molecular Weight (Da) | PI | Average of Hydropathicity |
|---|---|---|---|---|---|---|
| PgGRAS01-01 | comp10366_c0_seq1 | 1634 | 431 | 431 | 5.45 | −0.401 |
| PgGRAS02-01 | comp1049478_c0_seq1 | 338 | 34 | 3869.35 | 4.37 | −0.2 |
| PgGRAS03-01 | comp1105291_c0_seq1 | 259 | 51 | 5983 | 11.01 | −0.282 |
| PgGRAS04-01 | comp1121404_c0_seq1 | 312 | 85 | 9609.16 | 6.57 | −0.085 |
| PgGRAS05-01 | comp1387617_c0_seq1 | 239 | 58 | 6767.62 | 4.87 | −0.634 |
| PgGRAS06-01 | comp1596844_c0_seq1 | 258 | 78 | 8434.46 | 5.41 | −0.145 |
| PgGRAS07-01 | comp17248_c0_seq1 | 210 | 68 | 7836.98 | 10.03 | −0.35 |
| PgGRAS08-01 | comp17356_c0_seq1 | 2228 | 551 | 61,673.55 | 8.84 | −0.377 |
| PgGRAS08-02 | comp17356_c0_seq2 | 2145 | 551 | 61,673.55 | 8.84 | −0.377 |
| PgGRAS09-01 | comp1839771_c0_seq1 | 282 | 89 | 10,316.55 | 4.69 | −0.321 |
| PgGRAS10-01 | comp19390_c0_seq1 | 258 | 54 | 6011.81 | 6.16 | −0.369 |
| PgGRAS10-02 | comp19390_c0_seq2 | 221 | 39 | 4382.96 | 6.54 | −0.292 |
| PgGRAS11-01 | comp2100322_c0_seq1 | 251 | 78 | 8584.94 | 4.6 | 0.306 |
| PgGRAS12-01 | comp212859_c0_seq1 | 2198 | 552 | 61,408.99 | 4.73 | −0.291 |
| PgGRAS13-01 | comp22569_c0_seq1 | 1042 | 279 | 32,057.2 | 9.06 | −0.142 |
| PgGRAS14-01 | comp2261495_c0_seq1 | 210 | 55 | 5954.67 | 5.22 | −0.255 |
| PgGRAS15-01 | comp23356_c0_seq1 | 201 | 30 | 3357.14 | 10.31 | 0.023 |
| PgGRAS16-01 | comp25708_c0_seq1 | 865 | 266 | 29,992.15 | 5.74 | −0.197 |
| PgGRAS17-01 | comp26247_c0_seq1 | 694 | 200 | 22,426.1 | 4.88 | 0.31 |
| PgGRAS18-01 | comp2660386_c0_seq1 | 202 | 39 | 4228.04 | 7.98 | 0.667 |
| PgGRAS19-01 | comp267650_c0_seq1 | 1725 | 353 | 38,544.56 | 5.69 | −0.17 |
| PgGRAS20-01 | comp33784_c0_seq1 | 240 | 74 | 8432.86 | 8.8 | −0.15 |
| PgGRAS21-01 | comp37210_c0_seq1 | 692 | 120 | 14,029.28 | 9.16 | −0.512 |
| PgGRAS22-01 | comp38081_c0_seq1 | 1876 | 547 | 60,343.12 | 5.89 | −0.362 |
| PgGRAS23-01 | comp40091_c0_seq1 | 956 | 229 | 24,803.16 | 4.85 | 0.083 |
| PgGRAS24-03 | comp418104_c0_seq1 | 1633 | 437 | 49,927.34 | 9.22 | −0.45 |
| PgGRAS25-03 | comp44037_c0_seq3 | 782 | 72 | 8236.58 | 4.44 | 0.321 |
| PgGRAS26-01 | comp46234_c0_seq1 | 1723 | 527 | 60,102.48 | 4.61 | −0.422 |
| PgGRAS27-01 | comp472334_c0_seq1 | 733 | 218 | 24,669.44 | 4.52 | 0.139 |
| PgGRAS28-01 | comp479040_c0_seq1 | 578 | 172 | 19,639.29 | 9.01 | −0.61 |
| PgGRAS29-01 | comp50021_c0_seq1 | 652 | 129 | 14,932.18 | 6.19 | −0.156 |
| PgGRAS30-01 | comp51501_c0_seq1 | 2097 | 523 | 59,299.2 | 5.99 | −0.48 |
| PgGRAS31-01 | comp52182_c0_seq1 | 1105 | 244 | 26,552.45 | 4.9 | −0.454 |
| PgGRAS32-01 | comp522134_c0_seq1 | 756 | 236 | 25,687.72 | 6.37 | 0.198 |
| PgGRAS33-01 | comp53859_c0_seq1 | 1411 | 358 | 59,615.92 | 5.74 | −0.31 |
| PgGRAS34-10 | comp53978_c4_seq10 | 2272 | 541 | 59,954.61 | 5.71 | −0.249 |
| PgGRAS34-02 | comp53978_c4_seq2 | 2346 | 536 | 40,253.29 | 6.61 | −0.134 |
| PgGRAS34-05 | comp53978_c4_seq5 | 2194 | 541 | 59,615.92 | 5.74 | −0.31 |
| PgGRAS35-01 | comp546436_c0_seq1 | 540 | 109 | 12,690.7 | 8.46 | −0.376 |
| PgGRAS36-01 | comp54894_c0_seq1 | 2441 | 674 | 74,618.25 | 5.84 | −0.396 |
| PgGRAS36-02 | comp54894_c0_seq2 | 915 | 198 | 22,784.94 | −0.339 | |
| PgGRAS37-01 | comp564672_c0_seq1 | 400 | 107 | 12,078.8 | 4.73 | −0.128 |
| PgGRAS38-01 | comp572133_c0_seq1 | 458 | 141 | 15,201.03 | 5.07 | −0.227 |
| PgGRAS39-01 | comp57286_c0_seq1 | 704 | 148 | 15,923.56 | 4.53 | −0.376 |
| PgGRAS40-01 | comp578948_c0_seq1 | 309 | 91 | 10,079.48 | 11.16 | −0.552 |
| PgGRAS41-04 | comp59054_c0_seq4 | 930 | 168 | 19,216.92 | 5.11 | −0.066 |
| PgGRAS42-02 | comp59054_c1_seq2 | 345 | 76 | 8355.65 | 8.19 | −0.095 |
| PgGRAS43-01 | comp59228_c0_seq1 | 2069 | 564 | 62,183.25 | 5.99 | −0.521 |
| PgGRAS44-04 | comp59348_c0_seq4 | 1635 | 532 | 58,791.83 | 4.97 | −0.15 |
| PgGRAS45-01 | comp59447_c0_seq1 | 1622 | 413 | 45,985.08 | 7.7 | −0.094 |
| PgGRAS46-01 | comp61562_c0_seq1 | 2110 | 477 | 53,963.14 | 6.05 | −0.161 |
| PgGRAS47-01 | comp61661_c3_seq1 | 2420 | 542 | 60,731.41 | 5.54 | −0.335 |
| PgGRAS48-01 | comp62080_c0_seq1 | 2058 | 520 | 57,513.1 | 5.27 | −0.128 |
| PgGRAS49-01 | comp62792_c0_seq1 | 2000 | 493 | 55,553.44 | 5.91 | −0.126 |
| PgGRAS49-02 | comp62792_c0_seq2 | 2015 | 498 | 55,978.88 | 5.95 | −0.114 |
| PgGRAS49-03 | comp62792_c0_seq3 | 1385 | 375 | 42,219.01 | 5.37 | −0.177 |
| PgGRAS49-04 | comp62792_c0_seq4 | 1373 | 370 | 41,793.58 | 5.37 | −0.194 |
| PgGRAS49-05 | comp62792_c0_seq5 | 1891 | 493 | 55,612.53 | 5.84 | −0.129 |
| PgGRAS49-06 | comp62792_c0_seq6 | 1906 | 498 | 56,037.97 | 5.87 | −0.117 |
| PgGRAS50-01 | comp62989_c0_seq1 | 2491 | 580 | 63,896.36 | 5.04 | −0.243 |
| PgGRAS51-01 | comp63201_c1_seq1 | 1875 | 408 | 46,130.96 | 6 | −0.103 |
| PgGRAS51-02 | comp63201_c1_seq2 | 2487 | 608 | 67,328.55 | 6.14 | −0.225 |
| PgGRAS52-01 | comp63447_c0_seq1 | 1761 | 518 | 58,723.53 | 4.6 | −0.275 |
| PgGRAS52-02 | comp63447_c0_seq2 | 1747 | 518 | 58,723.53 | 4.6 | −0.275 |
| PgGRAS53-01 | comp63847_c0_seq1 | 2308 | 598 | 65,887.92 | 4.88 | −0.277 |
| PgGRAS54-01 | comp64175_c2_seq1 | 3053 | 798 | 86,476.95 | 5.92 | −0.284 |
| PgGRAS54-02 | comp64175_c2_seq2 | 3085 | 795 | 86,246.77 | 6 | −0.289 |
| PgGRAS55-01 | comp64629_c0_seq1 | 2043 | 541 | 60,690.52 | 6 | −0.343 |
| PgGRAS55-02 | comp64629_c0_seq2 | 1746 | 470 | 52,338.84 | 5.57 | −0.339 |
| PgGRAS56-02 | comp64837_c0_seq2 | 3009 | 578 | 64,507.79 | 5.87 | −0.421 |
| PgGRAS57-01 | comp65004_c0_seq1 | 2924 | 612 | 66,846.76 | 6.69 | −0.294 |
| PgGRAS58-01 | comp65321_c0_seq1 | 916 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-11 | comp65321_c0_seq11 | 1094 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-13 | comp65321_c0_seq13 | 1958 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-17 | comp65321_c0_seq17 | 1254 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-18 | comp65321_c0_seq18 | 1760 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-20 | comp65321_c0_seq20 | 1087 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-03 | comp65321_c0_seq3 | 928 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-35 | comp65321_c0_seq35 | 1123 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-37 | comp65321_c0_seq37 | 1109 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-04 | comp65321_c0_seq4 | 1009 | 143 | 16,880.14 | 6.51 | −0.483 |
| PgGRAS58-47 | comp65321_c0_seq47 | 449 | 123 | 14,595.54 | 5.93 | −0.464 |
| PgGRAS59-01 | comp65321_c2_seq1 | 435 | 131 | 15,178.62 | 9.23 | −0.41 |
| PgGRAS60-01 | comp65331_c0_seq1 | 2630 | 765 | 87,107.53 | 6.06 | −0.462 |
| PgGRAS60-02 | comp65331_c0_seq2 | 2791 | 765 | 87,107.53 | 6.06 | −0.462 |
| PgGRAS61-01 | comp65403_c0_seq1 | 2066 | 575 | 64,422.68 | 5.09 | −0.347 |
| PgGRAS61-02 | comp65403_c0_seq2 | 1021 | 294 | 33,240.21 | 6.03 | −0.14 |
| PgGRAS62-01 | comp65479_c0_seq1 | 2282 | 522 | 57,819.13 | 5.79 | −0.053 |
| PgGRAS62-02 | comp65479_c0_seq2 | 2265 | 443 | 48,548.35 | 5.63 | −0.03 |
| PgGRAS62-03 | comp65479_c0_seq3 | 3176 | 546 | 60,663.43 | 5.11 | −0.229 |
| PgGRAS62-04 | comp65479_c0_seq4 | 2137 | 443 | 48,548.35 | 5.63 | −0.03 |
| PgGRAS62-05 | comp65479_c0_seq5 | 3135 | 546 | 60,663.43 | 5.11 | −0.229 |
| PgGRAS62-06 | comp65479_c0_seq6 | 3025 | 380 | 42,404.63 | 5.49 | −0.03 |
| PgGRAS62-07 | comp65479_c0_seq7 | 2984 | 380 | 42,404.63 | 5.49 | −0.03 |
| PgGRAS62-08 | comp65479_c0_seq8 | 2154 | 522 | 57,819.13 | 5.79 | −0.053 |
| PgGRAS63-01 | comp66169_c0_seq1 | 3313 | 754 | 84,839.24 | 5.61 | −0.446 |
| PgGRAS63-02 | comp66169_c0_seq2 | 2999 | 754 | 84,839.24 | 5.16 | −0.446 |
| PgGRAS64-01 | comp66380_c0_seq1 | 2230 | 470 | 52,786.49 | 5.91 | −0.185 |
| PgGRAS64-02 | comp66380_c0_seq2 | 2291 | 465 | 52,285.89 | 5.94 | −0.221 |
| PgGRAS64-06 | comp66380_c0_seq6 | 2305 | 465 | 52,285.89 | 5.94 | −0.221 |
| PgGRAS64-08 | comp66380_c0_seq8 | 1013 | 317 | 35,871.94 | 6.17 | −0.38 |
| PgGRAS64-09 | comp66380_c0_seq9 | 2259 | 470 | 52,786.49 | 5.91 | −0.185 |
| PgGRAS65-01 | comp67093_c0_seq1 | 2855 | 753 | 84,264.99 | 5.32 | −0.437 |
| PgGRAS65-02 | comp67093_c0_seq2 | 3343 | 753 | 84,264.99 | 5.32 | −0.437 |
| PgGRAS65-03 | comp67093_c0_seq3 | 2904 | 753 | 84,264.99 | 5.32 | −0.437 |
| PgGRAS65-04 | comp67093_c0_seq4 | 3054 | 753 | 84,264.99 | 5.32 | −0.437 |
| PgGRAS66-02 | comp67249_c0_seq2 | 498 | 75 | 8053.39 | 9.22 | 0.505 |
| PgGRAS66-06 | comp67249_c0_seq6 | 484 | 50 | 5415.45 | 9.4 | 0.684 |
| PgGRAS67-12 | comp67428_c0_seq12 | 1917 | 569 | 63,367.08 | 5.03 | −0.123 |
| PgGRAS67-25 | comp67428_c0_seq25 | 1299 | 349 | 39,076.4 | 6.35 | 0.106 |
| PgGRAS67-26 | comp67428_c0_seq26 | 1891 | 569 | 63,367.08 | 5.04 | −0.123 |
| PgGRAS68-01 | comp67501_c0_seq1 | 2041 | 540 | 59,906.01 | 5.03 | −0.188 |
| PgGRAS68-02 | comp67501_c0_seq2 | 1733 | 361 | 39,883.54 | 5.83 | −0.057 |
| PgGRAS69-01 | comp67516_c0_seq2 | 2051 | 499 | 58,454.58 | 8.26 | −0.651 |
| PgGRAS69-02 | comp67516_c0_seq3 | 2633 | 726 | 83,814.41 | 5.91 | −0.623 |
| PgGRAS69-03 | comp67516_c0_seq4 | 2099 | 499 | 58,454.58 | 8.26 | −0.651 |
| PgGRAS69-04 | comp67516_c0_seq6 | 2538 | 671 | 77,840.89 | 6.02 | −0.652 |
| PgGRAS70-01 | comp67518_c0_seq1 | 1172 | 123 | 14,161.05 | 6.28 | −0.407 |
| PgGRAS70-02 | comp67518_c0_seq2 | 1081 | 129 | 15,078.18 | 7.63 | −0.416 |
| PgGRAS70-03 | comp67518_c0_seq3 | 1415 | 123 | 14,161.05 | 6.28 | −0.407 |
| PgGRAS70-04 | comp67518_c0_seq4 | 1055 | 129 | 15,078.18 | 7.63 | −0.416 |
| PgGRAS70-08 | comp67518_c0_seq8 | 1298 | 129 | 15,078.18 | 7.63 | −0.416 |
| PgGRAS71-01 | comp708716_c0_seq1 | 492 | 86 | 9590.63 | 5.49 | −0.316 |
| PgGRAS72-01 | comp726689_c0_seq1 | 415 | 119 | 12,725.35 | 5.31 | −0.224 |
| PgGRAS73-01 | comp753553_c0_seq1 | 363 | 78 | 7635.45 | 6.04 | 0.026 |
| PgGRAS74-01 | comp762664_c0_seq1 | 618 | 192 | 23,111.62 | 8.81 | −0.505 |
| PgGRAS75-01 | comp774347_c0_seq1 | 439 | 60 | 7033.28 | 11.22 | −0.522 |
| PgGRAS76-01 | comp866028_c0_seq1 | 430 | 68 | 7436.13 | 6.93 | −0.503 |
| PgGRAS77-01 | comp876245_c0_seq1 | 1036 | 209 | 23,803.37 | 6.09 | −0.293 |
| PgGRAS78-01 | comp913576_c0_seq1 | 443 | 58 | 6237.18 | 5.4 | −0.084 |
| PgGRAS79-01 | comp933760_c0_seq1 | 420 | 132 | 14,868.61 | 5.93 | −0.413 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Wang, K.; Li, S.; Jiang, Y.; Li, L.; Zhao, M.; Jiang, Y.; Zhu, L.; Wang, Y.; Su, Y.; et al. Transcriptome-Wide Identification, Evolutionary Analysis, and GA Stress Response of the GRAS Gene Family in Panax ginseng C. A. Meyer. Plants 2020, 9, 190. https://doi.org/10.3390/plants9020190
Wang N, Wang K, Li S, Jiang Y, Li L, Zhao M, Jiang Y, Zhu L, Wang Y, Su Y, et al. Transcriptome-Wide Identification, Evolutionary Analysis, and GA Stress Response of the GRAS Gene Family in Panax ginseng C. A. Meyer. Plants. 2020; 9(2):190. https://doi.org/10.3390/plants9020190
Chicago/Turabian StyleWang, Nan, Kangyu Wang, Shaokun Li, Yang Jiang, Li Li, Mingzhu Zhao, Yue Jiang, Lei Zhu, Yanfang Wang, Yingjie Su, and et al. 2020. "Transcriptome-Wide Identification, Evolutionary Analysis, and GA Stress Response of the GRAS Gene Family in Panax ginseng C. A. Meyer" Plants 9, no. 2: 190. https://doi.org/10.3390/plants9020190
APA StyleWang, N., Wang, K., Li, S., Jiang, Y., Li, L., Zhao, M., Jiang, Y., Zhu, L., Wang, Y., Su, Y., Wang, Y., & Zhang, M. (2020). Transcriptome-Wide Identification, Evolutionary Analysis, and GA Stress Response of the GRAS Gene Family in Panax ginseng C. A. Meyer. Plants, 9(2), 190. https://doi.org/10.3390/plants9020190




