Abstract
The genus Chlorophytum includes many economically important species well-known for medicinal, ornamental, and horticultural values. However, to date, few molecular genomic resources have been reported for this genus. Therefore, there is limited knowledge of phylogenetic studies, and the available chloroplast (cp) genome of Chlorophytum (C. rhizopendulum) does not provide enough information on this genus. In this study, we present genomic resources for C. comosum and C. gallabatense, which had lengths of 154,248 and 154,154 base pairs (bp), respectively. They had a pair of inverted repeats (IRa and IRb) of 26,114 and 26,254 bp each in size, separating the large single-copy (LSC) region of 84,004 and 83,686 bp from the small single-copy (SSC) region of 18,016 and 17,960 bp in C. comosum and C. gallabatense, respectively. There were 112 distinct genes in each cp genome, which were comprised of 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. The comparative analysis with five other selected species displayed a generally high level of sequence resemblance in structural organization, gene content, and arrangement. Additionally, the phylogenetic analysis confirmed the previous phylogeny and produced a phylogenetic tree with similar topology. It showed that the Chlorophytum species (C. comosum, C. gallabatense and C. rhizopendulum) were clustered together in the same clade with a closer relationship than other plants to the Anthericum ramosum. This research, therefore, presents valuable records for further molecular evolutionary and phylogenetic studies which help to fill the gap in genomic resources and resolve the taxonomic complexes of the genus.
1. Introduction
Chlorophytum Ker-Gawl. is a large and taxonomically complex genus [1], distributed in the Old World tropics, particularly, in Africa (possibly the center of origin), Australia, Asia, Madagascar, and India [2,3], that comprises about 215 species, six subspecies, and 14 varieties [4,5] in the subfamily Agavoideae in the Asparagaceae family [6]. They are perennial rhizomatous herbs, commonly known as spider plants, based on the mosaic form of C. comosum leaves [7]. The genus is well-known for its medicinal characteristics [8]. Therefore, it has attracted the attention of scientists due to pharmacologically relevant steroidal and triterpenoidal saponin contents in their roots [9]. Other pharmacological activities such as aphrodisiac, anticancer, larvicidal, and immunomodulatory were also mentioned by Deore et al., 2008, and Khanam et al., 2013, [10,11]. C. comosum is used to treat burns, bronchitis, fractures [12], and also has antitumor activity [13]. There are several well-documented studies on phytochemistry and the medicinal properties of different species in this genus [14]. The leaves of C. borivilianum and C. breviscapum are used as a vegetable [15]. Additionally, some of the species are cultivated for their ornamental values such as C. comosum and C. filipendulum subsp. amaniense, and are also effective in controlling soil erosion [1].
The increased economic values of the Chlorophytum species have contributed to unregulated harvesting and habitat degradation. In addition, poor germination and regeneration rates have resulted in the scarcity of these plants. This has culminated in some species being listed as vulnerable, threatened, or critically endangered. C. rhizopendulum is one of the critically endangered species according to the International Union for Conservation of Nature (IUCN) Red List (www.iucnredlist.org). Moreover, this genus is experiencing difficulties in classification, and different taxonomists have misidentified the species when using morphological features [16]. C. comosum and C. gallabatense were reported to have these delimitation problems [17]. This problem of species delimitation has not been solved, and it is highly desirable to find other methods to authenticate the species of Chlorophytum. Phylogenetic analysis based on the shared protein-coding genes or complete (cp) genomes could solve these problems; however, the Chlorophytum’s complete (cp) genomic data is very insufficient. To date, Chlorophytum genomic data is underrepresented in literature, only one complete chloroplast genome (C. rhizopendulum) has been published in this genus [18], regardless of the chloroplast being illustrated as important for plant genetic molecular studies [19]. Consequently, this limits further phylogenetic analysis of this genus.
Chloroplasts are important organelles in plants, where carbon fixation, photosynthesis, and pigment synthesis are carried out [20,21], and therefore they are the main plastid for green plants [22]. The cp is an independent genome (chloroplast DNA) with a preserved, circular structure and a low molecular weight [23]. It has a typical quadripartite structure, consisting of a pair of inverted repeat regions (IRs) of equal size, separating a large single copy (LSC) region from a small single copy (SSC) region [24]. The plastid size ranges from 107 kb to 218 kb [25,26]. It contains approximately 110 to 130 genes encoding ribosomal RNA (rRNA) genes, protein-coding genes (PCGs), and transfer RNAs genes (tRNAs) [27]. Chloroplast genomes are easier and efficient to use for evolutionary studies because of their self-replication and slow evolutionary process that is relatively independent [28,29]. Recently, complete (cp) genome sequencing and assembly have improved, becoming less costly and simpler [20,30], as the next-generation sequence technology has evolved rapidly [31]. Studying the structure and sequence information of the cp genome gives researchers very important information for species conservation, construction of phylogeny [28,32], understanding genomic evolution, and solving relationship issues at various taxonomic levels [28,33]. In addition, it provides information on the genetic structure, present genes, gene order, and nucleotide alignment according to Gao et al., 2009 [30]. This is possible because the chloroplast genome is basic, stable, and conservative [34].
Currently, there is little data available on the genetic structures of C. comosum and C. gallabatense, especially on their detailed phylogenetic placement or chloroplast genomes. Only one nucleotide sequence of Chlorophytum (C. rhizopendulum) has been published (GenBank no. KX931454). In this study, we sequenced the complete cp genome sequences of C. comosum and C. gallabatense for the first time. We aimed to elucidate the structural features in the cp genomes, codon usage, RNA editing, simple sequence repeat (SSR), highly divergent regions, and phylogenetic analyses of C. comosum and C. gallabatense, as well as compare them with five Asparagales species (four Asparagaceae and one Amaryllidaceae species). Our findings provide a basic genetic tool for species identification, evolution, genetic engineering, population genetics, and phylogenetic studies of the species in genus Chlorophytum. Moreover, our results help to fill the gap in genomic resources and resolve the taxonomic complexes of the genus.
2. Results and Discussion
2.1. C. comosum and C. gallabatense Chloroplast Genome Features
The cp genomes of C. comosum and C. gallabatense are 154,248 bp and 154,154 bp in size, respectively. The two sequences contained four regions; the LSC region of 84,004 bp and 83,686 bp length, the SSC regions of 18,016 bp and 17,960 bp length, and a pair of inverted repeat regions (IRa and IRb) of 26,114 bp and 26,254 bp each in length, in C. comosum and C. gallabatense, respectively (Figure 1 and Table 1). The overall guanine-cytosine (GC) ratio was 37.3% in the two cp genomes (Table 1). Both C. comosum and C. gallabatense cp genome features were similar to other reported species in the Asparagaceae family with respect to gene content, order, and orientation [35].
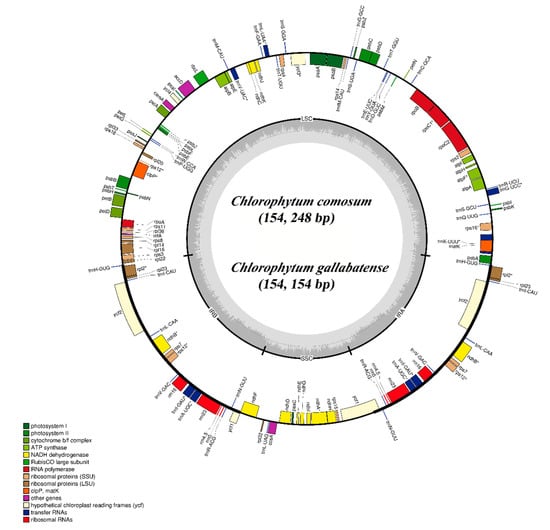
Figure 1.
Gene map of the C. comosum and C. gallabatense complete cp genomes. Genes drawn within the circle are transcribed in the clockwise direction, and genes drawn out are transcribed in the counterclockwise direction. Genes are color-filled, basing on different functions. Inverted repeat (IR), small single-copy (SSC), and large single-copy (LSC) regions are indicated.

Table 1.
Features of the chloroplast genomes of C. comosum and C. gallabatense and the related Asparagaceae species.
We detected a total of 112 unique genes in C. comosum and C. gallabatense cp genomes, which included 78 PCGs, four rRNA, and 30 tRNA genes (Table 1). Amongst them, 19 genes are replicated in the IR regions, comprising seven PCGs (ndhB, ycf1, ycf2, rpl2, rps7, rpl23, and rps12), four rRNA genes (rrn23, rrn4.5, rrn5, and rrn16), and eight tRNA genes (trnA-UGC, trnH-GUG, trnI-GAU, trnI-CAU, trnL-CAA, trnN-GUU, trnV-GAC, and trnR-ACG) (Table 2). There are fifteen intron-containing genes, of which thirteen have one intron (ndhA, atpF, ndhB, rpoC1, rpl2, rps16, rps12, trnV-UAC, trnl-GAU, trnL-UAA, trnK-UUU, trnG-UCC, and trnA-UGC), whereas two genes (ycf3 and clpP) contained two introns. In total, nine of these genes are PCGs and six are tRNA genes (Table 2). This result is similar with previous report by Sheng et al., 2017 [35].

Table 2.
Genes present in C. comosum and C. gallabatense chloroplast genomes.
The rps12 gene is unequally distributed (trans-splicing gene), with the 5′ exon positioned in the LCS part, and the 3′ exons are positioned in the IRa/IRb regions. A similar observation in cp genomes of other species has been previously documented [36,37]. The presence of introns in the plant plastomes can lead to this expression of exogenous genes at precise locations and times [38]. Moreover, two pseudogenes (rps2 and infA) were positioned in the LCS region. They are known as pseudogenes due to the reverse repetitive nature of the IR regions, and therefore not wholly duplicated, losing the capacity to encode whole proteins [39]. There were no significant rearrangements in gene order or inversions observed in comparisons of the two cp genomes of Chlorophytum with those of other selected genome sequences. Gene loss has been reported in some of the Asparagales species [40,41]. McKain et al., 2016 [18], confirmed that rps19 was either pseudogenized in various positions or absent. Comparative analysis of seven species of Asparagales showed that the rps19 gene was pseudogenized in all the species apart from the species of Chlorophytum, where rps19 was missing as reported by McKain et al., 2016 [18].
2.2. Codon Usage Analysis
Codons are useful for the transmission of genetic information used in the evolution of genomes because they connect proteins and nucleic acids [42]. Codon usage is the use of similar codons with which an organism encodes the amino acids in the genes of their proteins [43]. This codon usage, however, has been shown to vary across different species [44] due to different factors such as codon hydrophilicity, tRNA abundance, gene length, gene expression rate, and base group composition [45]. Codon usage bias signifies that each gene of a species has its own preferred amino acid codon [46]. In the two species, the similarity was found in the codon usage and amino acid frequencies. The PCGs contained 20,479 and 20,453 codons in C. comosum and C. gallabatense cp genomes, respectively (Table S1). A comparison was made with other selected species of Asparagales, and codon encoded the genes ranging from 19,986 (Asparagus officinalis) to 20,532 (A. ramosum), as shown in (Table S1). In addition, leucine with a percentage of 10.41% (2,132) and 10.43% (2,133), was the most frequent amino acid in the cp genomes, followed by isoleucine with 8.77% (1797) and 8.75% (1789) in C. comosum and C. gallabatense, respectively. Cysteine, however, encoded the least frequently used amino acids with only 1.11% (228) and 1.12% (230) in C. comosum and C. gallabatense, respectively (Figure 2 and Table S1). Cysteine are found less frequently due to their high sensitivity to changes in physiological and environmental conditions [47]. This was also similar to the rest of the Asparagaceae family (Table S1).
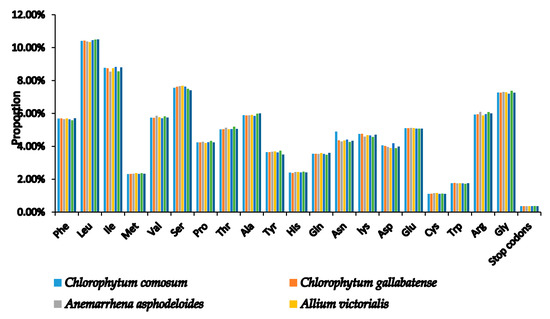
Figure 2.
Amino acid proportion in C. comosum and C. gallabatense protein-coding sequences.
If the relative value of synonymous codon usage (RSCU) is equal to one, the codon usage is not preferred (not biased), but highly preferred if the value is greater than one [36], indicating that the codon is used more often than expected [48] and less preferred with values of less than one. Almost all of the amino acid codons in C. comosum and C. gallabatense, cp genomes have preferences, which is due to the amino acid activity preventing error all through the transcription process, other than tryptophan (UGG) and methionine (AUG), which did not have a bias (RSCU values equal to one) (Table S1). The results, moreover, showed that all preferred synonymous codons (RSCU > 1) have an A or U position at the third codon, except for UUG (Table S1). In the cp genome, natural selection and direction of variation play a very significant role in influencing codon usage bias [49]. This analysis provides the required amino acids for protein biosynthesis in the cp genomes of C. comosum and C. gallabatense. These results are similar to cp genome reports of other terrestrial plants [50,51].
2.3. RNA Editing Sites
RNA editing is a common molecular process in the cp genomes of plants [52]. It has different functions that can change a transcribed RNA’s protein-coding sequence by altering, deleting, or inserting nucleotides during transcription [53]. The predictive plant RNA editor (PREP) software [54], which uses 35 genes as references for possible RNA editing sites by comparing the predicted genes to homologous protein genes from other plants, identified editing sites in 26 PCGs studied (Table S2). As a result, in C. comosum and C. gallabatense a total of 64 and 66 potential RNA editing sites were predicted, respectively. The ndhB gene had the highest number (12 sites) in the two cp genomes of RNA editing sites followed by ndhF, rpoB, and matK, which had more than five editing sites. The gene rpoC2, atpF, rpoC1, rps14, ycf3, accD, psbF, rpl20, clpP, petB, rpoA, rps8, rpl2, ccsA, ndhG, and ndhA had editing sites more than one. The remaining five genes (rps16, atpA, atpI, atpB, and petG) did not have RNA predicting sites. Additionally, variations in amino acid as a result of RNA editing occurred most frequently in serine to leucine which reflects a conversion into a hydrophobic amino acid from a hydrophilic, while arginine to cysteine occurred less frequently (Table S2). These general features of advanced RNA editing in the plants cp genomes have also been documented by previous studies [55,56] which found that important RNA editing sites direct amino acid to alter from polar to non-polar in order to form a fundamental organization of protein to enhance protein hydrophobicity [57].
2.4. Analysis of SSRs
Microsatellites or simple sequence repeats (SSRs) are tandemly repetitive DNA sequences, comprising of one to six (mono-, di-, tri-, tetra-, penta-, and hexa-) repeat nucleotide units. Also known as microsatellites, SSRs are good molecular markers often used in plant species phylogenetics, identification and population genetic studies [58], as they are highly reliable, reproductive, and highly polymorphic [37,44]. They are highly spread in the PCGs, introns, and intergenic regions. We detected a total of 111 SSRs in C. comosum and 90 SSRs in C. gallabatense (Figure 3 and Table 3). Likened to other Asparagales species, A. ramosum had the largest number of repeats with 73 SSRs, C. rhizopendulum with 71, Allium victorialis with 64 SSRs, Asparagus officianalis with 55 SSRs, and the least number of repeats in Anemarrhena asphodeloides with 49 SSRs. C. comosum was comprised of 86 mononucleotide repeats, 14 dinucleotide repeats, two trinucleotide repeats, and nine tetranucleotide repeats, and C. gallabatense included 68 repeats for mononucleotides, 12 repeats for dinucleotides, one repeat for trinucleotides, eight repeats for tetranucleotides, and one repeat for pentanucleotides. Ten hexanucleotides were only present in A. ramosum cp genome (Figure 4 and Table S3). The mononucleotide SSRs had the richest content in all the species, followed by dinucleotides and tetranucleotide. In the cp genomes, short polyadenine (poly A/T) repeats are the most frequent SSRs as compared with guanine (G) or cytosine (C) repeats [59]. In this research, the A/T mononucleotide repeats were the most abundant type in all the cp genomes. This result supports another study where the A/T repeats were abundant [60]. Genetic diversity is reflected in the regions with high mutation rates [61]. The identified loci, in this study, could be good molecular markers, useful in the study of population genetics of C. comosum and C. gallabatense and phylogenetic study in the future.
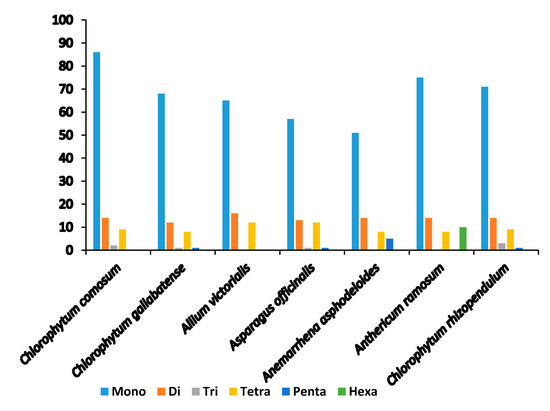
Figure 3.
Number of different types of SSRs.

Table 3.
Numbers of simple sequence repeats (SSRs) in the C. comosum and C. gallabatense chloroplast genomes.
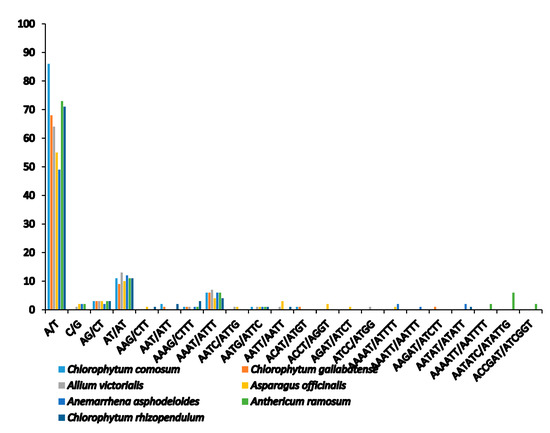
Figure 4.
Number of different repeat units of SSRs.
2.5. IR Expansion and Contraction
Variations in the size of the cp genome are primarily due to the contraction and expansion of the border regions [62]. Such modifications have long-term effects on the size of cp genomes. According to Zhang et al., 2017 [63], IR regions are possible distinctive features in most of the angiosperms. The arrangement of genes in the junctions of cp genomes differs from one species to another [64]. Most of the cp genomes tend to be rearranged due to the loss of the IR regions [65], although, Chumley et al., 2006 [25], discovered that the pelargonium x hortorum genome was rearranged even without the loss of the IR regions. A comparison was made among Asparagales species on the border regions and the adjacent genes and some remarkable variations were discovered (Figure 5). Similar to other typical cp genomes, these genomes showed clear variances at the junctions, but the general gene structures, contents, and orientations were the same. The rps19 gene was located at IRb, 37 bp, 49 bp, 53 bp, and 82 bp away from the junction of LSC/IRb in A. ramosum, A. asphodeloides, A. officinalis, and A. victorialis, respectively; however, this gene was missing in all three Chlorophytum species proving that the common ancestry lost this gene independently. The rpl2 gene was positioned at the IRb in all chloroplast genomes at variable extensions from the junction. Similarly, the psbA gene occurred in the LSC region in all the genome sequences 1 bp to 101 bp away from IRa/LSC boundary. The distance from the IRa/SSC junction to ndhF gene differed by 5 bp in A. asphodeloides to 70 bp in A. ramosum. Still, there was an overlap between ycf1/ndhF in A. victorialis, A. officinalis, C. comosum, C. gallabatense, and C. rhizopendulum. Interestingly, the ndhF gene overlapped the SSC and IRb junction with 2 bp in C. rhizopendulum and 31 bp in A. victorialis. The border region in the middle of IRa and SSC was positioned within the ycf1 gene, and the ψycf1 pseudogene situated at the IRa region ranging from 802 bp to 1174 bp in size. The length of the SSC regions ranged from 17,853 bp in A. victorialis to 18, 638 bp in A. officinalis. The ψycf1 gene was located at the junction between SSC and IRb. This pattern of IR expansion and contraction of partial copies of non-coding genes is a common occurrence in most terrestrial species [66]. These outcomes can provide insight into the evolutionary processes of chloroplast genomes, as well as being a source of DNA barcodes.

Figure 5.
Comparison of the junctions of LSC, SSC, and IR regions among seven chloroplast genomes.
2.6. Analysis of Nucleotide Diversity
To examine the divergence of sequence in the cp genomes, the nucleotide variability (Pi) value was calculated using the DnaSP v5.10 program. A comparison was made with the other randomly selected five Asparagales species on the two cp genomes of Chlorophytum. The Pi values of the seven Asparagales species ranged from 0 to 0.15897 (Figure 6). This indicates that the species in the order Asparagales could be undergoing rapid nucleotide substitution [36]. The IR regions, as predicted, had a much lower nucleotide variability than the SSC and LSC regions. Other nucleotide diversity analyses have also shown that the IR region has a lower value of nucleotide diversity than the SSC and LSC regions [67,68]. This shows that IRa/IRb regions are much more conserved as compared with the LSC and the SSC regions as a result of gene replication [69]. Six divergent regions revealed higher nucleotide diversity values (Pi values > 0.1). All these variable regions were found in the intergenic spacer. This indicated that the intergenic regions were more divergent than the coding regions. Four divergent regions are located in LSC (psbK/psbI, trnS/trnG, psbM/trnD, and rps12/clpP), one in SSC (rps15/ycf1), and one in IR (trnV/rps12). Some of these regions have been previously reported in other species cp genomes [37,67]. Such highly variable regions can provide potential molecular markers for the authentication of plants and assist in phylogenetic analysis studies in this genus [70].
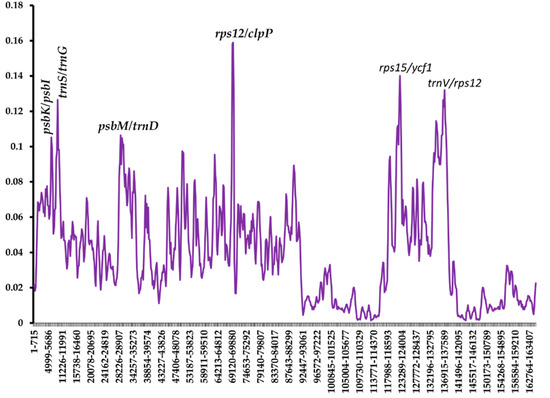
Figure 6.
Nucleotide diversity of different regions of Asparagales chloroplast genomes.
2.7. Comparative Genome Analysis
Comparative genomic analysis and the available DNA sequences make it possible to have a comprehensive view of a genus [71]. The cp genome sequences of C. comosum and C. gallabatense were compared to those of C. rhizopendulum, A. victorialis, A. asphodeloides, A. officinalis, and A. ramosum using the mVISTA program (Figure 7) and MAUVE program (Figure 8). Among the cp genomes compared, there were no significant rearrangements observed, except for the slight variations in size and gene positioning (Figure 8). A. asphodeloides (156,917 bp) was the largest, followed by A. officinalis (156,699 bp), then A. ramosum (155,812 bp), then C. comosum (154,248 bp), then C. gallabatense (154,154 bp), then A. victorialis (154,074 bp), and finally the shortest was C. rhizopendulum (153,504 bp). The results revealed that the genomes with some level of divergence were greatly conserved. The non-coding regions were less conserved as compared with the coding regions. The LSC and the SSC regions were equally less conserved than the IR regions. In addition, it was also detected that the intergenic spacer regions had the highest divergent regions. Some of the high divergent regions included psbI-trnG-UCC, matK-rps16, psbZ-rps14, rps16-psbI, atpH-atpI, trnC-GCA-petN, and trnS-GGA- trnL-UAA (Figure 7). The comparative study showed the conservation of gene order, signifying the evolutionary conservation of these species [71]. These regions can be used as unique barcodes for DNA and also give phylogenetic information.
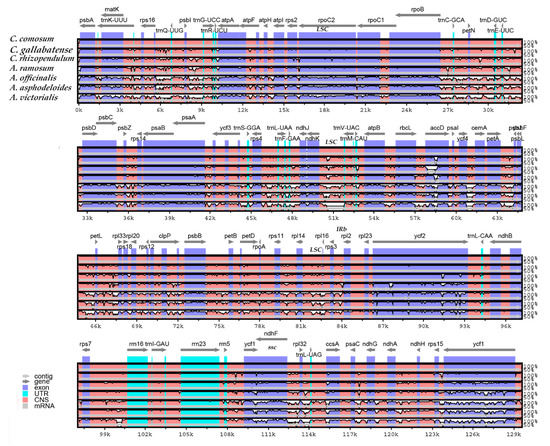
Figure 7.
Comparison of seven cp genomes using mVISTA alignment program. Genome regions are color-coded as protein-coding, rRNA coding, tRNA coding, or conserved non-coding sequences. The vertical scale shows the percentage of identity, varying from 50% to 100%.
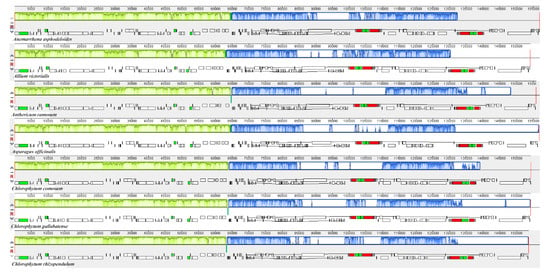
Figure 8.
Comparison of the genome structure of seven Asparagales species using Mauve program. The DNA sequences above the line are presented in a clockwise direction, and those below the line in a counterclockwise direction.
2.8. Phylogenetic Analysis
Due to the rapid development of the sequencing technologies, cp genomes, which are valuable genomic resources, have been used to a greater extent in the reconstruction of plant phylogenies and evolutionary relationships [72]. The phylogeny of Asparagales was reconstructed using whole chloroplast genome sequences of 48 species belonging to the family Asparagaceae (thirty-eight species), the family Amaryllidaceae (eight species), and Iridaceae (two species) as outgroup. All the 46 cp genomes were downloaded from GenBank (Table S4), and the 48 cp genomes were aligned by MAFFT v7.308 [73]. Then, the phylogeny was reconstructed using the maximum likelihood (ML) approach to reveal the evolutionary position of C. comosum and C. gallabatense and the relationship with other Asparagales species. Almost all of the nodes had high bootstrap support values (BP). The result indicated that genus Chlorophytum is closely related to genus Anthericum. The Chlorophytum species (C. comosum, C. gallabatense, and C. rhizopendulum) were clustered together in the same clade with a bootstrap value of 100%. Bjora et al., 2008 [17], in their study, stated that C. gallabatense clusters together with C. comosum, C. filipendulum, and C. macrophyllum. These species had a closer relationship to the Anthericum ramosum which formed an independent clade (Figure 9). This shows the affinity relationship among the species. During the early classification system, genus Chlorophytum was classified to be under the family Anthericaceae together with Anthericum. After several revisionary works, the two genera were separated using morphological characters (stamen filament ornamentation) and some of the species of the Anthericum were transferred into genus Chlorophytum [74], hence, the close relationship in the phylogenetic tree. These findings show that information from the whole cp genome sequences can give more reliable phylogenetic outcomes, therefore, more species of Chlorophytum can be sequenced in order to have a more comprehensive phylogenetic tree in this genus and give insight into the Asparagaceae relationship. Our results confirm the position of the selected Asparagales species supporting the previous whole genome and multigene-based analyses [35,75,76] (Figure 9). Therefore, this study has provided an in-depth molecular analysis within the family Asparagaceae and can be used in the taxonomy of the family especially using the molecular markers upon further analysis.
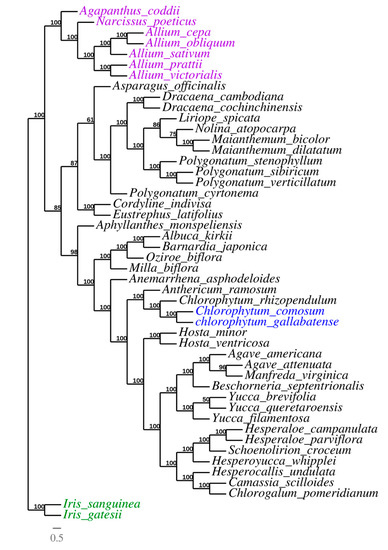
Figure 9.
Phylogenetic trees of Asparagales species based on whole chloroplast genome sequences, with two species from family Iridaceae used as outgroup.
3. Materials and Methods
3.1. DNA Isolation and Chloroplast Genome Sequencing
Young, fresh, and healthy leaf samples of C. comosum and C. gallabatense were collected from Kenya (C. comosum SAJIT-004032 and C. gallabatense SAJIT-006319) and dried in silica gel to preserve the DNA [77]. All the samples were taken to the Herbarium of Wuhan Botanical Garden (HIB) and stored at −80 °C until DNA extraction. The genomic DNA was isolated from 100 micrograms of leaves using a modified cetyltrimethylammonium bromide (CTAB) technique [78] and sequenced using the Illumina platform at Novogene Company (Beijing, China).
3.2. Genome Assembly and Annotation
The clean data obtained after filtering the low-quality data and adaptors were assembled using GetOrganelle version 1.6.2 software [79], and then, manually corrected. The assembled chloroplasts were used for gene annotation by the GeSeq online tool (https://chlorobox.mpimp-golm.mpg.de/geseq.Html) with default settings [80]. Manual verification was done using Sequin Viewer to relate the cp genomes of Chlorophytum species with those of Asparagaceae. Annotations of tRNAs were confirmed using the tRNAscan-SE [81]. The boundaries of the introns and the start and stop codons were manually corrected. Gene maps of complete cp genome sequences were drawn using OrganellarGenome DRAW software [82] (Figure 1). The annotated cp genomes of the C. comosum and C. gallabatense were submitted to the GenBank (GenBank numbers: C. comosum: MT076065 and C. gallabatense: MT036265).
3.3. Genome Structure and Comparison
The genome features of C. comosum and C. gallabatense were determined using MEGA7 [83] by comparing the five other available cp genomes of Asparagales which were downloaded from NCBI (Table S4). The PCGs from the Chlorophytum species and the four Asparagales species were extracted manually and the Predictive RNA Editor for plants (PREP) suite online software was used to identify the RNA editing sites [54] where the cutoff value was set to 0.8. PREP server. The default settings used 35 genes to compare and predict the potential RNA editing sites.
For comparative analysis, the cp genomes of C. comosum and C. gallabatense with the five randomly selected Asparagales species was constructed using the mVISTA program [84]. Furthermore, in the rearrangement analysis, the cp genomes were aligned and constructed in MAUVE program [85]. The expansion and contraction of IRs were analyzed using the IRscope online program (https://irscope.shinyapps.io/irapp/), then manually modified. To detect the divergence hotspots, the complete chloroplast genomes of the selected seven Asparagales species were aligned by MAFFT v7.308 [73]. DnaSP v5.10 was used to calculate the nucleotide divergence values of the chloroplast genome sequence alignment of seven analyzed species using the sliding window method [86] with a window length of 600 bp and a 200 bp step size.
3.4. Codon Usage
The codon usage frequency in each of the seven Asparagales species was analyzed for all the PCGs using MEGA7 [83]. Synonymous codon usage and the relative synonymous codon usage (RSCU) was conducted to determine if the plastid genes were under selection.
3.5. Simple Sequence Repeats
Simple sequence repeats (SSRs) present in cp genomes of C. comosum and C. gallabatense were analyzed using the software MISA (MicroSAtellite) [87] with the settings of ten repeat units for mononucleotides, five for dinucleotides and trinucleotides, and three for tetranucleotides, pentanucleotides, and hexanucleotide repeats.
3.6. Phylogenetic Analysis
To confirm the phylogenetic positions of C. comosum and C. gallabatense within the family Asparagaceae, we downloaded forty-six cp genomes representing three families of Asparagales (Amaryllidaceae, Asparagaceae, and Iridaceae as outgroup) from the NCBI database (Table S4) for analysis. Multiple sequence alignment of the forty-eight complete cp genome sequences was done using MAFFT v7.308 [73] with default parameters and the maximum likelihood (ML) tree with default settings and 1000 bootstrap replicates was reconstructed using the program IQ-TREE v6.10 with the best fit model TVM + I + G4. TreeDyn [88,89] was used to visualize and refine the tree.
4. Conclusions
Chlorophytum is one of the essential genera in the family Asparagaceae as it is used for its medicinal properties, however, its cp genomes are not well studied. In this study, we analyzed the complete cp genomes of C. comosum and C. gallabatense of the family Asparagaceae for the first time. These genomes provided a basic genetic tool for species identification within the genus. We compared the cp genomes of C. comosum and C. gallabatense with five randomly selected Asparagales species. The results showed that the gene size, content, and order were all similar. The loss of rps19 gene in the cp genomes of Chlorophytum, displays a distinctive feature of evolution within precise characteristics. The codon usage, RNA editing, and microsatellites were determined. The ndhB and ndhF genes had the highest number of editing sites. These results provide useful insights regarding editing sites that were gained or lost during the evolution of the Angiosperm genomes. Additionally, such findings provide genetic information for the genus Chlorophytum for the creation of molecular markers and for research into genetic diversity. The contraction and expansion of the boundary regions showed the genome size variation in the selected Asparagales species. Nucleotide analysis revealed the highest nucleotide diversity was in rps12/clpP and rps15/ycf1 regions, which are highly variable regions that can be used as potential markers in family Asparagaceae for species identification and phylogeny. Additionally, the phylogenetic analysis confirmed the previous phylogeny and produced a phylogenetic tree with similar topology. It showed that the Chlorophytum species (C. comosum, C. gallabatense, and C. rhizopendulum) were clustered together in the same clade with a closer relationship than other plants to the A. ramosum. However, more complete cp genomes on Chlorophytum species should be sequenced which would help solve the relationship within Asparagaceae. Generally, this study provides valuable genetic information of Chlorophytum which can aid in further phylogenetic studies, species identification, and evolutionary relationships between Chlorophytum and Anthericum.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/3/296/s1, Table S1: Codon usage, Table S2: RNA editing, Table S3: Simple sequence repeats, and Table S4: NCBI accession number.
Author Contributions
Formal analysis was performed by J.N.M. and X.D.; drafting the manuscript and preparation of figures and tables was done by J.N.M.; revision and manuscript editing was done by J.-X.Y., E.M.M., V.O.W., M.A.O., and J.K.S.; conceptualization and supervision was done by P.M.M. and G.-W.H.; resources and funding acquisition was provided by G.-W.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Backbone Talents Project of Wuhan Botanical Garden, CAS (Y655301M01), Sino-Africa Joint Research Center, CAS (SAJC201614) and the National Natural Science Foundation of China (31970211).
Acknowledgments
Sincere thanks to John Nzei and Virginia Mwanzia for providing helpful suggestions and support related to the analysis performed. We also thank the Managing Editor Harry Fu and the anonymous reviewers for their useful comments and precious remarks on the manuscript.
Conflicts of Interest
The authors affirm no conflict of interest.
References
- Basu, S.; Jha, T.B. Cytogenetic studies in four species of Chlorophytum Ker-Gawl. (Liliaceae). The Nucleus 2011, 54, 123–132. [Google Scholar] [CrossRef]
- Govaerts, R.; Zonneveld, B.; Zona, S. World Checklist of Asparagaceae; Facilitated by the Royal Botanic Gardens: Kew, UK, 2012; Available online: http://apps.kew.org/wcsp/ (accessed on 26 February 2020).
- Malpure, N.V.; Yadav, S. Chlorophytum gothanense, a new species of Anthericaceae from the Western Ghats of India. Kew Bull. 2009, 64, 739–741. [Google Scholar] [CrossRef]
- Govaerts, R. World Checklist of Selected Plant Families Asparagaceae; Royal Botanic Gardens: Kew, UK, 2015. [Google Scholar]
- Lekhak, M.; Adsul, A.; Yadav, S. Cytotaxonomical investigations into the genus Chlorophytum from India. Kew Bull. 2012, 67, 285–292. [Google Scholar] [CrossRef]
- Group, A.P. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar]
- Adsul, A.A. Taxonomic Revision of Genus Chlorophytum Ker Gawl for India. Ph.D. Thesis, Shivaji University, Kolhapur India, 2015. [Google Scholar]
- Kumar, M.; Meena, P.; Verma, S.; Kumar, M.; Kumar, A. Anti-tumour, anti-mutagenic and chemomodulatory potential of Chlorophytum borivilianum. Asian Pac. J. Cancer Prev. 2010, 11, 327–334. [Google Scholar]
- Kalra, S.; Kumar, S.; Lakhanpal, N.; Kaur, J.; Singh, K. Characterization of squalene synthase gene from Chlorophytum borivilianum (Sant. and Fernand.). Mol. Biotechnol. 2013, 54, 944–953. [Google Scholar] [CrossRef]
- Deore, S.; Khadabadi, S. Standardisation and pharmaceutical evaluation of Chlorophytum borivilianum mucilage. Rasayan J. Chem. 2008, 1, 887–892. [Google Scholar]
- Khanam, Z.; Singh, O.; Singh, R.; Bhat, I.U.H. Safed musli (Chlorophytum borivilianum): A review of its botany, ethnopharmacology and phytochemistry. J. Ethnopharmacol. 2013, 150, 421–441. [Google Scholar] [CrossRef]
- Kaushik, N. Saponins of Chlorophytum species. Phytochem. Rev. 2005, 4, 191–196. [Google Scholar] [CrossRef]
- Haque, R.; Saha, S.; Bera, T. A peer reviewed literature on medicinal activity of Chlorophytum borivilianum commercial medicinal plant. Int. J. Drug Dev. Res. 2011, 3, 1116, 1130. [Google Scholar]
- O’Donnell, G.; Bucar, F.; Gibbons, S. Phytochemistry and antimycobacterial activity of Chlorophytum inornatum. Phytochemistry 2006, 67, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Geetha, K. Characterization, genetic improvement and cultivation of Chlorophytum borivilianum—an important medicinal plant of India. Plant Genet. Resour. 2005, 3, 264–272. [Google Scholar] [CrossRef]
- Adsul, A.A.; Lekhak, M.M.; Yadav, S.R. Chlorophytum sharmae (Asparagaceae): A new species from Kerala, India. Kew Bull. 2014, 69. [Google Scholar] [CrossRef]
- Bjorå, C.S.; Hemp, A.; Hoell, G.; Nordal, I. A taxonomic and ecological analysis of two forest Chlorophytum taxa (Anthericaceae) on Mount Kilimanjaro, Tanzania. Plant Syst. Evol. 2008, 274, 243–253. [Google Scholar] [CrossRef]
- McKain, M.R.; McNeal, J.R.; Kellar, P.R.; Eguiarte, L.E.; Pires, J.C.; Leebens-Mack, J. Timing of rapid diversification and convergent origins of active pollination within Agavoideae (Asparagaceae). Am. J. Bot. 2016, 103, 1717–1729. [Google Scholar] [CrossRef]
- Deguilloux, M.-F.; Pemonge, M.-H.l.N.; Petit, R.M.J. Use of chloroplast microsatellites to differentiate oak populations. Ann. For. Sci. 2004, 61, 825–830. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Neuhaus, H.; Emes, M. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef]
- Zhou, T.; Wang, J.; Li, W.; Zhang, X.; Xu, Y.; Xu, F.; Zhu, H.; Wang, X. The complete chloroplast genome of Euphrasia regelii, pseudogenization of ndh genes and the phylogenetic relationships within Orobanchaceae. Front. Genet. 2019, 10, 444. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Bendich, A.J. Circular chloroplast chromosomes: The grand illusion. Plant Cell 2004, 16, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium× hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-P.; Huang, J.-P.; Wu, C.-S.; Hsu, C.-Y.; Chaw, S.-M. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol. Evol. 2010, 2, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 84. [Google Scholar] [CrossRef]
- Gao, L.; Yi, X.; Yang, Y.-X.; Su, Y.-J.; Wang, T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: Insights into evolutionary changes in fern chloroplast genomes. BMC Evol. Biol. 2009, 9, 130. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.; Cai, Q.; Lin, F.; Huang, P.; Zheng, Y. Development of chloroplast genomic resources for Akebia quinata (Lardizabalaceae). Conserv. Genet. Resour. 2016, 8, 447–449. [Google Scholar] [CrossRef]
- Sun, M.; Li, J.; Li, D.; Shi, L. Complete chloroplast genome sequence of the medical fern Drynaria roosii and its phylogenetic analysis. Mitochondrial Dna Part B 2017, 2, 7–8. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.-H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef]
- Sheng, W.; Chai, X.; Rao, Y.; Tu, X.; Du, S. The Complete Chloroplast Genome Sequence of Asparagus (Asparagus officinalis L.) and its phylogenetic positon within Asparagales. Int. J. Plant Biol. Res. 2017, 5, 121–128. [Google Scholar]
- Cui, Y.; Nie, L.; Sun, W.; Xu, Z.; Wang, Y.; Yu, J.; Song, J.; Yao, H. Comparative and Phylogenetic Analyses of Ginger (Zingiber officinale) in the Family Zingiberaceae Based on the Complete Chloroplast Genome. Plants 2019, 8, 283. [Google Scholar] [CrossRef] [PubMed]
- Iram, S.; Hayat, M.Q.; Tahir, M.; Gul, A.; Ahmed, I. Chloroplast Genome Sequence of Artemisia scoparia: Comparative Analyses and Screening of Mutational Hotspots. Plants 2019, 8, 476. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Feng, D.; Song, G.; Wei, X.; Chen, L.; Wu, X.; Li, X.; Zhu, Z. The first intron of rice EPSP synthase enhances expression of foreign gene. Sci. China Ser. C: Life Sci. 2003, 46, 561. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, Z.; Huang, S.; An, W.; Li, J.; Zheng, X. Comprehensive Analysis of Rhodomyrtus tomentosa Chloroplast Genome. Plants 2019, 8, 89. [Google Scholar] [CrossRef]
- Meerow, A.W. Convergence or reticulation? Mosaic evolution in the canalized American Amaryllidaceae. In Diversity, Phylogeny and Evolution in the Monocotyledons; Seberg, O., Petersen, G., Barfod, A.S., Davis, J.I., Eds.; Aarhus University Press: Aarhus, Denmark, 2010; pp. 145–168. [Google Scholar]
- Steele, P.R.; Hertweck, K.L.; Mayfield, D.; McKain, M.R.; Leebens-Mack, J.; Pires, J.C. Quality and quantity of data recovered from massively parallel sequencing: Examples in Asparagales and Poaceae. Am. J. Bot. 2012, 99, 330–348. [Google Scholar] [CrossRef]
- Liu, Q.; Dou, S.; Ji, Z.; Xue, Q. Synonymous codon usage and gene function are strongly related in Oryza sativa. Biosystems 2005, 80, 123–131. [Google Scholar] [CrossRef]
- Campbell, W.H.; Gowri, G. Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol. 1990, 92, 1–11. [Google Scholar] [CrossRef]
- Srivastava, D.; Shanker, A. Identification of simple sequence repeats in chloroplast genomes of Magnoliids through bioinformatics approach. Interdiscip. Sci. Comput. Life Sci. 2016, 8, 327–336. [Google Scholar] [CrossRef]
- Li, Y.; Kuang, X.J.; Zhu, X.X.; Zhu, Y.; Sun, C. Codon usage bias of Catharanthus roseus. Zhongguo Zhong Yao Za Zhi = Zhongguo Zhongyao Zazhi = China J. Chin. Mater. Med. 2016, 41, 4165–4168. [Google Scholar]
- Wu, X.-M.; Wu, S.-F.; Ren, D.-M.; Zhu, Y.-P.; He, F.-C. The analysis method and progress in the study of codon bias. Yi Chuan = Hered. 2007, 29, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.M.; Gladyshev, V.N. Analysis and functional prediction of reactive cysteine residues. J. Biol. Chem. 2012, 287, 4419–4425. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.-H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xue, Q. Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. J. Genet. 2005, 84, 55–62. [Google Scholar] [CrossRef]
- Park, I.; Kim, W.; Yeo, S.-M.; Choi, G.; Kang, Y.-M.; Piao, R.; Moon, B. The complete chloroplast genome sequences of Fritillaria ussuriensis Maxim. and Fritillaria cirrhosa D. Don, and comparative analysis with other Fritillaria species. Molecules 2017, 22, 982. [Google Scholar] [CrossRef]
- Zuo, L.-H.; Shang, A.-Q.; Zhang, S.; Yu, X.-Y.; Ren, Y.-C.; Yang, M.-S.; Wang, J.-M. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 2017, 12, e0171264. [Google Scholar] [CrossRef]
- Freyer, R.; Hoch, B.; Neckermann, K.; Maier, R.M.; Kössel, H. RNA editing in maize chloroplasts is a processing step independent of splicing and cleavage to monocistronic mRNAs. Plant J. 1993, 4, 621–629. [Google Scholar] [CrossRef]
- Tang, W.; Luo, C. Molecular and functional diversity of RNA editing in plant mitochondria. Mol. Biotechnol. 2018, 60, 935–945. [Google Scholar] [CrossRef]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef]
- Corneille, S.; Lutz, K.; Maliga, P. Conservation of RNA editing between rice and maize plastids: Are most editing events dispensable? Mol. Gen. Genet. 2000, 264, 419–424. [Google Scholar] [CrossRef]
- Jain, B.P.; Chauhan, P.; Tanti, G.K.; Singarapu, N.; Ghaskadbi, S.; Goswami, S.K. Tissue specific expression of SG2NA is regulated by differential splicing, RNA editing and differential polyadenylation. Gene 2015, 556, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Taylor, F.; Coates, D. The code within the codons. Biosystems 1989, 22, 177–187. [Google Scholar] [CrossRef]
- Qi, W.; Lin, F.; Liu, Y.; Huang, B.; Cheng, J.; Zhang, W.; Zhao, H. High-throughput development of simple sequence repeat markers for genetic diversity research in Crambe abyssinica. BMC Plant Biol. 2016, 16, 139. [Google Scholar] [CrossRef] [PubMed]
- Kuang, D.-Y.; Wu, H.; Wang, Y.-L.; Gao, L.-M.; Zhang, S.-Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, X.; Cui, Y.; Sun, W.; Li, Y.; Wang, Y.; Song, J.; Yao, H. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. Int. J. Mol. Sci. 2017, 18, 1839. [Google Scholar] [CrossRef]
- Qi, W.-H.; Jiang, X.-M.; Yan, C.-C.; Zhang, W.-Q.; Xiao, G.-S.; Yue, B.-S.; Zhou, C.-Q. Distribution patterns and variation analysis of simple sequence repeats in different genomic regions of bovid genomes. Sci. Rep. 2018, 8, 14407. [Google Scholar] [CrossRef]
- Wang, W.; Messing, J. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS ONE 2011, 6, e24670. [Google Scholar] [CrossRef]
- Zhang, H.; Hall, N.; McElroy, J.S.; Lowe, E.K.; Goertzen, L.R. Complete plastid genome sequence of goosegrass (Eleusine indica) and comparison with other Poaceae. Gene 2017, 600, 36–43. [Google Scholar] [CrossRef]
- Yang, J.-B.; Tang, M.; Li, H.-T.; Zhang, Z.-R.; Li, D.-Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef]
- Strauss, S.H.; Palmer, J.D.; Howe, G.T.; Doerksen, A.H. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proc. Natl. Acad. Sci. USA 1988, 85, 3898–3902. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cui, L.; Feng, K.; Deng, P.; Du, X.; Wan, F.; Weining, S.; Nie, X. Comparative analysis of Asteraceae chloroplast genomes: Structural organization, RNA editing and evolution. Plant Mol. Biol. Report. 2015, 33, 1526–1538. [Google Scholar] [CrossRef]
- Yu, X.; Tan, W.; Zhang, H.; Gao, H.; Wang, W.; Tian, X. Complete Chloroplast Genomes of Ampelopsis humulifolia and Ampelopsis japonica: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis. Plants 2019, 8, 410. [Google Scholar] [CrossRef] [PubMed]
- De Souza, U.J.B.; Nunes, R.; Targueta, C.P.; Diniz-Filho, J.A.F.; de Campos Telles, M.P. The complete chloroplast genome of Stryphnodendron adstringens (Leguminosae-Caesalpinioideae): Comparative analysis with related Mimosoid species. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef]
- Ma, P.-F.; Zhang, Y.-X.; Zeng, C.-X.; Guo, Z.-H.; Li, D.-Z. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst. Biol. 2014, 63, 933–950. [Google Scholar] [CrossRef]
- Lin, H.; Yu, M.; Wang, X.; Zhang, X.-H. Comparative genomic analysis reveals the evolution and environmental adaptation strategies of vibrios. BMC Genom. 2018, 19, 135. [Google Scholar] [CrossRef]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kunth, C. Melanthaceae [sic]. Enumeratio Plant. 1843, 4, 1. [Google Scholar]
- Wang, Y.; Sun, J.; Yuan, Q.; Guo, L. Complete chloroplast genome sequence of Dracaena cochinchinensis: Genome structure and genomic resources. Mitochondrial DNA Part B 2019, 4, 1750–1751. [Google Scholar] [CrossRef]
- Pfanzelt, S.; Albach, D.C.; von Hagen, K.B. Extremely low levels of chloroplast genome sequence variability in Astelia pumila (Asteliaceae, Asparagales). PeerJ 2019, 7, e6244. [Google Scholar] [CrossRef]
- Chase, M.W.; Hills, H.H. Silica gel: An ideal material for field preservation of leaf samples for DNA studies. Taxon 1991, 40, 215–220. [Google Scholar] [CrossRef]
- Doyle, J. DNA protocols for plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv 2018, 256479. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M. Phylogeny. fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Chevenet, F.; Brun, C.; Bañuls, A.-L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).