Differential Regulation of Drought Responses in Two Phaseolus vulgaris Genotypes
Abstract
1. Introduction
2. Results
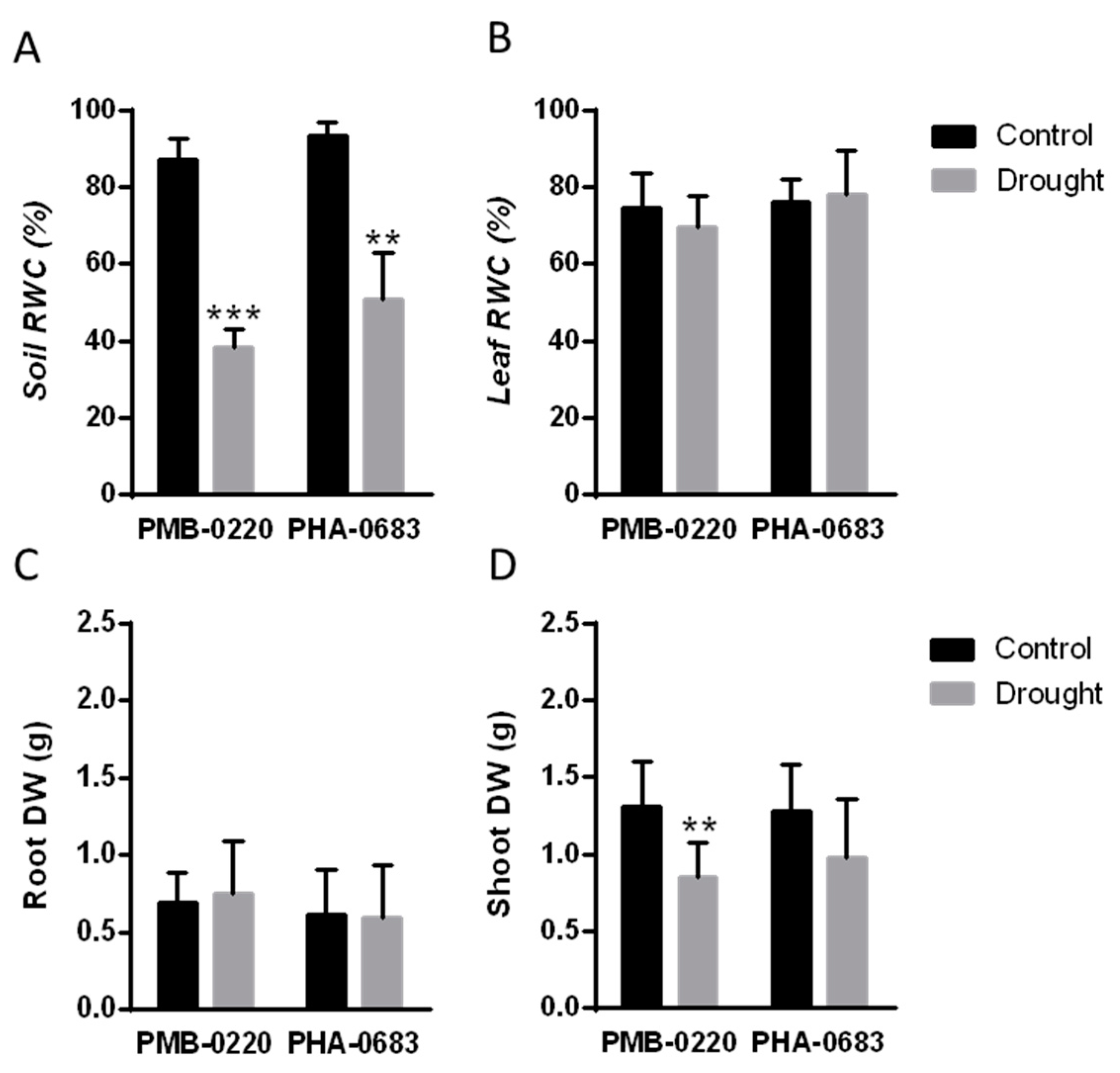
2.1. Physiological Effects of Drought in Two Common Bean Drought Tolerant Plants
2.2. Changes in Total Chlorophyll Contents in Response to Drought
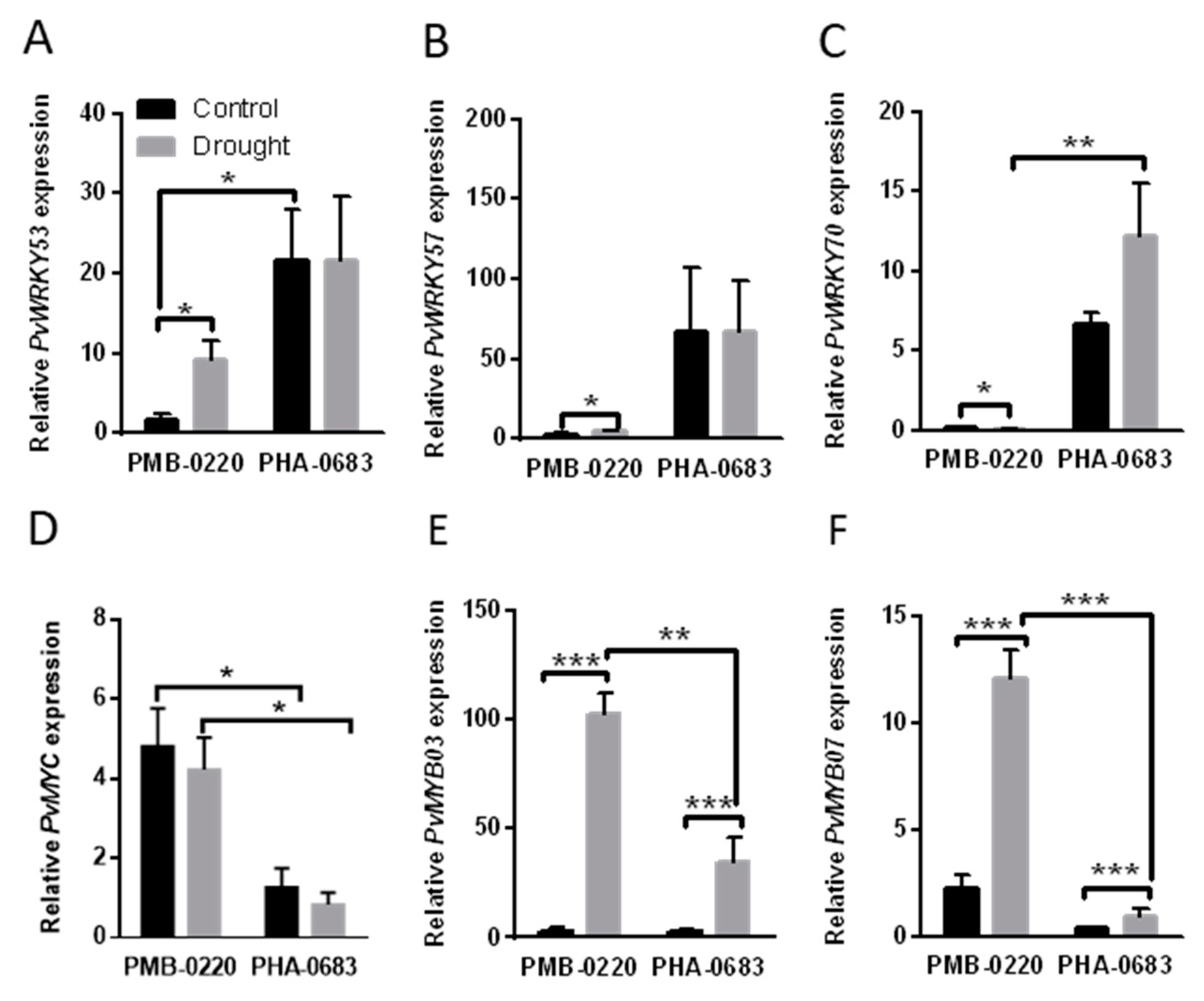
2.3. Analysis of Genes Expression Related to ABA-Mediated Response
2.4. Analysis of Genes Expression Related to Senescence in Response to Drought
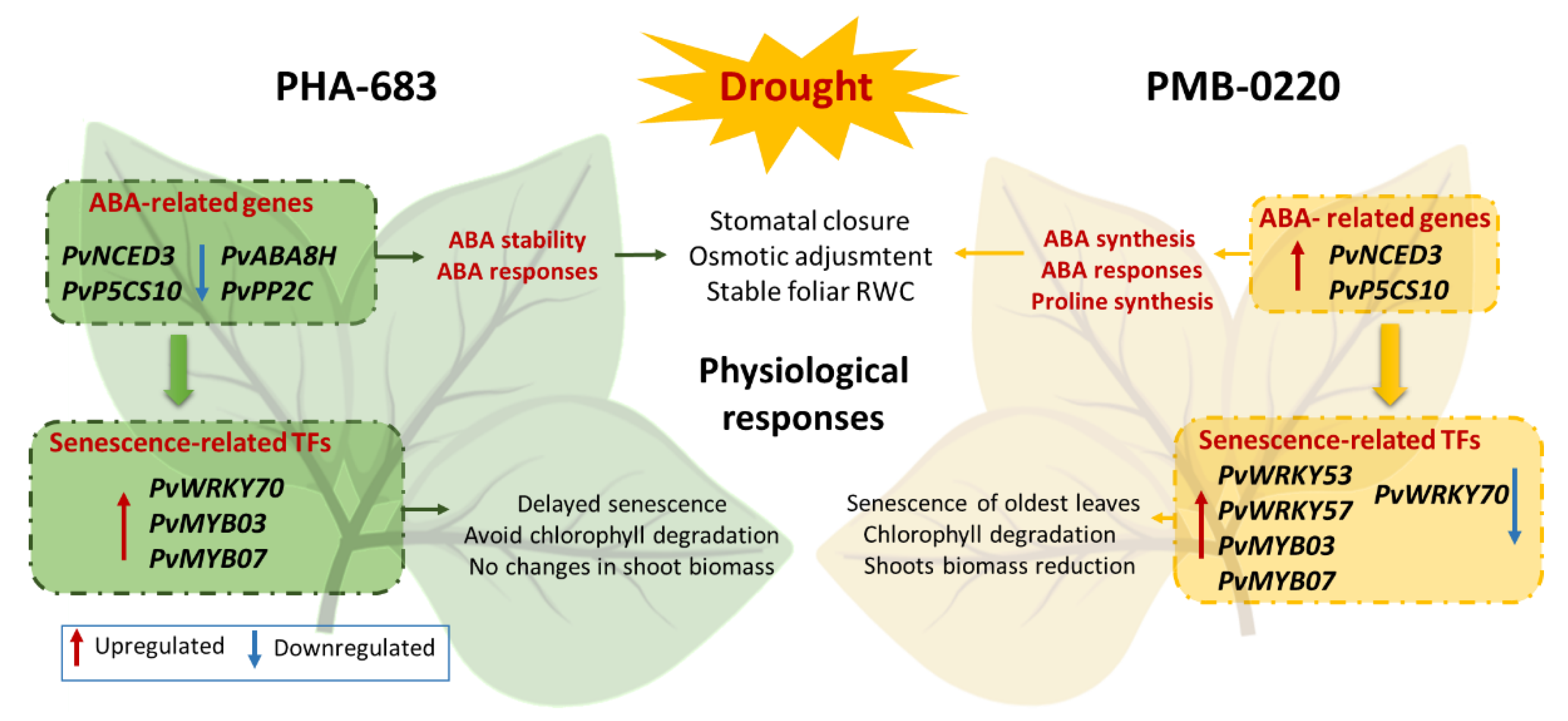
3. Discussion
4. Materials and Methods
4.1. Growth Conditions and Plant Material
4.2. Physiological Analysis
4.3. Chlorophyll Determination
4.4. Determination of Catalase and Superoxide Dismutase Activities
4.5. Nucleic Acid Isolation and Quantification
4.6. Analysis of Gene Expression
4.7. Analysis of Promotor Regulatory Motives in WRKY Coding Genes
4.8. Experimental Design and Statistical Analysis of Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Broughton, W.J.; Hernández, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.)—Model food legumes. Plant Soil. 2003, 252, 55–128. [Google Scholar] [CrossRef]
- De Ron, A.M.; Papa, R.; Bitocchi, E.; González, A.M.; Debouck, D.G.; Brick, M.A.; Fourie, D.; Marsolais, F.; Beaver, J.; Geffroy, V.; et al. Common Bean. In Grain Legumes, Handbook of Plant Breeding; Springer: New York, NY, USA, 2015; pp. 1–36. [Google Scholar]
- Beebe, S.; Rao, I.M.; Blair, M.W.; Acosta-Gallegos, J.A. Phenotyping common beans for adaptation to drought. Front. Physiol. 2013, 4, 35. [Google Scholar] [CrossRef]
- Long, S.P.; Ort, D.R. More than taking the heat: Crops and global change. Curr. Opin. Plant Biol. 2010, 13, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Atkins, C.A. Ammonia assimilation and export of nitrogen from the legume nodule. In Biology and Biochemistry of Nitrogen Fixation; Dilworth, M.J., Glenn, A.R., Eds.; Elsevier Science Publishers: Amsterdam, The Netherlands, 1991; pp. 293–319. [Google Scholar]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and Purine Biosynthesis and Degradation In Plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, T.R.; Serraj, R. Legume nitrogen fixation and drought. Nat. Cell Biol. 1995, 378, 344. [Google Scholar] [CrossRef]
- King, C.A.; Purcell, L.C. Inhibition of N2 Fixation in Soybean Is Associated with Elevated Ureides and Amino Acids1. Plant Physiol. 2005, 137, 1389–1396. [Google Scholar] [CrossRef]
- Gil-Quintana, E.; Larrainzar, E.; Seminario, A.; Díaz-Leal, J.L.; Alamillo, J.M.; Pineda, M.; Arrese-Igor, C.; Wienkoop, S.; González, E.M. Local inhibition of nitrogen fixation and nodule metabolism in drought-stressed soybean. J. Exp. Bot. 2013, 64, 2171–2182. [Google Scholar] [CrossRef]
- Alamillo, J.M.; Díaz-Leal, J.L.; Pineda, M.; Sánchez-Moran, M.V. Molecular analysis of ureide accumulation under drought stress in Phaseolus vulgaris L. Plant Cell Environ. 2010, 33, 1828–1837. [Google Scholar] [CrossRef]
- Coleto, I.; Pineda, M.; Rodiño, A.P.; De Ron, A.M.; Alamillo, J.M. Comparison of inhibition of N2 fixation and ureide accumulation under water deficit in four common bean genotypes of contrasting drought tolerance. Ann. Bot. 2014, 113, 1071–1082. [Google Scholar] [CrossRef]
- Takagi, H.; Ishiga, Y.; Watanabe, S.; Konishi, T.; Egusa, M.; Akiyoshi, N.; Matsuura, T.; Mori, I.C.; Hirayama, T.; Kaminaka, H.; et al. Allantoin, a stress-related purine metabolite, can activate jasmonate signaling in a MYC2-regulated and abscisic acid-dependent manner. J. Exp. Bot. 2016, 67, 2519–2532. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Matsumoto, M.; Hakomori, Y.; Takagi, H.; Shimada, H.; Sakamoto, A. The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ. 2013, 37, 1022–1036. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, A.; Paul, S.; Basu, S. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013, 32, 985–1006. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Koiwai, H.; Akaba, S.; Komano, T.; Oritani, T.; Kamiya, Y.; Koshiba, T. Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J. 2000, 23, 481–488. [Google Scholar] [CrossRef]
- Seo, M.; Peeters, A.J.M.; Koiwai, H.; Oritani, T.; Marion-Poll, A.; Zeevaart, J.A.D.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and Drought Stress Signal Transduction Inplants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Farquhar, G.D.; Sharkey, T.D. Stomatal Conductance and Photosynthesis. Annu. Rev. Plant Physiol. 1982, 33, 317–345. [Google Scholar] [CrossRef]
- Rivero, R.M.; Kojima, M.; Gepstein, A.; Sakakibara, H.; Mittler, R.; Gepstein, S.; Blumwald, E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA 2007, 104, 19631–19636. [Google Scholar] [CrossRef]
- Zabadal, T.J. A Water Potential Threshold for the Increase of Abscisic Acid in Leaves. Plant Physiol. 1974, 53, 125–127. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1069. [Google Scholar] [CrossRef]
- Park, S.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA- binding START proteins. Science 2010, 324, 1068–1071. [Google Scholar] [CrossRef]
- Rodrigues, A.; Adamo, M.; Crozet, P.; Margalha, L.; Confraria, A.; Martinho, C.; Elias, A.; Rabissi, A.; Lumbreras, V.; González-Guzmán, M.; et al. ABI1 and PP2CA Phosphatases Are Negative Regulators of Snf1-Related Protein Kinase1 Signaling in Arabidopsis. Plant Cell 2013, 25, 3871–3884. [Google Scholar] [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 Protein Kinases are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In vitro reconstitution of an abscisic acid signalling pathway. Nat. Cell Biol. 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC Gene Regulating Senescence Improves Grain Protein, Zinc, and Iron Content in Wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Riov, J.; Dagan, E.; Goren, R.; Yang, S.F. Characterization of Abscisic Acid-Induced Ethylene Production in Citrus Leaf and Tomato Fruit Tissues. Plant Physiol. 1990, 92, 48–53. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Liu, L.; Xu, W.; Hu, X.; Liu, H.; Lin, Y. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016, 6, 20881. [Google Scholar] [CrossRef]
- Zou, X.; Seemann, J.R.; Neuman, D.; Shen, Q.J. A WRKY Gene from Creosote Bush Encodes an Activator of the Abscisic Acid Signaling Pathway. J. Biol. Chem. 2004, 279, 55770–55779. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, J.; Wang, L.; Wang, S. Genome-Wide Investigation of WRKY Transcription Factors Involved in Terminal Drought Stress Response in Common Bean. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.A.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Hummel, M.; Hallahan, B.F.; Brychkova, G.; Ramirez-Villegas, J.; Guwela, V.; Chataika, B.; Curley, E.; McKeown, P.C.; Morrison, L.; Talsma, E.F.; et al. Reduction in nutritional quality and growing area suitability of common bean under climate change induced drought stress in Africa. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Frahm, M.A.; Rosas, J.C.; Mayek-Pérez, N.; López-Salinas, E.; Acosta-Gallegos, J.A.; Kelly, J.D.; Aggarwal, V.D.; Pastor-Corrales, M.A.; Chirwa, R.M.; Buruchara, R.A. Breeding beans for resistance to terminal drought in the Lowland tropics. Euphytica 2004, 136, 223–232. [Google Scholar] [CrossRef]
- Riveiro, M. Tolerancia de Variedades de Judía a Estrés Hídrico Estacional e Implicaciones en la Fijación Simbiótica de Nitrógeno. Ph.D. Thesis, Universidad de Santiago de Compostela, Santiago de Compostela, Spain, 2012. [Google Scholar]
- Muñoz-Perea, C.G.; Terán, H.; Allen, R.G.; Wright, J.L.; Westermann, D.T.; Singh, S.P. Selection for Drought Resistance in Dry Bean Landraces and Cultivars. Crop. Sci. 2006, 46, 2111–2120. [Google Scholar] [CrossRef]
- Terán, H.; Singh, S.P. Comparison of Sources and Lines Selected for Drought Resistance in Common Bean. Crop. Sci. 2002, 42, 64–70. [Google Scholar] [CrossRef]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and Comparative Analysis of Differential Gene Expression in Soybean Leaf Tissue under Drought and Flooding Stress Revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef]
- Mashaki, K.M.; Garg, V.; Ghomi, A.A.N.; Kudapa, H.; Chitikineni, A.; Nezhad, K.Z.; Yamchi, A.; Soltanloo, H.; Varshney, R.K.; Thudi, M. RNA-Seq analysis revealed genes associated with drought stress response in kabuli chickpea (Cicer arietinum L.). PLoS ONE 2018, 13, e0199774. [Google Scholar] [CrossRef]
- Morgil, H.; Tardu, M.; Cevahir, G.; Kavakli, I.H. Comparative RNA-seq analysis of the drought-sensitive lentil (Lens culinaris) root and leaf under short- and long-term water deficits. Funct. Integr. Genom. 2019, 19, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, L.; Li, L.; Wang, S. De Novo Assembly of the Common Bean Transcriptome Using Short Reads for the Discovery of Drought-Responsive Genes. PLoS ONE 2014, 9, e109262. [Google Scholar] [CrossRef] [PubMed]
- Jorge, J.G.; Villalobos-López, M.A.; Chavarría-Alvarado, K.L.; Ríos-Meléndez, S.; López-Meyer, M.; Arroyo-Becerra, A. Genome-wide transcriptional changes triggered by water deficit on a drought-tolerant common bean cultivar. BMC Plant Biol. 2020, 20, 525. [Google Scholar] [CrossRef]
- López, C.M.; Pineda, M.; Alamillo, J.M. Transcriptomic Response to Water Deficit Reveals a Crucial Role of Phosphate Acquisition in a Drought-Tolerant Common Bean Landrace. Plants 2020, 9, 445. [Google Scholar] [CrossRef]
- Bai, G.; Xie, H.; Yao, H.; Li, F.; Chen, X.; Zhang, Y.; Xiao, B.; Yang, J.; Li, Y.; Yang, D.-H. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nicotiana tabacum. BMC Genom. 2019, 20, 575. [Google Scholar] [CrossRef]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [CrossRef]
- Zhou, Q.-Y.; Tian, A.-G.; Zou, H.-F.; Xie, Z.-M.; Lei, G.; Huang, J.; Wang, C.-M.; Wang, H.-W.; Zhang, J.; Chen, S. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Singh, S.P. Drought Resistance in the Race Durango Dry Bean Landraces and Cultivars. Agron. J. 2007, 99, 1219–1225. [Google Scholar] [CrossRef]
- Lizana, C.; Wentworth, M.; Martinez, J.P.; Villegas, D.; Meneses, R.; Murchie, E.H.; Pastenes, C.; Lercari, B.; Vernieri, P.; Horton, P.; et al. Differential adaptation of two varieties of common bean to abiotic stress. J. Exp. Bot. 2006, 57, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zeevaart, J.A.D. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 1999, 96, 15354–15361. [Google Scholar] [CrossRef] [PubMed]
- Seo, M. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Krochko, J.E.; Abrams, G.D.; Loewen, M.K.; Abrams, S.R.; Cutler, A.J. (+)-Abscisic Acid 8′-Hydroxylase Is a Cytochrome P450 Monooxygenase. Plant Physiol. 1998, 118, 849–860. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic Acid Biosynthesis and Catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Roychoudhury, A.; Banerjee, A.; Lahiri, V. Metabolic and molecular-genetic regulation of proline signaling and itscross-talk with major effectors mediates abiotic stress tolerance in plants. Turk. J. Bot. 2015, 39, 887–910. [Google Scholar] [CrossRef]
- Kishor, P.B.K.; Sreenivasulu, N. Is proline accumulation perse correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 2013, 37, 300–311. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Székely, G.; Abraham, E.; Cseplo, A.; Rigor, G.; Zsigmond, L.; Csiszã¡r, J.; Ayaydin, F.; Strizhov, N.; Jã¡sik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, S.; Yabor, L.; Díez, M.J.; Prohens, J.; Boscaiu, M.; Vicente, O. The Use of Proline in Screening for Tolerance to Drought and Salinity in Common Bean (Phaseolus vulgaris L.) Genotypes. Agronomy 2020, 10, 817. [Google Scholar] [CrossRef]
- Hossain, M.A.; Hoque, A.; Burritt, D.J.; Fujita, M. Proline Protects Plants Against Abiotic Oxidative Stress. In Oxidative Damage to Plants; Elsevier BV: Amsterdam, The Netherlands, 2014; Volume 2014, pp. 477–522. [Google Scholar]
- Gan, S.; Amasino, R.M. Inhibition of Leaf Senescence by Autoregulated Production of Cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef]
- Yang, J.C.; Zhang, J.; Wang, Z.Q.; Zhu, Q.S.; Liu, L.J. Involvement of abscisic acid and cytokinins in the senescence and remobilization of carbon reserves in wheat subjected to water stress during grain filling. Plant Cell Environ. 2003, 26, 1621–1631. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, G.; Yu, D. Activated Expression of WRKY57 Confers Drought Tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S.-S. AtMYB2 Regulates Whole Plant Senescence by Inhibiting Cytokinin-Mediated Branching at Late Stages of Development in Arabidopsis. Plant Physiol. 2011, 156, 1612–1619. [Google Scholar] [CrossRef]
- Zentgraf, U.; Doll, J. Arabidopsis WRKY53, a Node of Multi-Layer Regulation in the Network of Senescence. Plants 2019, 8, 578. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Gil Nam, H. Leaf Senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Hörtensteiner, S. Chlorophyll Degradation During Senescence. Annu. Rev. Plant Biol. 2006, 57, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Christ, B.; Hörtensteiner, S. Mechanism and Significance of Chlorophyll Breakdown. J. Plant Growth Regul. 2014, 33, 4–20. [Google Scholar] [CrossRef]
- Volaire, F.; Norton, M. Summer Dormancy in Perennial Temperate Grasses. Ann. Bot. 2006, 98, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.D.; Hosfield, G.L.; Varner, G.V.; Uebersax, M.A.; Taylor, J. Registration of “Matterhorn” Great Northern Bean. Crop. Sci. 1999, 39, 589–590. [Google Scholar] [CrossRef]
- Rigaud, J.; Puppo, A. Indole-3-acetic Acid Catabolism by Soybean Bacteroids. J. Gen. Microbiol. 1975, 88, 223–228. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. In Methods in Enzymology; Academic Press: Orlando, FL, USA, 1987; Volume 148, pp. 183–350. [Google Scholar]
- Konieczny, R.; Banaś, A.K.; Surówka, E.; Michalec, Ż.; Miszalski, Z.; Libik-Konieczny, M. Pattern of antioxidant enzyme activities and hydrogen peroxide content during developmental stages of rhizogenesis from hypocotyl explants of Mesembryanthemum crystallinum L. Plant Cell Rep. 2013, 33, 165–177. [Google Scholar] [CrossRef][Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
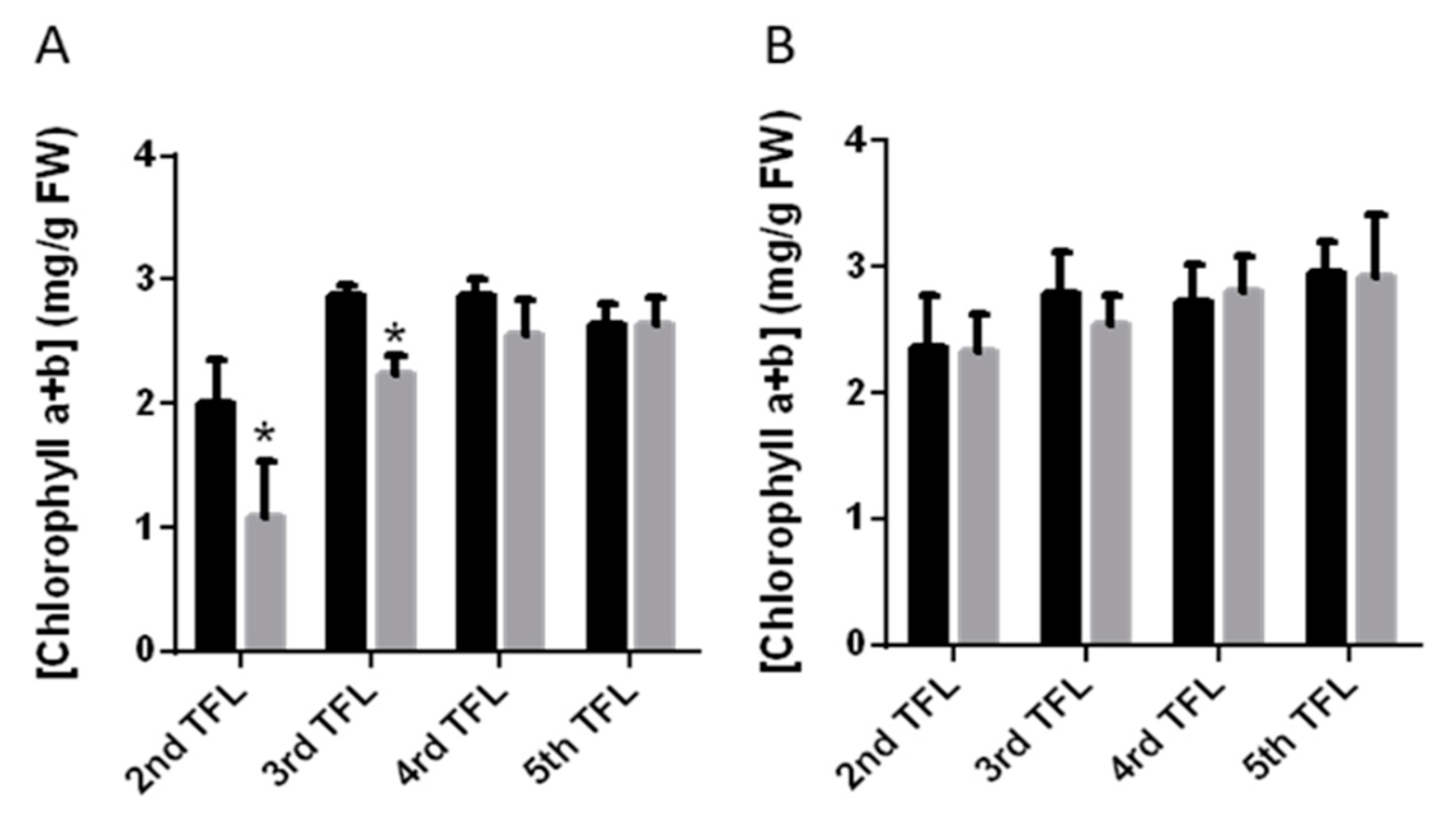

| [Chlorophyll a] | [Chlorophyll b] | [Chlorophyll a + b] | |||||
|---|---|---|---|---|---|---|---|
| PMB-0220 | PHA-0683 | PMB-0220 | PMB-0683 | PMB-0220 | PHA-0683 | ||
| 2nd TFL | Control | 1.270 ± 0.209 | 1.475 ± 0.279 | 0.634 ± 0.115 | 0.732 ± 0.151 | 2.013 ± 0.336 | 2.362 ± 0.417 |
| Drought | 0.501 ± 0.144 | 1.485 ± 0.201 | 0.429 ± 0.207 | 0.634 ± 0.070 | 1.078 ± 0.451 | 2.329 ± 0.294 | |
| 3rd TFL | Control | 1.664 ± 0.225 | 1.673 ± 0.160 | 0.789 ± 0.146 | 0.873 ± 0.148 | 2.880 ± 0.085 | 2.795 ± 0.317 |
| Drought | 1.452 ± 0.195 | 1.589 ± 0.186 | 0.704 ± 0.071 | 0.729 ± 0.044 | 2.246 ± 0.140 | 2.546 ± 0.230 | |
| 4rd TFL | Control | 1.599 ± 0.215 | 1.669 ± 0.172 | 0.891 ± 0.082 | 0.877 ± 0.125 | 2.867 ± 0.142 | 2.798 ± 0.267 |
| Drought | 1.546 ± 0.251 | 1.657 ± 0.148 | 0.778 ± 0.118 | 0.906 ± 0.148 | 2.551 ± 0.294 | 2.813 ± 0.267 | |
| 5th TFL | Control | 1.587 ± 0.178 | 1.797 ± 0.174 | 0.687 ± 0.156 | 0.886 ± 0.125 | 2.685 ± 0.166 | 2.950 ± 0.251 |
| Drought | 1.628 ± 0.134 | 1.776 ± 0.160 | 0.779 ± 0.191 | 0.889 ± 0.297 | 2.644 ± 0.206 | 2.926 ± 0.487 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López, C.M.; Pineda, M.; Alamillo, J.M. Differential Regulation of Drought Responses in Two Phaseolus vulgaris Genotypes. Plants 2020, 9, 1815. https://doi.org/10.3390/plants9121815
López CM, Pineda M, Alamillo JM. Differential Regulation of Drought Responses in Two Phaseolus vulgaris Genotypes. Plants. 2020; 9(12):1815. https://doi.org/10.3390/plants9121815
Chicago/Turabian StyleLópez, Cristina María, Manuel Pineda, and Josefa M. Alamillo. 2020. "Differential Regulation of Drought Responses in Two Phaseolus vulgaris Genotypes" Plants 9, no. 12: 1815. https://doi.org/10.3390/plants9121815
APA StyleLópez, C. M., Pineda, M., & Alamillo, J. M. (2020). Differential Regulation of Drought Responses in Two Phaseolus vulgaris Genotypes. Plants, 9(12), 1815. https://doi.org/10.3390/plants9121815






