Genetic Analysis and Molecular Mapping of the Quantitative Trait Loci Governing Low Phytic Acid Content in a Novel LPA Rice Mutant, PLM11
Abstract
1. Introduction
2. Results
2.1. Identification of the Novel LPA Mutant
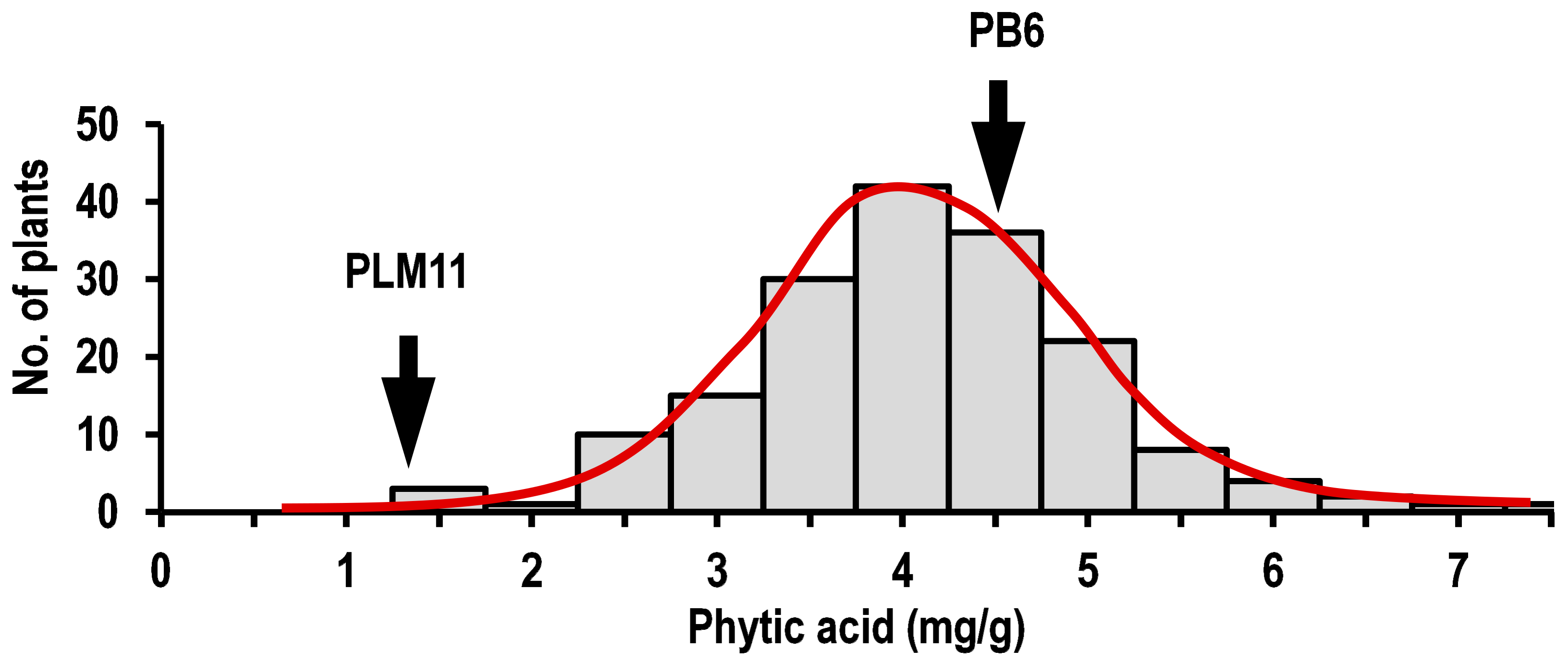
2.2. Phenotypic Variation in Parents and the F2:3 Population
2.3. Development of Linkage Map Using Simple Sequence Repeat (SSR) Markers
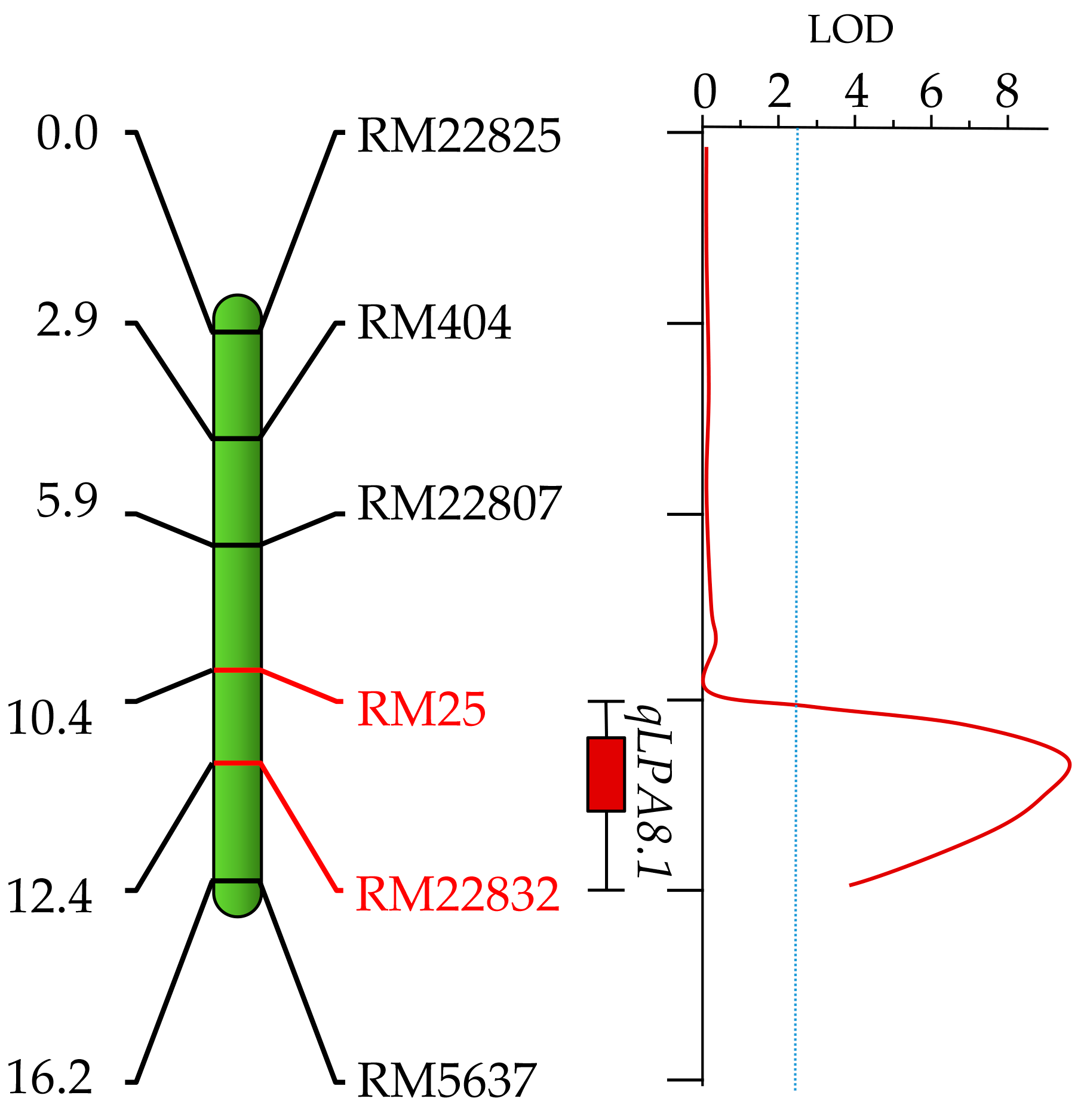
2.4. Mapping the QTL for LPA Content and In Silico Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. HIP Assay for Screening of EMS Mutants
4.3. Phenotyping of Mapping Population
4.4. Construction of Molecular Linkage Map
4.5. QTL Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- White, P.J.; Broadley, M.R. Biofortification of crops with seven mineral elements often lacking in human diets—Iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 2009, 182, 49–84. [Google Scholar] [CrossRef] [PubMed]
- Bollinedi, H.; Vinod, K.K.; Bisht, K.; Chauhan, A.; Krishnan, S.G.; Bhowmick, P.K.; Nagarajan, M.; Rao, D.S.; Ellur, R.K.; Singh, A.K. Characterising the diversity of grain nutritional and physico-chemical quality in Indian rice landraces by multivariate genetic analyses. Indian J. Genet. 2020, 80, 26–38. [Google Scholar]
- Bollinedi, H.; Yadav, A.K.; Vinod, K.K.; Gopala Krishnan, S.; Bhowmick, P.K.; Nagarajan, M.; Neeraja, C.N.; Ellur, R.K.; Singh, A.K. Genome-wide association study reveals novel marker-trait associations (MTAs) governing the localization of Fe and Zn in the rice grain. Front. Genet. 2020, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Alwan, N.A.; Hamamy, H. Prenatal exposures and short and long term developmental outcomes: Maternal iron status in pregnancy and long-term health outcomes in the offspring. J. Pediatr. Genet. 2015, 4, 111. [Google Scholar] [PubMed]
- Hojyo, S.; Fukada, T. Roles of zinc signaling in the immune system. J. Immunol. Res. 2016, 2016, 1–21. [Google Scholar] [CrossRef]
- Graham, R.D.; Welch, R.M.; Bouis, H.E. Addressing micronutrient malnutrition through enhancing the nutritional quality of staple foods: Principles, perspectives and knowledge gaps. Adv. Agron. 2001, 70, 77–142. [Google Scholar] [CrossRef]
- Raboy, V. The ABCs of low-phytate crops. Nat. Biotechnol. 2007, 25, 874–875. [Google Scholar] [CrossRef]
- Qamar, Z.U.; Hameed, A.; Ashraf, M.; Rizwan, M.; Akhtar, M. Development and molecular characterization of low phytate Basmati rice through induced mutagenesis, hybridization, backcross, and marker assisted breeding. Front. Plant Sci. 2019, 10, 1525. [Google Scholar] [CrossRef]
- Zhao, H.-J.; Liu, Q.-L.; Ren, X.-L.; Wu, D.-X.; Shu, Q. Gene identification and allele-specific marker development for two allelic low phytic acid mutations in rice (Oryza sativa L.). Mol. Breed. 2008, 22, 603–612. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop. J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Larson, S.R.; Rutger, J.N.; Young, K.A.; Raboy, V. Isolation and genetic mapping of a non-lethal rice (Oryza sativa), low phytic acid 1 mutation. Crop Sci. 2000, 40, 1397–1405. [Google Scholar] [CrossRef]
- Frank, T.; Habernegg, R.; Yuan, F.-J.; Shu, Q.; Engel, K.-H. Assessment of the contents of phytic acid and divalent cations in low phytic acid (lpa) mutants of rice and soybean. J. Food Compos. Anal. 2009, 22, 278–284. [Google Scholar] [CrossRef]
- Frank, T.; Meuleye, B.S.; Miller, A.; Shu, Q.; Engel, K.-H. Metabolite profiling of two low phytic acid (lpa) rice mutants. J. Agric. Food Chem. 2007, 55, 11011–11019. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.D.; Casati, P.; Walbot, V.A. Multidrug resistance–associated protein involved in anthocyanin transport in Zea mays. Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef]
- Liu, Q.-L.; Xu, X.-H.; Ren, X.-L.; Fu, H.-W.; Wu, D.-X.; Shu, Q. Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.). Theor. Appl. Genet. 2007, 114, 803–814. [Google Scholar] [CrossRef]
- Perera, I.; Fukushima, A.; Akabane, T.; Horiguchi, G.; Seneweera, S.; Hirotsu, N. Expression regulation of myo-inositol 3-phosphate synthase 1 (INO1) in determination of phytic acid accumulation in rice grain. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Li, H.; Ye, G.; Wang, J. A modified algorithm for the improvement of composite interval mapping. Genetics 2007, 175, 361–374. [Google Scholar] [CrossRef]
- Stangoulis, J.C.; Huynh, B.-L.; Welch, R.M.; Choi, E.-Y.; Graham, R.D. Quantitative trait loci for phytate in rice grain and their relationship with grain micronutrient content. Euphytica 2007, 154, 289–294. [Google Scholar] [CrossRef]
- Perera, I.; Fukushima, A.; Arai, M.; Yamada, K.; Nagasaka, S.; Seneweera, S.; Hirotsu, N. Identification of low phytic acid and high Zn bioavailable rice (Oryza sativa L.) from 69 accessions of the world rice core collection. J. Cereal Sci. 2019, 85, 206–213. [Google Scholar] [CrossRef]
- Singh, V.P.; Singh, A.K.; Atwal, S.S.; Joseph, M.; Mohapatra, T. Pusa 1121: A rice line with exceptionally high cooked kernel elongation and basmati quality. Int. Rice Res. Notes 2002, 27, 25–26. [Google Scholar]
- Murray, M.; Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Wang, J.-K. Inclusive Composite Interval Mapping of quantitative trait genes. Acta Agron. Sin. 2009, 35, 239–245. [Google Scholar] [CrossRef]
- McCouch, S.R.; Cho, G.Y.; Yano, M. Report on QTL nomenclature. Rice Genet Newsl. 1997, 14, 11–13. [Google Scholar]


| Chromosome | Total No. of Markers | No. of Polymorphic Markers Used for Genotyping F2s | Percent Polymorphism | Map Length (cM) | Marker Density (cM/marker) |
|---|---|---|---|---|---|
| 1 | 39 | 10 | 25.6 | 169.8 | 17.0 |
| 2 | 42 | 10 | 23.8 | 91.1 | 9.1 |
| 3 | 24 | 6 | 25.0 | 129.6 | 21.6 |
| 4 | 51 | 10 | 19.6 | 76.7 | 7.7 |
| 5 | 31 | 6 | 19.4 | 17.2 | 2.9 |
| 6 | 26 | 7 | 26.9 | 115.2 | 16.5 |
| 7 | 13 | 6 | 46.2 | 43.8 | 7.3 |
| 8 | 29 | 6 | 20.7 | 16.2 | 2.7 |
| 9 | 28 | 6 | 21.4 | 49.2 | 8.2 |
| 10 | 15 | 4 | 26.7 | 84.5 | 21.1 |
| 11 | 22 | 4 | 18.2 | 27.7 | 6.9 |
| 12 | 11 | 3 | 27.3 | 49.1 | 16.4 |
| Total | 331 | 78 | 25.1 | 870.2 | 11.4 |
| Method | QTL | Chr | Position (cM) | Left Marker | Right Marker | LOD | PVE (%) | Additive Effect * | Dominance Effect * |
|---|---|---|---|---|---|---|---|---|---|
| ICIM | qLPA8.1 | 8 | 12 | RM25 | RM22832 | 9.06 | 22.21 | 0.7 | 0.084 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gyani, P.C.; Bollinedi, H.; Gopala Krishnan, S.; Vinod, K.K.; Sachdeva, A.; Bhowmick, P.K.; Ellur, R.K.; Nagarajan, M.; Singh, A.K. Genetic Analysis and Molecular Mapping of the Quantitative Trait Loci Governing Low Phytic Acid Content in a Novel LPA Rice Mutant, PLM11. Plants 2020, 9, 1728. https://doi.org/10.3390/plants9121728
Gyani PC, Bollinedi H, Gopala Krishnan S, Vinod KK, Sachdeva A, Bhowmick PK, Ellur RK, Nagarajan M, Singh AK. Genetic Analysis and Molecular Mapping of the Quantitative Trait Loci Governing Low Phytic Acid Content in a Novel LPA Rice Mutant, PLM11. Plants. 2020; 9(12):1728. https://doi.org/10.3390/plants9121728
Chicago/Turabian StyleGyani, Prem Chand, Haritha Bollinedi, Subbaiyan Gopala Krishnan, Kunnummal Kurungara Vinod, Archana Sachdeva, Prolay Kumar Bhowmick, Ranjith Kumar Ellur, Mariappan Nagarajan, and Ashok Kumar Singh. 2020. "Genetic Analysis and Molecular Mapping of the Quantitative Trait Loci Governing Low Phytic Acid Content in a Novel LPA Rice Mutant, PLM11" Plants 9, no. 12: 1728. https://doi.org/10.3390/plants9121728
APA StyleGyani, P. C., Bollinedi, H., Gopala Krishnan, S., Vinod, K. K., Sachdeva, A., Bhowmick, P. K., Ellur, R. K., Nagarajan, M., & Singh, A. K. (2020). Genetic Analysis and Molecular Mapping of the Quantitative Trait Loci Governing Low Phytic Acid Content in a Novel LPA Rice Mutant, PLM11. Plants, 9(12), 1728. https://doi.org/10.3390/plants9121728






