RING-Type E3 Ubiqitin Ligase Barley Genes (HvYrg1–2) Control Characteristics of Both Vegetative Organs and Seeds as Yield Components
Abstract
1. Introduction
2. Results and Discussion
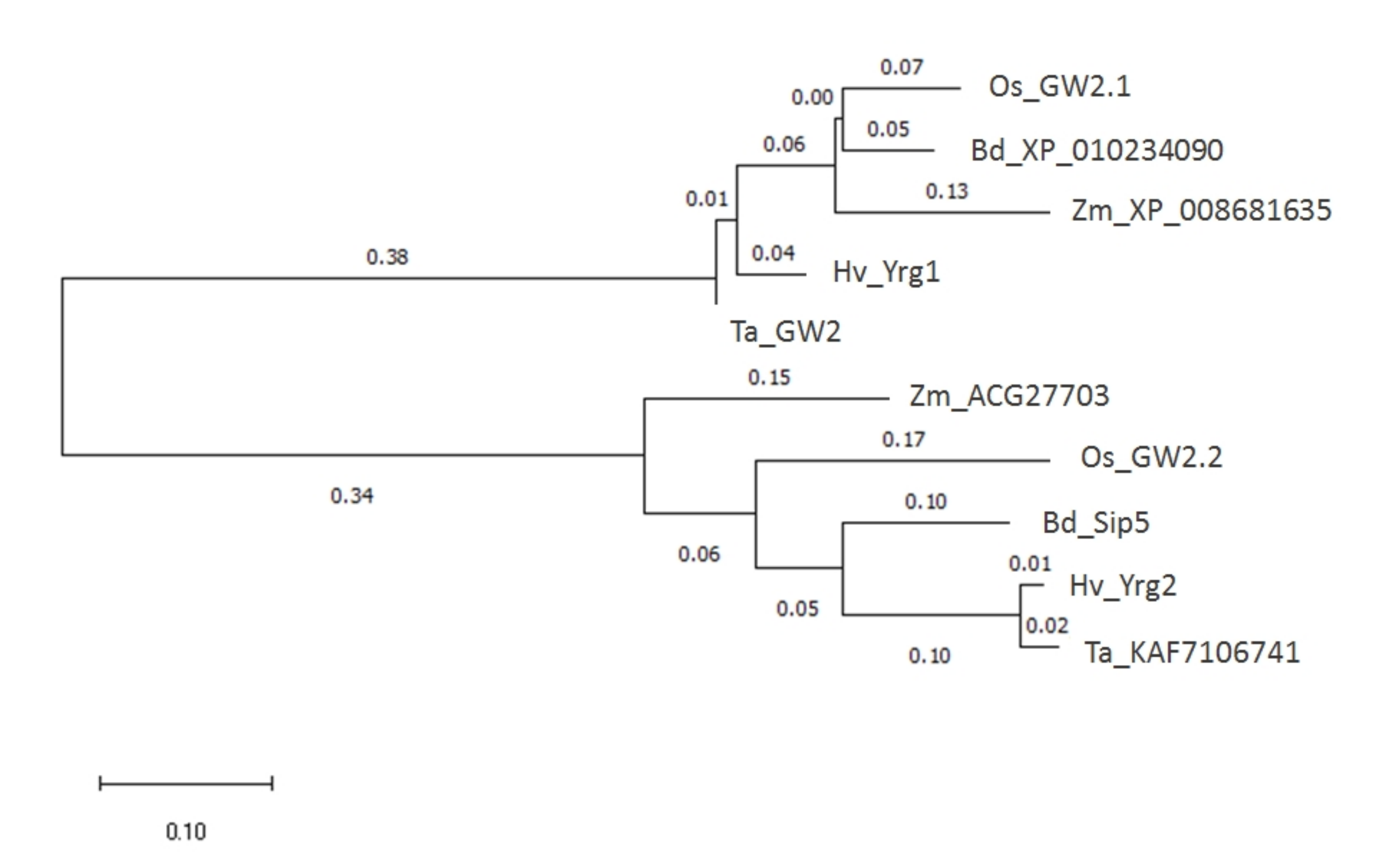
2.1. Two Homologs of HvYrg Genes Encoding RING-Type Protein as E3 Ubiquitin Ligases in Barley
2.2. Production of T2 Generation Transgenic Barley Plants with Silenced Expression of the HvYrg Gene Variants
2.3. Altered Vegetative Organ Parameters in Transgenic Barley Plants with Silenced HvYrg Genes
2.4. Down-Regulation of Barley HvYrg Genes Modulates Growth and Developmental Characteristics of Transgenic Plants
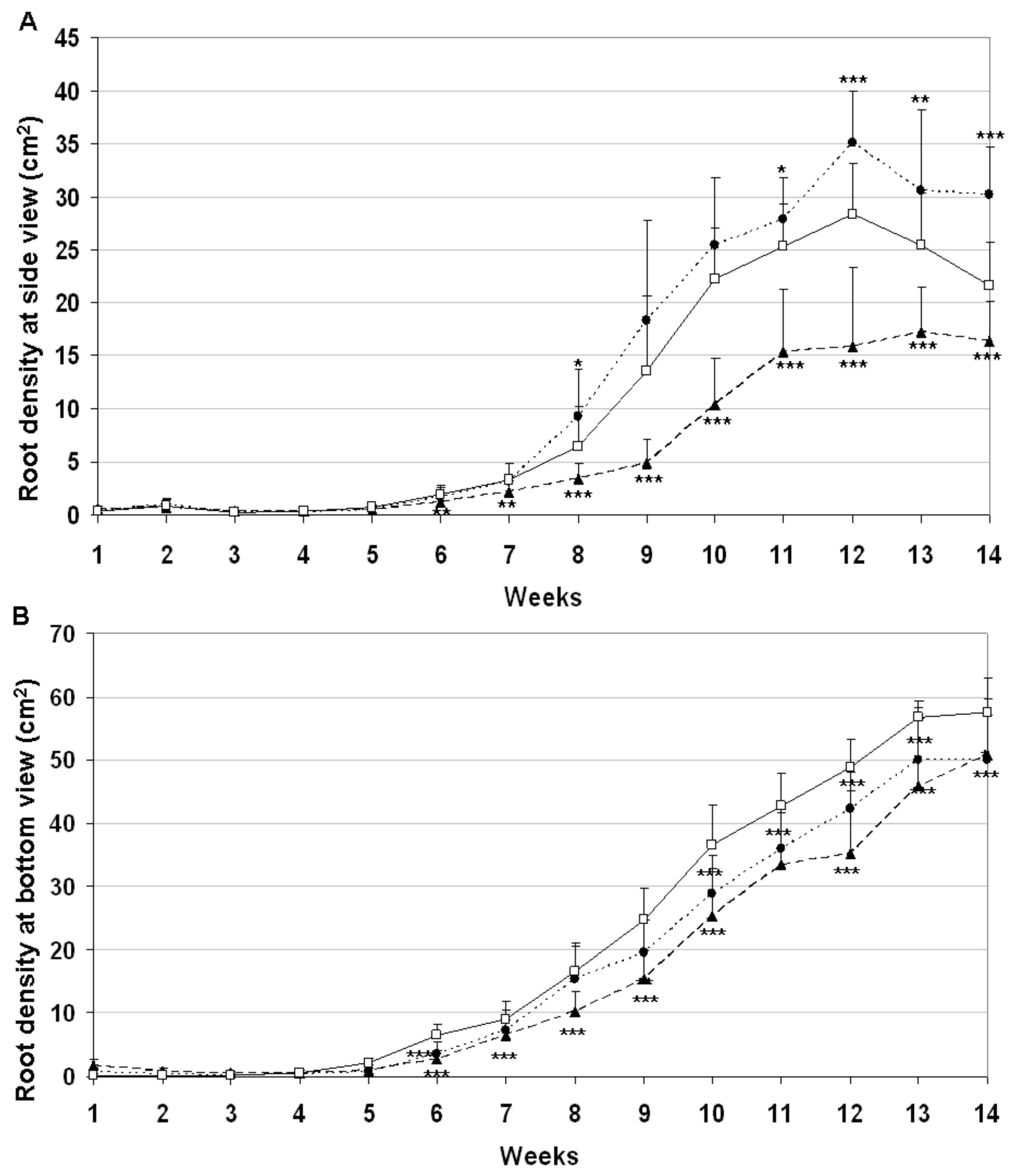
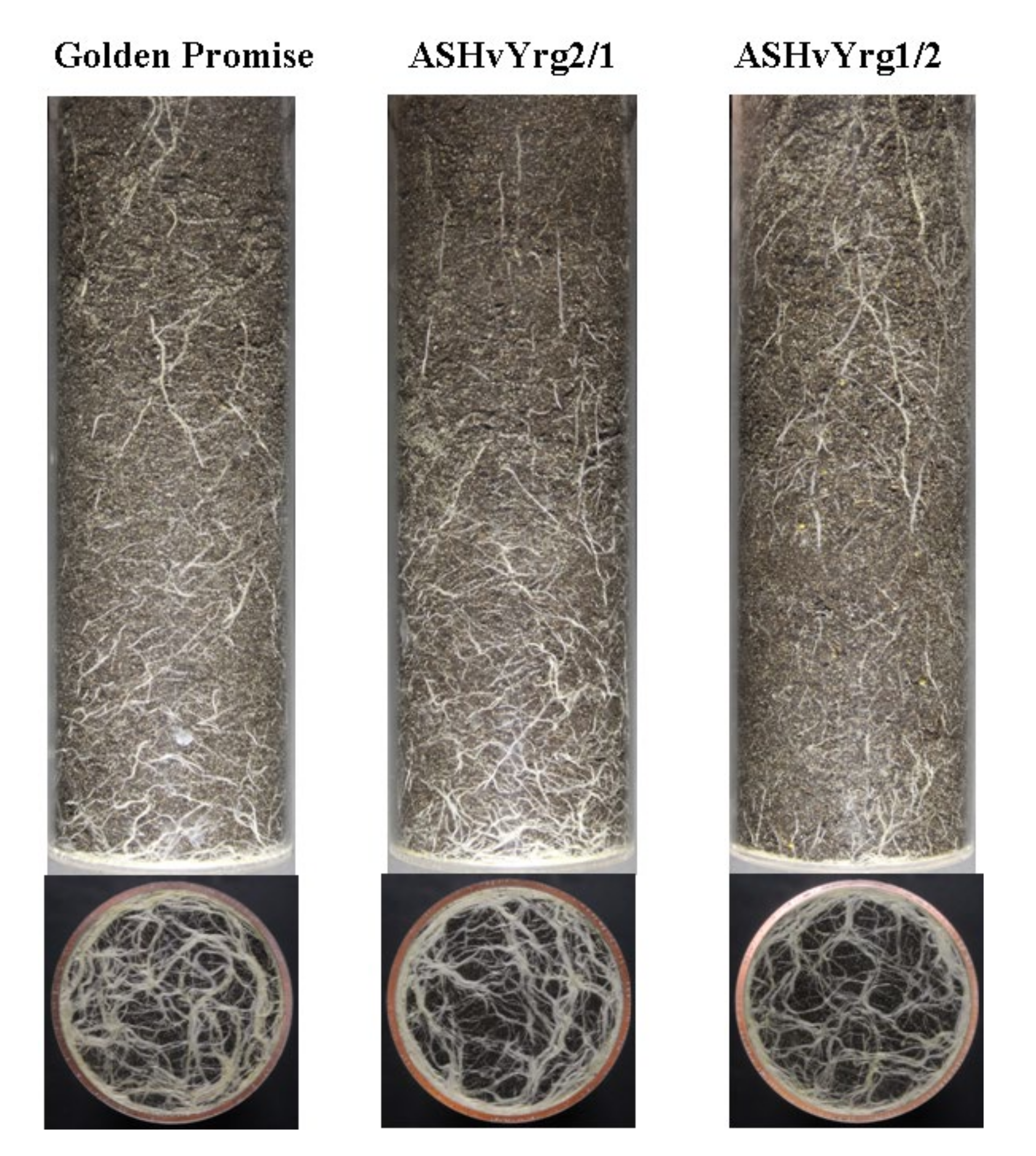
2.5. Down-Regulation of the HvYrg Gene Variants Can Differentially Modify Development of Root System Monitored in a Semi-Automated Phenotyping Platform
2.6. Silencing the HvYrg Barley Genes Can Alter Seed Parameters in Antisense Transgenic Plants
3. Materials and Methods
3.1. Construction of Antisense HvYrg RING-Type E3 Expression Vectors and Barley Transformation
3.2. Growth Conditions for Barley Plants in Greenhouse or Semi-Automatic Phenotyping Platform
3.3. Quantitative Real-Time PCR (qRT-PCR) for Measurement of Expression of HvYrg1 and HvYrg2 Gene Variants in Transgenic Barley Plants
3.4. Data Management
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nadolska-Orczyk, A.; Rajchel, I.K.; Orczyk, W.; Gasparis, S. Major genes determining yield-related traits in wheat and barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Wang, X.D.; Zhang, H.; Yang, F.S.; Yao, Z.P.; Shi, Q.H. Cloning Of Hordeum Vulgare yield related gene Hvyrg1 and construction of its plant RNA interfere vector. Bull. Bot. Res. 2009, 29, 445–452. [Google Scholar]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Sun, J.; Sun, Y.; Ahmed, R.I.; Ren, A.; Xie, A.M. Research progress on plant RING-finger proteins. Genes 2019, 10, 973. [Google Scholar] [CrossRef]
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant. Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Kelley, D.R. E3 ubiquitin ligases: Key regulators of hormone signaling in plants. Mol. Cell. Prot. 2018, 17, 1047–1054. [Google Scholar] [CrossRef]
- Qing, L.; Yang, X.; Bai, G.; Warburton, M.L.; Mahuku, G.; Gore, M.; Dai, J.; Li, J.; Yan, J. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. App. Gen. 2010, 120, 753–763. [Google Scholar]
- Bednarek, J.; Boulaflous, A.; Girousse, C.; Ravel, C.; Tassy, C.; Barret, P.; Bouzidi, M.F.; Mouzeyar, S. Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 2012, 63, 5945–5955. [Google Scholar] [CrossRef]
- Yang, Z.B.; Bai, Z.Y.; Li, X.L.; Wang, P.; Wu, Q.X.; Yang, L.; Li, L.Q.; Li, X.J. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor. App. Gen. 2012, 125, 1057–1068. [Google Scholar] [CrossRef]
- Du, D.F.; Gao, X.; Geng, J.; Li, Q.Y.; Li, L.Q.; Lu, Q.; Li, X.J. Identification of key proteins and networks related to grain development in wheat (Triticum aestivum L.) by comparative transcription and proteomic analysis of allelic variants in TaGW2-6A. Front. Plant. Sci. 2016, 7, 922. [Google Scholar] [CrossRef]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; Del Blanco, A.; Dubcovsky, J.; Uauy, C. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. App. Gen. 2016, 129, 1099–1112. [Google Scholar] [CrossRef]
- Zhai, H.; Feng, Z.; Du, X. A novel allele of TaGW2-A1 is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.). Theor. App. Gen. 2018, 131, 539–553. [Google Scholar] [CrossRef]
- Lee, K.H.; Park, S.W.; Kim, Y.J.; Koo, Y.J.; Song, J.T.; Seo, H.S. Grain width 2 (GW2) and its interacting proteins regulate seed development in rice (Oryza sativa L.). Bot. Stud. 2018, 59, 23. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.S.; Kim, Y.J.; Markkandan, K.I.D.; Koo, Y.J.; Song, J.T.; Seo, H.S. GW2 Functions as an E3 Ubiquitin Ligase for Rice Expansin-Like 1. Int. J. Mol. Sci. 2018, 19, 1904. [Google Scholar] [CrossRef]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-Type ubiquitin ligase family of Arabidopsis. Plant. Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef]
- Fagard, M.; Vaucheret, H. (Trans)gene silencing in plants: How Many Mechanisms? Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 2000, 51, 167–194. [Google Scholar] [CrossRef]
- Christensen, A.H.; Quail, P.H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef]
- Cseri, A.; Sass, L.; Törjék, O.; Pauk, J.; Vass, I.; Dudits, D. Monitoring drought responses of barley genotypes with semi-robotic phenotyping platform and association analysis between recorded traits and allelic variants of some stress genes. Aust. J. Crop. Sci. 2013, 7, 1560–1570. [Google Scholar]
- Fehér-Juhász, E.; Majer, P.; Sass, L.; Lantos, C.; Csiszár, J.; Turóczy, Z.; Mihály, R.; Mai, A.; Horváth, V.G.; Vass, I.; et al. Phenotyping shows improved physiological traits and seed yield of transgenic wheat plants expressing the alfalfa aldose reductase under permanent drought stress. Acta Physiol. Plant. 2014, 36, 663–673. [Google Scholar] [CrossRef]
- Hussein, A.; Tavakol, E.; Horner, D.S.; Muńoz-Amatriaín, M.; Muehlbauer, G.J.; Rossini, L. Genetics of Tillering in Rice and Barley. Plant. Genome 2014, 7, 1–20. [Google Scholar] [CrossRef]
- Liller, C.B.; Neuhaus, R.; von Korff, M.; Koornneef, M.; van Esse, W. Mutations in Barley Row Type Genes Have Pleiotropic Effects on Shoot Branching. PLoS ONE 2015, 10, e0140246. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cheng, Z.; Liu, M.; Yang, X.; Qiu, D. C3HC4-Type RING Finger Protein NbZFP1 is involved in growth and fruit development in Nicotiana benthamiana. PLoS ONE 2014, 9, e99352. [Google Scholar] [CrossRef]
- Boden, S.A.; Weiss, D.; Ross, J.J.; Davies, N.W.; Trevaskis, B.; Chandler, P.M.; Swain, S.M. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant. Cell 2014, 26, 1557–1569. [Google Scholar] [CrossRef]
- Yang, Y.; Fu, D.; Zhu, C.; He, Y.; Zhang, H.J.; Liu, T.; Li, X.; Wu, C. The RING-Finger Ubiquitin Ligase HAF1 Mediates Heading date 1 Degradation during Photoperiodic Flowering in Rice. Plant. Cell 2015, 27, 2455–2468. [Google Scholar] [CrossRef]
- Wu, X.; Tang, Y.; Li, C.; Wu, C. Characterization of the rate and duration of grain filling in wheat in southwestern China. Plant. Prod. Sci. 2018, 21, 358–369. [Google Scholar] [CrossRef]
- Youssef, H.M.; Hansson, M. Crosstalk among hormones in barley spike contributes to the yield. Plant. Cell Rep. 2019, 38, 1013–1016. [Google Scholar] [CrossRef]
- Walter, A.; Liebisch, F.; Hund, A. Plant phenotyping: From bean weighing to image analysis. Plant. Methods 2015, 11, 14. [Google Scholar] [CrossRef]
- Koiwai, H.; Tagiri, A.; Katoh, S.; Katoh, E.; Ichikawa, H.; Minami, E.; Nishizawa, Y. RING-H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. Plant. J. 2007, 51, 92–104. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Nagy, B.; Majer, P.; Mihály, R.; Pauk, J.; Horváth, V.G. Stress tolerance of transgenic barley accumulating the alfalfa aldose reductase in the cytoplasm and the chloroplast. Phytochemistry 2016, 129, 14–23. [Google Scholar] [CrossRef]
- Hausmann, L.; Toepfer, R. Entwicklung von Plasmidvektoren. Vorträge für Pflanzenzüchtung 1999, 45, 153–171. [Google Scholar]
- Hensel, G.; Kumlehn, J. Genetic transformation of barley (Hordeum vulgare L.) by co-culture of immature embryos with Agrobacteria. In Transgenic Crops World; Curtis, I.S., Ed.; Springer: Dordrecht, The Netherlands, 2004; pp. 35–44. [Google Scholar]
- Dudits, D.; Török, K.; Cseri, A.; Paul, K.; Nagy, A.V.V.; Nagy, B.; Sass, L.; Ferenc, G.; Vankova, R.; Dobrev, P.; et al. Response of Organ Structure and Physiology to Autotetraploidization in Early Development of Energy Willow Salix viminalis. Plant. Physiol. 2016, 170, 1504–1523. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Lendvai, Á.; Pettkó -Szandtner, A.; Csordás-Tóth, É.; Miskolczi, P.; Horváth, V.G.; Györgyey, J.; Dudits, D. Dicot and monocot plants differ in retinoblastoma-related protein subfamilies. J. Exp. Bot. 2007, 58, 1663–1675. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



 ) and PCR− plant (N° 10: = 28.07 mg,
) and PCR− plant (N° 10: = 28.07 mg,  ) segregants of ASHvYrg1/1 line.
) segregants of ASHvYrg1/1 line.
 ) and PCR− plant (N° 10: = 28.07 mg,
) and PCR− plant (N° 10: = 28.07 mg,  ) segregants of ASHvYrg1/1 line.
) segregants of ASHvYrg1/1 line.
| Genotype | Golden Promise | ASHvYrg 1/1 | ASHvYrg 1/2 | ASHvYrg 1/3 | ASHvYrg 1/4 | ASHvYrg 2/1 | ASHvYrg 2/2 | |
|---|---|---|---|---|---|---|---|---|
| Transcription | Normalized | 1 | 0.86 | 0.49 | 0.69 | 0.49 | 0.73 | 0.80 |
| Weight of 3 Flag Leaves (g) | MEAN | 0.15 | 0.19 *** | 0.19 ** | 0.17 | 0.17 | 0.25 | 0.15 |
| SD | 0.003 | 0.04 | 0.04 | 0.05 | 0.06 | 0.07 | 0.04 | |
| p-value | <0.001 | 0.001 | 0.111 | 0.085 | 0.75 | 0.638 | ||
| Shoot Number (main) | MEAN | 4.33 | 3.64 | 2.5 | 2.44 *** | 2.31 | 1.25 *** | 2.68 |
| SD | 0.98 | 1.45 | 0.798 | 1.54 | 1.5 | 0.46 | 1.15 | |
| p-value | 0.849 | 0.666 | <0.001 | 0.057 | <0.001 | 0.75 | ||
| Shoot Number (side) | MEAN | 1.88 | 2.50 | 3.58 ** | 5.39 *** | 4.38 *** | 3.62 ** | 3.33 ** |
| SD | 0.27 | 1.30 | 1.16 | 2.93 | 2.06 | 1.06 | 1.37 | |
| p-value | 0.208 | 0.0037 | <0.001 | <0.001 | 0.004 | 0.005 | ||
| Plant Weight (g) | MEAN | 9.02 | 11.02 ** | 8.90 | 8.61 | 10.17 * | 9.38 | 9.03 |
| SD | 0.92 | 1.39 | 1.07 | 1.76 | 2.05 | 1.41 | 1.7 | |
| p-value | 0.005 | 0.32 | 0.369 | 0.034 | 0.456 | 0.907 | ||
| Plant Height (cm) | MEAN | 65.06 | 70.47 ** | 60.92 | 70.50 | 70.75 * | 67.52 | 55.88 |
| SD | 11.26 | 6.21 | 4.70 | 7.25 | 7.30 | 7.67 | 7.74 | |
| p-value | 0.013 | 0.155 | 0.087 | 0.033 | 0.86 | 0.758 |
| Leaf Width (mm) | |||
|---|---|---|---|
| Genotypes | Mean | SD | p-Value |
| Golden Promise | 9.99 | 0.73 | - |
| ASHvYrg1/2 | 9.94 | 0.57 | 0.21 |
| ASHvYrg1/4 | 8.94 | 0.57 | 0.16 |
| ASHvYrg2/1 | 11.30 *** | 0.77 | <0.001 |
| ASHvYrg2/2 | 11.69 *** | 0.87 | <0.001 |
| Genotypes | Origin of Spikes | Kernel No/Spike | Kernel Weight/Spike (g) | Thousand Kernel Weight (g) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | p-Value | Mean | SD | p-Value | Mean | SD | p-Value | |||
| Golden Promise | main | 29.30 | 2.63 | 0.95 | 0.07 | 32.85 | 2.6 | ||||
| side | 23.50 | 4.03 | 0.47 | 0.11 | 21.26 | 3.25 | |||||
| ASHvYrg1/1 | main | PCR+ | 28.47 * | 2.27 | 0.94 * | 0.07 | 33.01 | 2.76 | |||
| PCR− | 26.00 | 1.87 | 0.039 | 0.85 | 0.05 | 0.017 | 33.09 | 3.77 | 0.962 | ||
| side | PCR+ | 19.57 | 4.86 | 0.54 | 0.14 | 27.8 *** | 3.80 | <0.001 | |||
| PCR− | 21.16 | 3.28 | 0.514 | 0.62 | 0.07 | 0.314 | 29.64 | 2.01 | 0.340 | ||
| ASHvYrg1/2 | main | PCR+ | 30.44 | 2.28 | 0.90 | 0.10 | 29.81 | 3.58 | |||
| PCR− | 30.50 | 0.94 | 0.958 | 0.84 | 0.02 | 0.170 | 27.85 | 0.47 | 0.248 | ||
| side | PCR+ | 25.73 | 2.05 | 0.57 | 0.11 | 26.39 *** | 4.50 | 0.002 | |||
| PCR− | 25.03 | 2.72 | 0.56 | 0.55 | 0.07 | 0.680 | 22.20 | 2.11 | 0.046 | ||
| ASHvYrg1/4 | main | PCR+ | 22.64 * | 4.08 | 0.77 | 0.09 | 32.07 * | 3.90 | |||
| PCR− | 28.13 | 0.01 | 0.012 | 0.78 | 0.04 | 0.803 | 28.09 | 0.87 | 0.04 | ||
| side | PCR+ | 18.16 | 4.91 | 0.48 | 0.14 | 28.87 *** | 4.94 | <0.001 | |||
| PCR− | 21.16 | 3.38 | 0.24 | 0.54 | 0.06 | 0.395 | 24.10 | 5.57 | 0.094 | ||
| Kernel Length (mm) | Kernel Width (mm) | |||||
|---|---|---|---|---|---|---|
| Genotypes | Mean | SD | p-Value | Mean | SD | p-Value |
| Golden Promise | 10.95 | 0.31 | 3.69 | 0.1 | ||
| ASHvYrg1/2 | 10.77 | 0.27 | 0.051 | 3.57 *** | 0.08 | <0.001 |
| ASHvYrg1/3 | 11.18 * | 0.37 | 0.041 | 3.85 *** | 0.12 | <0.001 |
| ASHvYrg1/4 | 12.66 *** | 0.57 | <0.001 | 4.03 *** | 0.15 | <0.001 |
| ASHvYrg2/1 | 11.62 *** | 0.34 | <0.001 | 3.93 *** | 0.12 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zombori, Z.; Nagy, B.; Mihály, R.; Pauk, J.; Cseri, A.; Sass, L.; Horváth V., G.; Dudits, D. RING-Type E3 Ubiqitin Ligase Barley Genes (HvYrg1–2) Control Characteristics of Both Vegetative Organs and Seeds as Yield Components. Plants 2020, 9, 1693. https://doi.org/10.3390/plants9121693
Zombori Z, Nagy B, Mihály R, Pauk J, Cseri A, Sass L, Horváth V. G, Dudits D. RING-Type E3 Ubiqitin Ligase Barley Genes (HvYrg1–2) Control Characteristics of Both Vegetative Organs and Seeds as Yield Components. Plants. 2020; 9(12):1693. https://doi.org/10.3390/plants9121693
Chicago/Turabian StyleZombori, Zoltán, Bettina Nagy, Róbert Mihály, János Pauk, András Cseri, László Sass, Gábor Horváth V., and Dénes Dudits. 2020. "RING-Type E3 Ubiqitin Ligase Barley Genes (HvYrg1–2) Control Characteristics of Both Vegetative Organs and Seeds as Yield Components" Plants 9, no. 12: 1693. https://doi.org/10.3390/plants9121693
APA StyleZombori, Z., Nagy, B., Mihály, R., Pauk, J., Cseri, A., Sass, L., Horváth V., G., & Dudits, D. (2020). RING-Type E3 Ubiqitin Ligase Barley Genes (HvYrg1–2) Control Characteristics of Both Vegetative Organs and Seeds as Yield Components. Plants, 9(12), 1693. https://doi.org/10.3390/plants9121693





