Response of Horticultural Soil Microbiota to Different Fertilization Practices
Abstract
1. Introduction
2. Results
2.1. Soil Bacterial Community Diversity
2.2. Soil Microbial Composition Correlation to Experimental Factors
2.3. Soil Prokaryotic Taxa Differences between Fertilization Practices
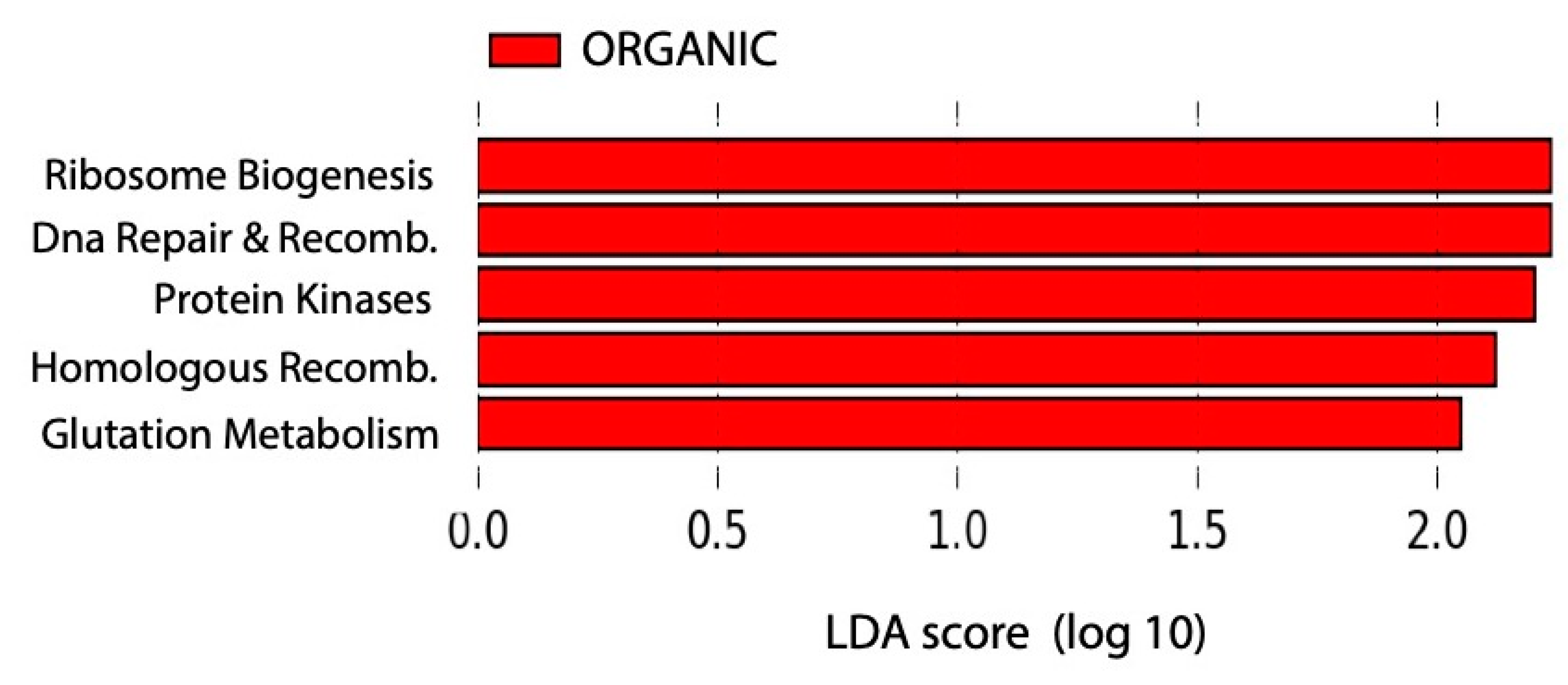
2.4. Predictions of Soil Microbial Communities’ Functions
3. Discussion
4. Materials and Methods
4.1. Field Experimental Design
4.2. Soil Sampling
4.3. DNA Extraction, PCR Amplification, and 16S rDNA Sequencing
4.4. Sequence Data Processing and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pingali, P.L. Green Revolution: Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. USA 2012, 109, 12302–12308. [Google Scholar] [CrossRef] [PubMed]
- Reganold, J.P.; Wachter, J.M. Organic agriculture in the twenty–First century. Nat. Plants 2016, 2, 15221. [Google Scholar] [CrossRef] [PubMed]
- Gomiero, T. Food quality assessment in organic vs. conventional agricultural produce: Findings and issues. Appl. Soil Ecol. 2018, 123, 714–728. [Google Scholar] [CrossRef]
- Reganold, J.P.; Andrews, P.K.; Reeve, J.R.; Carpenter-Boggs, L.; Schadt, C.W.; Alldredge, J.R.; Ross, C.F.; Davies, N.M.; Zhou, J. Fruit and Soil Quality of Organic and Conventional Strawberry Agroecosystems. PLoS ONE 2010, 5, e12346. [Google Scholar] [CrossRef]
- Lupatini, M.; Korthals, G.W.; de Hollander, M.; Janssens, T.K.S.; Kuramae, E.E. Soil Microbiome Is More Heterogeneous in Organic than in Conventional Farming System. Front. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Chen, H.; Xia, Q.; Yang, T.; Shi, W. Eighteen-Year Farming Management Moderately Shapes the Soil Microbial Community Structure but Promotes Habitat-Specific Taxa. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Pan, Y.; Cassman, N.; de Hollander, M.; Mendes, L.W.; Korevaar, H.; Geerts, R.H.E.M.; van Veen, J.A.; Kuramae, E.E. Impact of long-term N, P, K, and NPK fertilization on the composition and potential functions of the bacterial community in grassland soil. FEMS Microbiol. Ecol. 2014, 90, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Khafipour, E.; Krause, D.O.; Entz, M.H.; de Kievit, T.R.; Fernando, W.G.D. Pyrosequencing Reveals the Influence of Organic and Conventional Farming Systems on Bacterial Communities. PLoS ONE 2012, 7, e51897. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Kumar Bhatt, M.; Labanya, R.; Joshi, H.C. Influence of Long-term Chemical fertilizers and Organic Manures on Soil Fertility—A Review. Univers. J. Agric. Res. 2019, 7, 177–188. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine—Associated Microbiota. MBio 2015, 6, e02527-14. [Google Scholar] [CrossRef]
- Allard, S.M.; Walsh, C.S.; Wallis, A.E.; Ottesen, A.R.; Brown, E.W.; Micallef, S.A. Solanum lycopersicum (tomato) hosts robust phyllosphere and rhizosphere bacterial communities when grown in soil amended with various organic and synthetic fertilizers. Sci. Total Environ. 2016, 573, 555–563. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, X.; Wei, D.; Zhao, B.; Ma, M.; Chen, S.; Cao, F.; Shen, D.; Guan, D.; Li, J. Consistent effects of nitrogen fertilization on soil bacterial communities in black soils for two crop seasons in China. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Pershina, E.; Valkonen, J.; Kurki, P.; Ivanova, E.; Chirak, E.; Korvigo, I.; Provorov, N.; Andronov, E. Comparative Analysis of Prokaryotic Communities Associated with Organic and Conventional Farming Systems. PLoS ONE 2015, 10, e0145072. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Tu, C.; Hu, S.; Gumpertz, M.; Ristaino, J.B. Effect of organic, sustainable, and conventional management strategies in grower fields on soil physical, chemical, and biological factors and the incidence of Southern blight. Appl. Soil Ecol. 2007, 37, 202–214. [Google Scholar] [CrossRef]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef]
- Esperschütz, J.; Gattinger, A.; Mäder, P.; Schloter, M.; Fließbach, A. Response of soil microbial biomass and community structures to conventional and organic farming systems under identical crop rotations: Response of soil microbial biomass and community structures. FEMS Microbiol. Ecol. 2007, 61, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Gonthier, D.J.; Ennis, K.K.; Farinas, S.; Hsieh, H.-Y.; Iverson, A.L.; Batary, P.; Rudolphi, J.; Tscharntke, T.; Cardinale, B.J.; Perfecto, I. Biodiversity conservation in agriculture requires a multi-scale approach. Proc. R. Soc. B Biol. Sci. 2014, 281, 20141358. [Google Scholar] [CrossRef]
- Liñero, O.; Cidad, M.; Carrero, J.A.; Nguyen, C.; de Diego, A. Accumulation and Translocation of Essential and Nonessential Elements by Tomato Plants (Solanum lycopersicum) Cultivated in Open-Air Plots under Organic or Conventional Farming Techniques. J. Agric. Food Chem. 2015, 63, 9461–9470. [Google Scholar] [CrossRef]
- Liñero, O.; Cidad, M.; Carrero, J.A.; Nguyen, C.; de Diego, A. Partitioning of nutrients and non-essential elements in Swiss chards cultivated in open-air plots. J. Food Compos. Anal. 2017, 59, 179–187. [Google Scholar] [CrossRef]
- St-Pierre, B.; Wright, A.-D.G. Comparative metagenomic analysis of bacterial populations in three full-scale mesophilic anaerobic manure digesters. Appl. Microbiol. Biotechnol. 2014, 98, 2709–2717. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, X.-X.; Guo, X.; Wang, D.; Chu, H. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 2015, 88, 9–18. [Google Scholar] [CrossRef]
- Lin, W.; Lin, M.; Zhou, H.; Wu, H.; Li, Z.; Lin, W. The effects of chemical and organic fertilizer usage on rhizosphere soil in tea orchards. PLoS ONE 2019, 14, e0217018. [Google Scholar] [CrossRef]
- Bartram, A.K.; Jiang, X.; Lynch, M.D.J.; Masella, A.P.; Nicol, G.W.; Dushoff, J.; Neufeld, J.D. Exploring links between pH and bacterial community composition in soils from the Craibstone Experimental Farm. FEMS Microbiol. Ecol. 2014, 87, 403–415. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Hartman, W.H.; Richardson, C.J.; Vilgalys, R.; Bruland, G.L. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc. Natl. Acad. Sci. USA 2008. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340. [Google Scholar] [CrossRef]
- Yang, S.-X.; Liao, B.; Li, J.; Guo, T.; Shu, W.-S. Acidification, heavy metal mobility and nutrient accumulation in the soil–plant system of a revegetated acid mine wasteland. Chemosphere 2010, 80, 852–859. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Lyu, S.; Wei, X.; Chen, J.; Wang, C.; Wang, X.; Pan, D. Titanium as a Beneficial Element for Crop Production. Front. Plant Sci. 2017, 8, 597. [Google Scholar] [CrossRef]
- Gosling, P.; Hodge, A.; Goodlass, G.; Bending, G.D. Arbuscular mycorrhizal fungi and organic farming. Agric. Ecosyst. Environ. 2006, 113, 17–35. [Google Scholar] [CrossRef]
- Lehmann, A.; Rillig, M.C. Arbuscular mycorrhizal contribution to copper, manganese and iron nutrient concentrations in crops—A meta-analysis. Soil Biol. Biochem. 2015, 81, 147–158. [Google Scholar] [CrossRef]
- Miransari, M. Interactions between arbuscular mycorrhizal fungi and soil bacteria. Appl. Microbiol. Biotechnol. 2011, 89, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, H.; Feng, Y.; Wang, L.; Xiao, X.; Xi, Y.; Luo, X.; Sun, R.; Ye, X.; Huang, Y.; et al. Consistent responses of the microbial community structure to organic farming along the middle and lower reaches of the Yangtze River. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Ding, G.-C.; Bai, M.; Han, H.; Li, H.; Ding, X.; Yang, H.; Xu, T.; Li, J. Microbial taxonomic, nitrogen cycling and phosphorus recycling community composition during long-term organic greenhouse farming. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef]
- Li, H.; Cai, X.; Gong, J.; Xu, T.; Ding, G.; Li, J. Long-Term Organic Farming Manipulated Rhizospheric Microbiome and Bacillus Antagonism Against Pepper Blight (Phytophthora capsici). Front. Microbiol. 2019, 10, 342. [Google Scholar] [CrossRef]
- Sugiyama, A.; Vivanco, J.M.; Jayanty, S.S.; Manter, D.K. Pyrosequencing assessment of soil microbial communities in organic and conventional potato farms. Plant Dis. 2010, 94, 1329–1335. [Google Scholar] [CrossRef]
- Bull, C.T.; Shetty, K.G.; Subbarao, K.V. Interactions between Myxobacteria, Plant Pathogenic Fungi, and Biocontrol Agents. Plant Dis. 2002, 86, 889–896. [Google Scholar] [CrossRef]
- Chan, Y.-K.; McCormick, W.A.; Ma, B.L. Effects of inorganic fertilizer and manure on soil archaeal abundance at two experimental farms during three consecutive rotation-cropping seasons. Appl. Soil Ecol. 2013, 68, 26–35. [Google Scholar] [CrossRef]
- He, J.; Shen, J.; Zhang, L.; Zhu, Y.; Zheng, Y.; Xu, M.; Di, H. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 2007, 9, 2364–2374. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, L.; Zhu, Y.; Zhang, J.; He, J. Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam: Abundance and composition of AOB and AOA communities. Environ. Microbiol. 2008, 10, 1601–1611. [Google Scholar] [CrossRef]
- Carbonetto, B.; Rascovan, N.; Álvarez, R.; Mentaberry, A.; Vázquez, M.P. Structure, Composition and Metagenomic Profile of Soil Microbiomes Associated to Agricultural Land Use and Tillage Systems in Argentine Pampas. PLoS ONE 2014, 9, e99949. [Google Scholar] [CrossRef]
- Hossain, M.A.; Mostofa, M.G.; Diaz-Vivancos, P.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. (Eds.) Glutathione in Plant Growth, Development, and Stress Tolerance; Springer International Publishing: Cham, Switzerland, 2017; ISBN 978-3-319-66681-5. [Google Scholar]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.H. Vegan: Community Ecology Package, R Package Version 2.0-7. 2013. Available online: http://CRAN.R-project.org/package=vegan (accessed on 10 December 2017).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
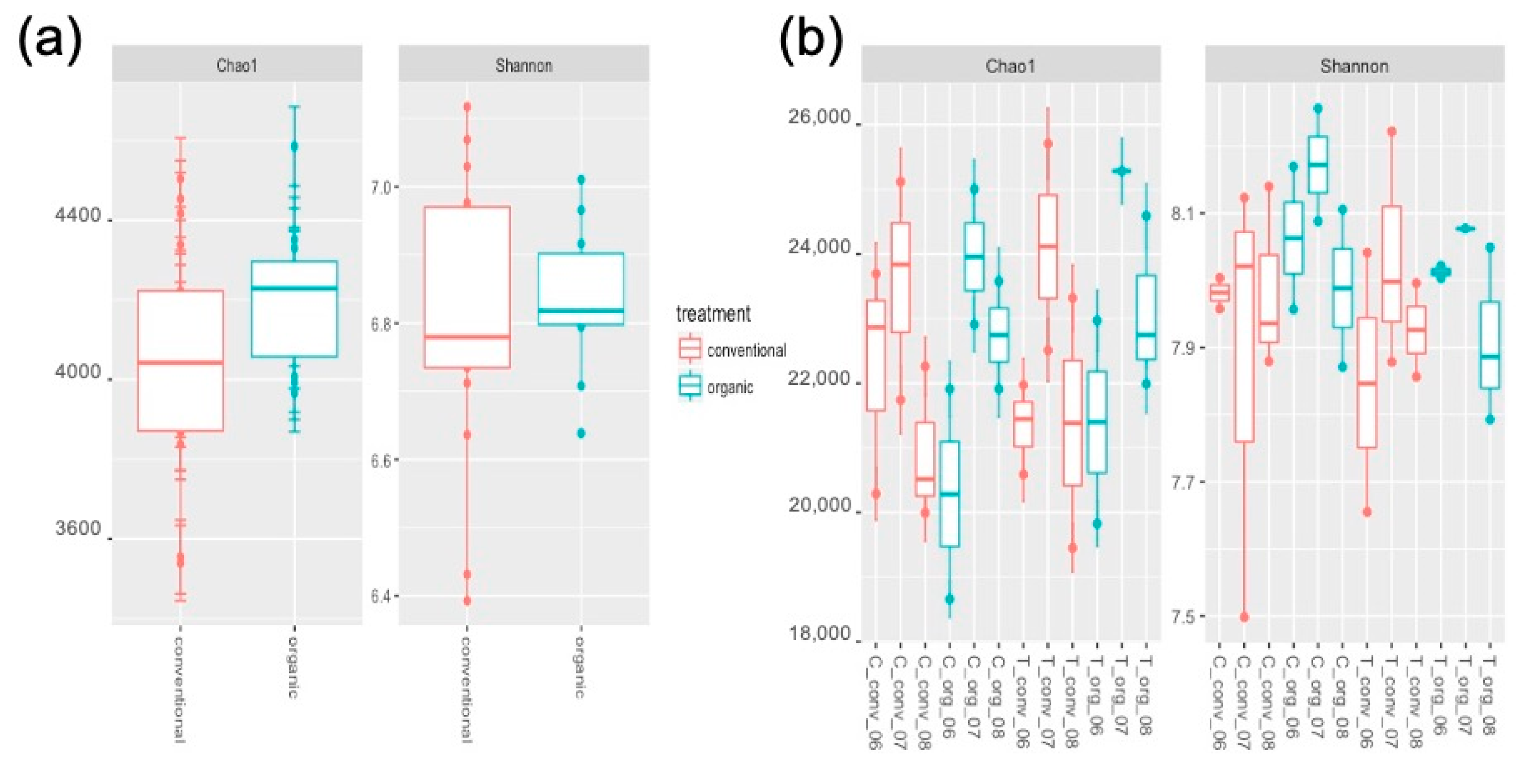
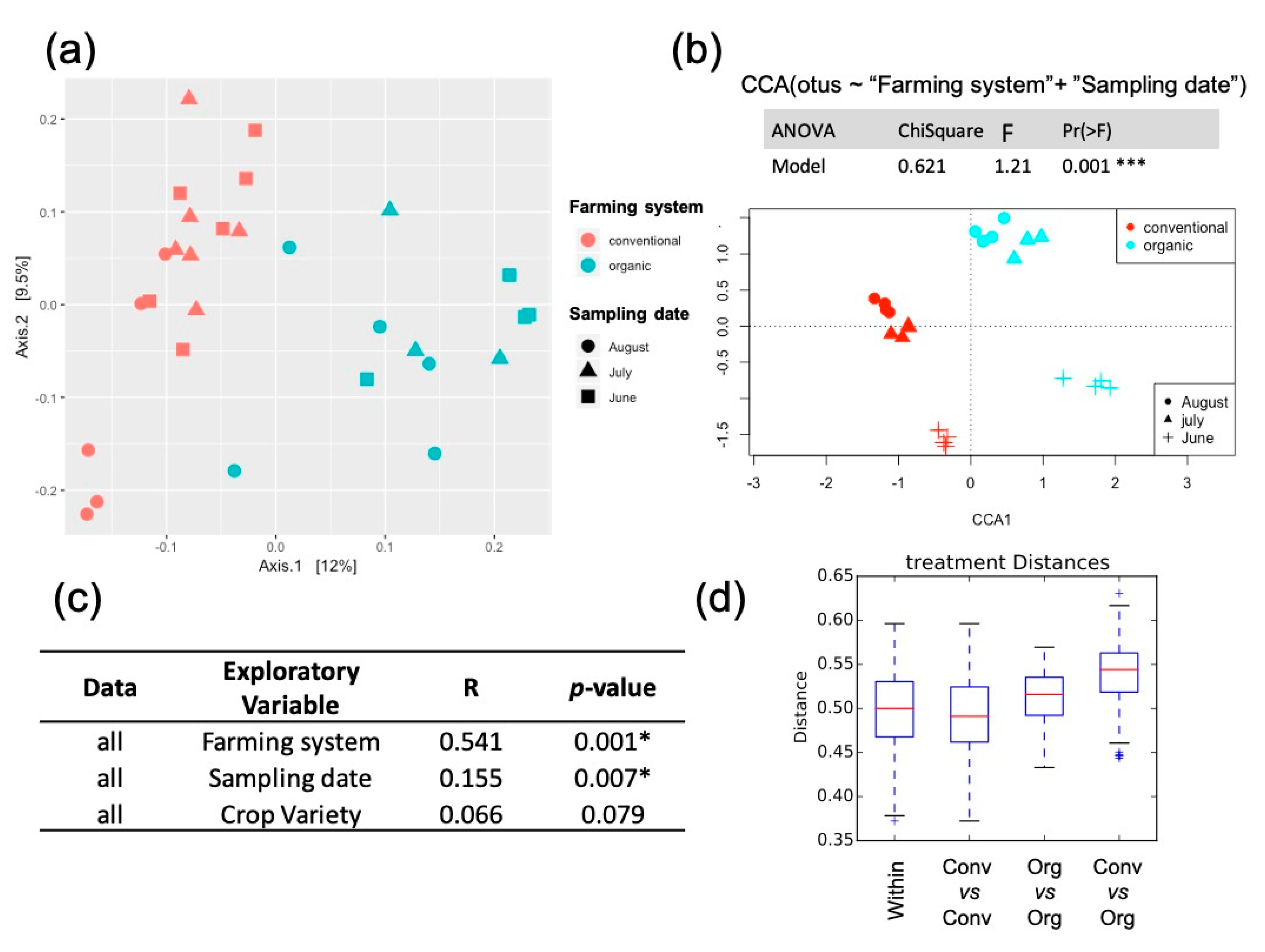
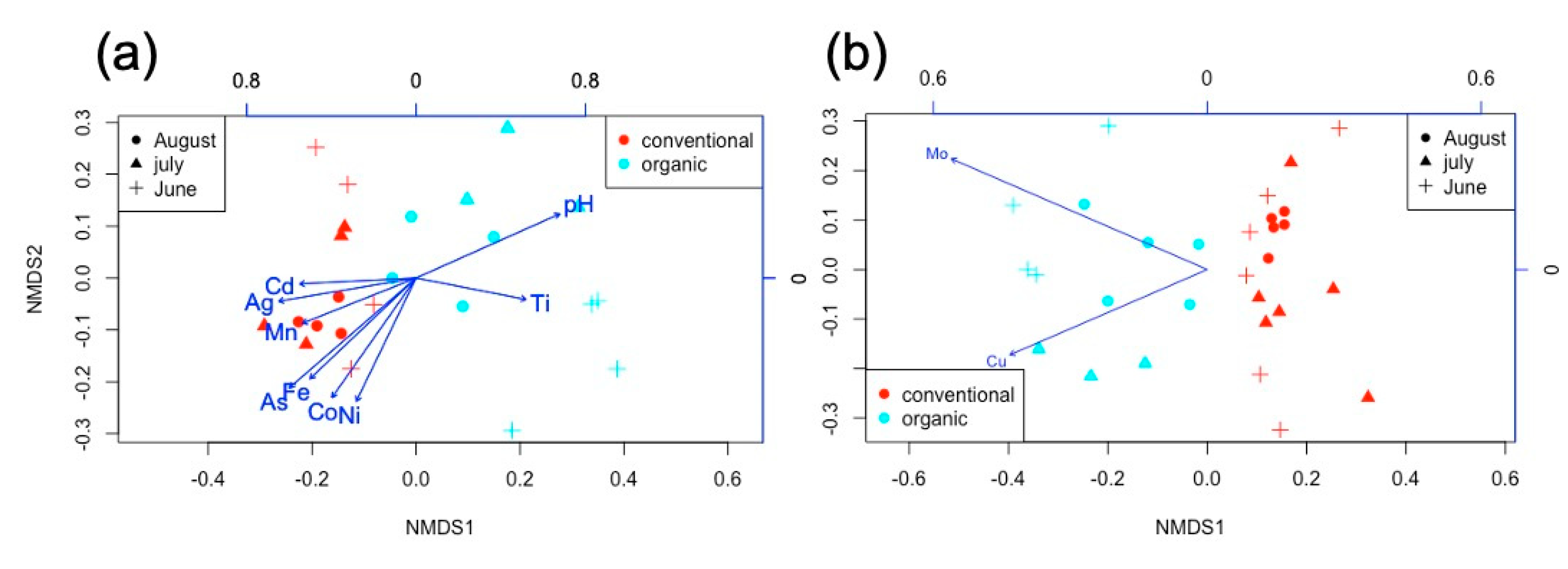
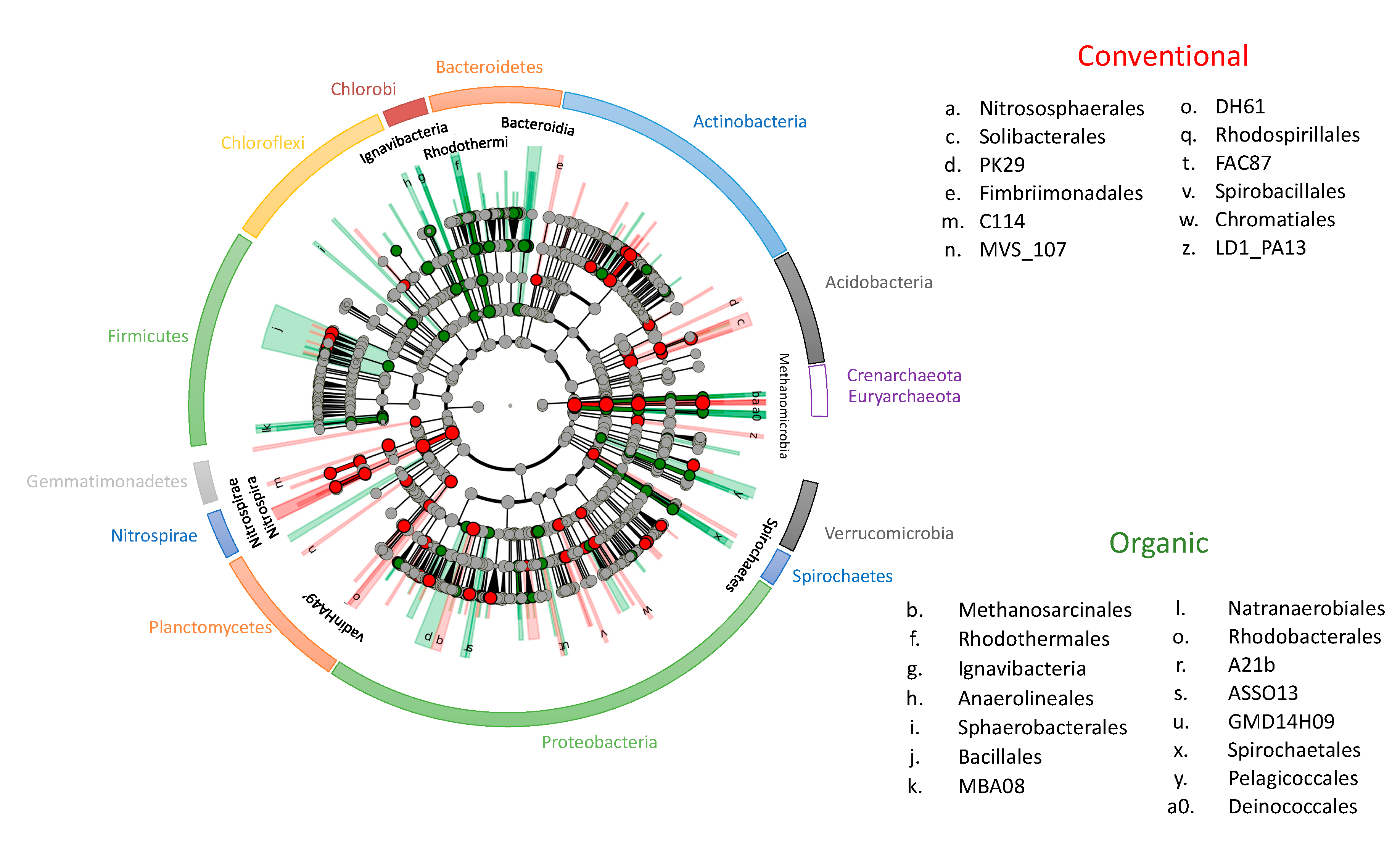
| Phyla | Class | Conventional Mean ± SD% | Organic Mean ± SD% |
|---|---|---|---|
| Firmicutes | Bacilli | 1.62 ± 1.04 a | 0.68 ± 0.30 |
| Acidobacteria | Solibacteres | 1.11 ± 0.40 a | 0.69 ± 0.14 |
| Planctomycetes | C6 | 0.06 ± 0.03 a | 0.02 ± 0.02 |
| Planctomycetia | 1.71 ± 0.24 a | 1.39 ± 0.29 | |
| vadinHA49 | 0.03 ± 0.02 a | 0.02 ± 0.01 | |
| Euryarchaeota | Methanomicrobia | 0.00 ± 0.00 | 0.00 ± 0.01 b |
| Proteobacteria | Deltaproteobacteria | 8.77 ± 1.62 | 11.93 ± 2.85 b |
| Chloroflexi | Anaerolineae | 0.35 ± 0.08 | 1.36 ± 0.97 b |
| Verrucomicrobia | Opitutae | 0.20 ± 0.10 | 0.47 ± 0.20 b |
| Fibrobacteres | Fibrobacteria | 0.04 ± 0.05 | 0.07 ± 0.03 b |
| Thermi | Deinococci | 0.00 ± 0.00 | 0.01 ± 0.01 b |
| Spirochaetes | Spirochaetes | 0.00 ± 0.01 | 0.03 ± 0.04 b |
| Bacteroidetes | Bacteroidia | 0.00 ± 0.00 | 0.13 ± 0.20 b |
| Rhodothermi | 0.00 ± 0.00 | 0.05 ± 0.04 b | |
| Chlorobi | Ignavibacteria | 0.00 ± 0.00 | 0.01 ± 0.01 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarraonaindia, I.; Martínez-Goñi, X.S.; Liñero, O.; Muñoz-Colmenero, M.; Aguirre, M.; Abad, D.; Baroja-Careaga, I.; Diego, A.d.; Gilbert, J.A.; Estonba, A. Response of Horticultural Soil Microbiota to Different Fertilization Practices. Plants 2020, 9, 1501. https://doi.org/10.3390/plants9111501
Zarraonaindia I, Martínez-Goñi XS, Liñero O, Muñoz-Colmenero M, Aguirre M, Abad D, Baroja-Careaga I, Diego Ad, Gilbert JA, Estonba A. Response of Horticultural Soil Microbiota to Different Fertilization Practices. Plants. 2020; 9(11):1501. https://doi.org/10.3390/plants9111501
Chicago/Turabian StyleZarraonaindia, Iratxe, Xabier Simón Martínez-Goñi, Olaia Liñero, Marta Muñoz-Colmenero, Mikel Aguirre, David Abad, Igor Baroja-Careaga, Alberto de Diego, Jack A. Gilbert, and Andone Estonba. 2020. "Response of Horticultural Soil Microbiota to Different Fertilization Practices" Plants 9, no. 11: 1501. https://doi.org/10.3390/plants9111501
APA StyleZarraonaindia, I., Martínez-Goñi, X. S., Liñero, O., Muñoz-Colmenero, M., Aguirre, M., Abad, D., Baroja-Careaga, I., Diego, A. d., Gilbert, J. A., & Estonba, A. (2020). Response of Horticultural Soil Microbiota to Different Fertilization Practices. Plants, 9(11), 1501. https://doi.org/10.3390/plants9111501







