Grapevine–Downy Mildew Rendezvous: Proteome Analysis of the First Hours of an Incompatible Interaction
Abstract
1. Introduction
2. Results and Discussion
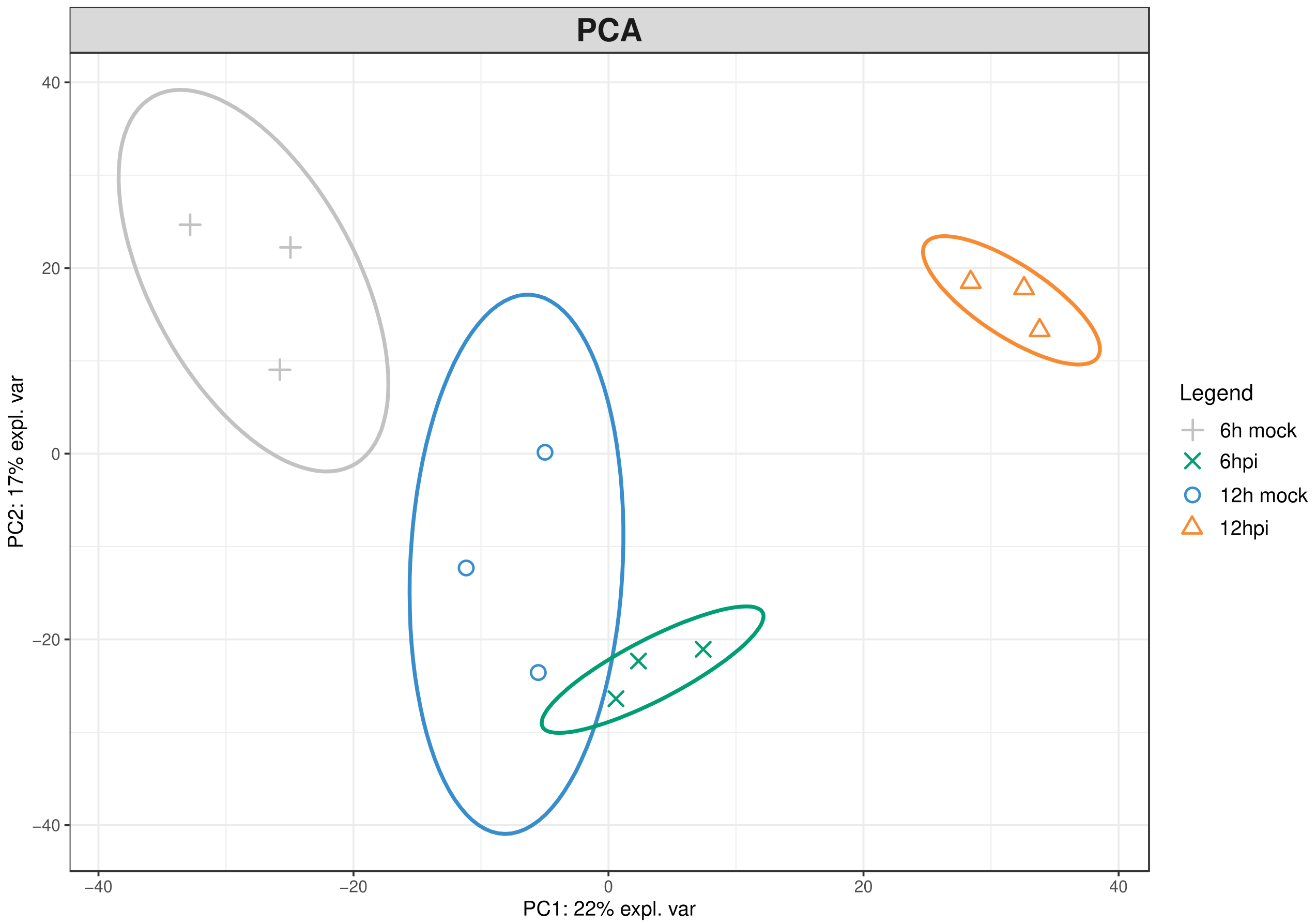
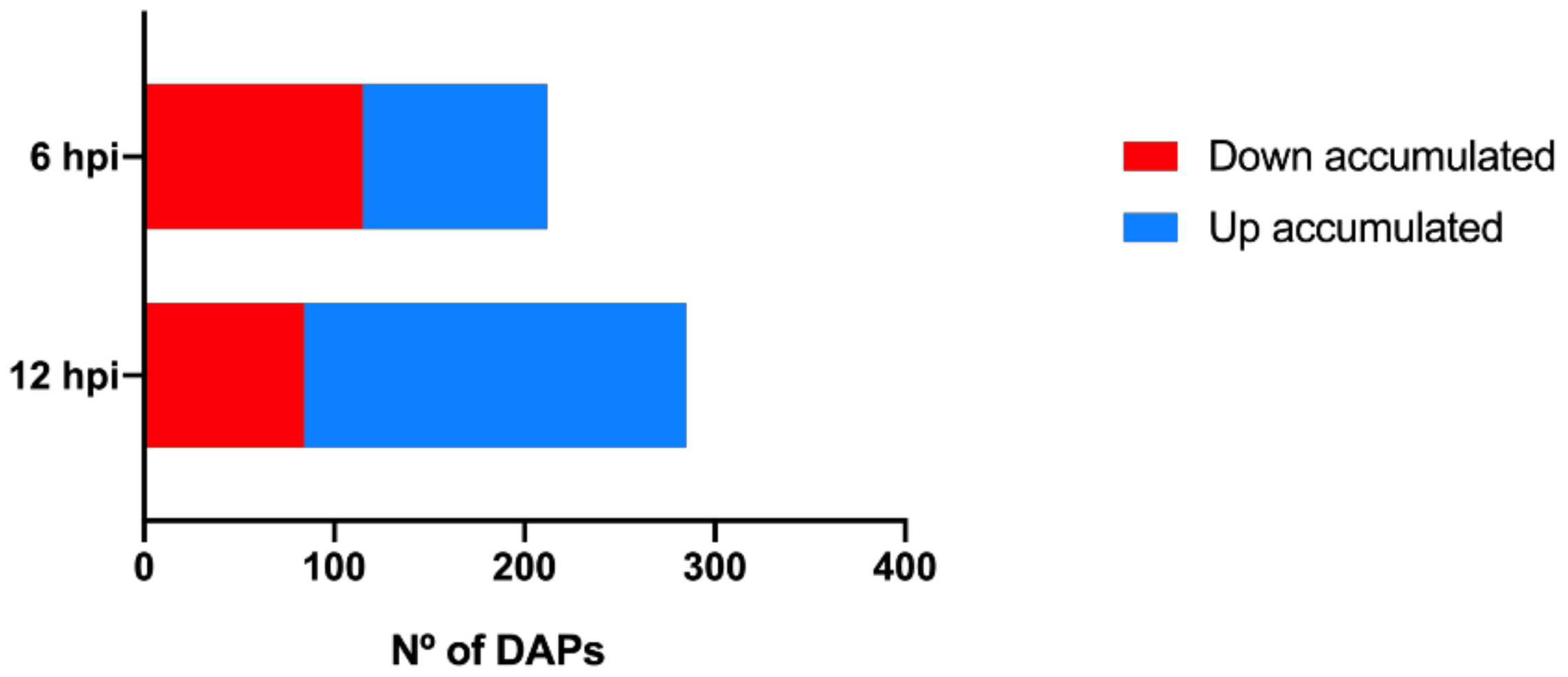
2.1. Proteomic Modulation in Grapevine Leaves in the First Hours after P. viticola Inoculation
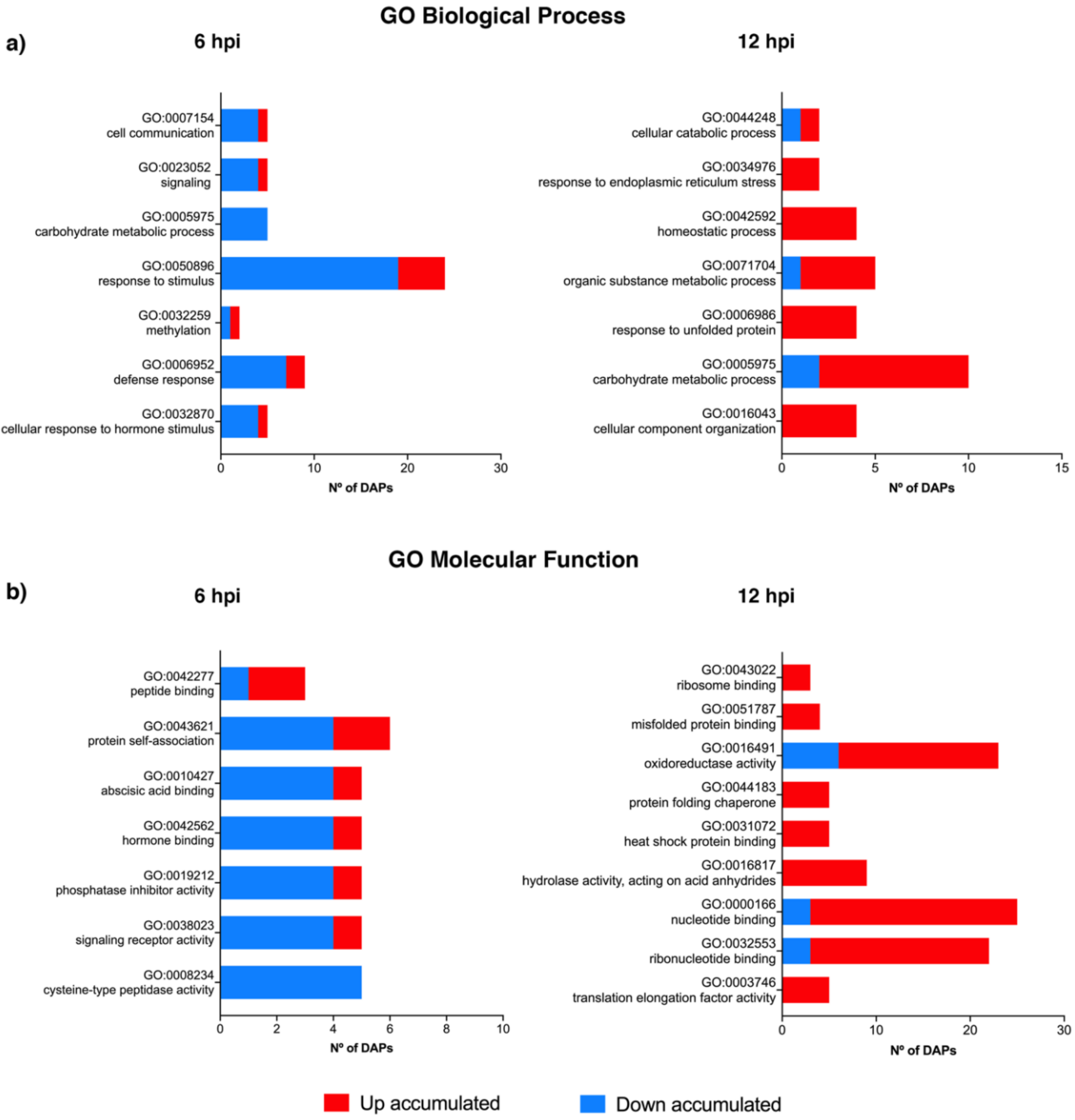
2.2. Translation and Protein Transport Processes Are Initially Boosted but Become Repressed at 12 hpi
2.3. Stress Related Proteins Are Negatively Affected at 6 hpi but Accumulated at 12 hpi
2.4. Several DAPs Are Significantly Repressed at Both Time Points
2.5. The DAPs Accumulated at Both Time Points of Grapevine–P. viticola Interaction Reflect Chloroplast Translation and Defense Response
3. Materials and Methods
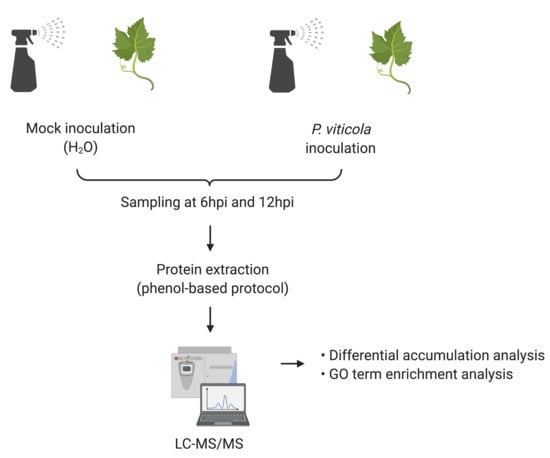
3.1. Plant Material and Inoculation Experiments
3.2. Sample Preparation
3.3. Liquid Chromatography Mass Spectrometry-Based Proteomics
3.4. Differential Accumulation Analysis
3.5. GO Term Enrichment Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ingle, R.A.; Carstens, M.; Denby, K.J. PAMP recognition and the plant-pathogen arms race. BioEssays 2006, 28, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Hein, I.; Gilroy, E.M.; Armstrong, M.R.; Birch, P.R.J.J. The zig-zag-zig in oomycete-plant interactions. Mol. Plant Pathol. 2009, 10, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2013, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- de Vries, S.; de Vries, J.; von Dahlen, J.K.; Gould, S.B.; Archibald, J.M.; Rose, L.E.; Slamovits, C.H. On plant defense signaling networks and early land plant evolution. Commun. Integr. Biol. 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fürst-Jansen, J.M.R.; de Vries, S.; de Vries, J. Evo-physio: On stress responses and the earliest land plants. J. Exp. Bot. 2020, 71, 3254–3269. [Google Scholar] [CrossRef] [PubMed]
- International Organization of Vine and Wine. 2019 Statistical Report on World Vitiviniculture; International Organization of Vine and Wine: Paris, France, 2019. [Google Scholar]
- Armijo, G.; Schlechter, R.; Agurto, M.; Muñoz, D.; Nuñez, C.; Arce-Johnson, P. Grapevine pathogenic microorganisms: Understanding infection strategies and host response scenarios. Front. Plant Sci. 2016, 7, 1–18. [Google Scholar] [CrossRef]
- Gessler, C.; Pertot, I.; Perazzolli, M. Plasmopara viticola: A review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 2011, 50, 3–44. [Google Scholar] [CrossRef]
- Peressotti, E.; Wiedemann-Merdinoglu, S.; Delmotte, F.; Bellin, D.; Di Gaspero, G.; Testolin, R.; Merdinoglu, D.; Mestre, P. Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 2010, 10, 147. [Google Scholar] [CrossRef]
- Delmotte, F.; Mestre, P.; Schneider, C.; Kassemeyer, H.H.; Kozma, P.; Richart-Cervera, S.; Rouxel, M.; Delière, L. Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: Evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect. Genet. Evol. 2014, 27, 500–508. [Google Scholar] [CrossRef]
- Bove, F.; Bavaresco, L.; Caffi, T.; Rossi, V. Assessment of Resistance Components for Improved Phenotyping of Grapevine Varieties Resistant to Downy Mildew. Front. Plant Sci. 2019, 10, 1559. [Google Scholar] [CrossRef]
- Casagrande, K.; Falginella, L.; Castellarin, S.D.; Testolin, R.; Di Gaspero, G. Defence responses in Rpv3-dependent resistance to grapevine downy mildew. Planta 2011, 234, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Díez-Navajas, A.M.; Wiedemann-Merdinoglu, S.; Greif, C.; Merdinoglu, D. Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology 2008, 98, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Polesani, M.; Bortesi, L.; Ferrarini, A.; Zamboni, A.; Fasoli, M.; Zadra, C.; Lovato, A.; Pezzotti, M.; Delledonne, M.; Polverari, A. General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genom. 2010, 11, 117. [Google Scholar] [CrossRef]
- Di Gaspero, G.; Cipriani, G.; Adam-Blondon, A.F.; Testolin, R. Linkage maps of grapevine displaying the chromosomal locations of 420 microsatellite markers and 82 markers for R-gene candidates. Theor. Appl. Genet. 2007, 114, 1249–1263. [Google Scholar] [CrossRef] [PubMed]
- Buonassisi, D.; Colombo, M.; Migliaro, D.; Dolzani, C.; Peressotti, E.; Mizzotti, C.; Velasco, R.; Masiero, S.; Perazzolli, M.; Vezzulli, S. Breeding for grapevine downy mildew resistance: A review of “omics” approaches. Euphytica 2017, 213, 1–21. [Google Scholar] [CrossRef]
- Guillier, C.; Gamm, M.; Lucchi, G.; Truntzer, C.; Pecqueur, D.; Ducoroy, P.; Adrian, M.; Héloir, M.C. Toward the identification of two glycoproteins involved in the stomatal deregulation of downy mildew-infected grapevine leaves. Mol. Plant-Microbe Interact. 2015, 28, 1227–1236. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Milli, A.; Cecconi, D.; Bortesi, L.; Persi, A.; Rinalducci, S.; Zamboni, A.; Zoccatelli, G.; Lovato, A.; Zolla, L.; Polverari, A. Proteomic analysis of the compatible interaction between Vitis vinifera and Plasmopara viticola. J. Proteom. 2012, 75, 1284–1302. [Google Scholar] [CrossRef]
- Rossin, G.; Villalta, D.; Martelli, P.; Cecconi, D.; Polverari, A.; Zoccatelli, G. Grapevine Downy Mildew Plasmopara viticola Infection Elicits the Expression of Allergenic Pathogenesis-Related Proteins. Int. Arch. Allergy Immunol. 2015, 168, 90–95. [Google Scholar] [CrossRef]
- Xu, P.; Liu, Y.; Qin, H.; Ai, J.; Fan, S.; Yang, Y.; Zhao, Y.; Li, X.; Li, X. Proteomic Analysis of the Resistant Responses of Two Vitis amurensis Cultivars to Plasmopara viticola Infections. Curr. Proteom. 2015, 12, 63–68. [Google Scholar] [CrossRef]
- Figueiredo, A.; Martins, J.; Sebastiana, M.; Guerreiro, A.; Silva, A.; Matos, A.R.; Monteiro, F.; Pais, M.S.; Roepstorff, P.; Coelho, A.V. Specific adjustments in grapevine leaf proteome discriminating resistant and susceptible grapevine genotypes to Plasmopara viticola. J. Proteom. 2017, 152, 48–57. [Google Scholar] [CrossRef]
- Nascimento-Gavioli, M.C.A.; Agapito-Tenfen, S.Z.; Nodari, R.O.; Welter, L.J.; Sanchez Mora, F.D.; Saifert, L.; da Silva, A.L.; Guerra, M.P. Proteome of Plasmopara viticola -infected Vitis vinifera provides insights into grapevine Rpv1/Rpv3 pyramided resistance to downy mildew. J. Proteom. 2017, 151, 264–274. [Google Scholar] [CrossRef]
- Valledor, L.; Jorrín, J. Back to the basics: Maximizing the information obtained by quantitative two dimensional gel electrophoresis analyses by an appropriate experimental design and statistical analyses. J. Proteom. 2011, 74, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Monteiro, F.; Fortes, A.M.; Bonow-Rex, M.; Zyprian, E.; Sousa, L.; Pais, M.S. Cultivar-specific kinetics of gene induction during downy mildew early infection in grapevine. Funct. Integr. Genom. 2012, 12, 379–386. [Google Scholar] [CrossRef]
- Warner, J.R.; McIntosh, K.B. How Common Are Extraribosomal Functions of Ribosomal Proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef]
- Solano-De La Cruz, M.T.; Adame-García, J.; Gregorio-Jorge, J.; Jiménez-Jacinto, V.; Vega-Alvarado, L.; Iglesias-Andreu, L.G.; Escobar-Hernández, E.E.; Luna-Rodríguez, M. Functional categorization of de novo transcriptome assembly of Vanilla planifolia Jacks. potentially points to a translational regulation during early stages of infection by Fusarium oxysporum f. sp. vanillae. BMC Genom. 2019, 20, 826. [Google Scholar] [CrossRef] [PubMed]
- Di, R.; Tumer, N.E. Expression of a truncated form of ribosomal protein L3 confers resistance to pokeweed antiviral protein and the Fusarium mycotoxin deoxynivalenol. Mol. Plant-Microbe Interact. 2005, 18, 762–770. [Google Scholar] [CrossRef]
- Shimizu, T.; Satoh, K.; Kikuchi, S.; Omura, T. The repression of cell wall- and plastid-related genes and the induction of defense-related genes in rice plants infected with Rice dwarf virus. Mol. Plant-Microbe Interact. 2007, 20, 247–254. [Google Scholar] [CrossRef]
- Kirstein-Miles, J.; Scior, A.; Deuerling, E.; Morimoto, R.I. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 2013, 32, 1451–1468. [Google Scholar] [CrossRef]
- Elagamey, E.; Narula, K.; Sinha, A.; Ghosh, S.; Abdellatef, M.A.E.; Chakraborty, N.; Chakraborty, S. Quantitative Extracellular Matrix Proteomics Suggests Cell Wall Reprogramming in Host-Specific Immunity During Vascular Wilt Caused by Fusarium oxysporum in Chickpea. Proteomics 2017, 17, 1600374. [Google Scholar] [CrossRef]
- Hajheidari, M.; Abdollahian-Noghabi, M.; Askari, H.; Heidari, M.; Sadeghian, S.Y.; Ober, E.S.; Salekdeh, G.H. Proteome analysis of sugar beet leaves under drought stress. Proteomics 2005, 5, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wu, J.; Xue, A.; Li, W.; Chen, W.; Wei, L.; Lv, H.; Lin, S.; Fan, S.; Li, N.; et al. Differentially Expressed Genes of Soybean During Infection by Phytophthora sojae. J. Integr. Agric. 2012, 11, 368–377. [Google Scholar] [CrossRef]
- Lemaître-Guillier, C.; Hovasse, A.; Schaeffer-Reiss, C.; Recorbet, G.; Poinssot, B.; Trouvelot, S.; Daire, X.; Adrian, M.; Héloir, M.C. Proteomics towards the understanding of elicitor induced resistance of grapevine against downy mildew. J. Proteom. 2017, 156, 113–125. [Google Scholar] [CrossRef]
- Sireesha, Y.; Velazhahan, R. Analysis of defense genes expression in maize upon infection with Peronosclerospora sorghi. Cereal Res. Commun. 2017, 45, 272–283. [Google Scholar] [CrossRef][Green Version]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant-Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Khorramdelazad, M.; Bar, I.; Whatmore, P.; Smetham, G.; Bhaaskaria, V.; Yang, Y.; Bai, S.H.; Mantri, N.; Zhou, Y.; Ford, R. Transcriptome profiling of lentil (Lens culinaris) through the first 24 hours of Ascochyta lentis infection reveals key defence response genes. BMC Genom. 2018, 19, 108. [Google Scholar] [CrossRef]
- Sierla, M.; Hõrak, H.; Overmyer, K.; Waszczak, C.; Yarmolinsky, D.; Maierhofer, T.; Vainonen, J.P.; Salojärvi, J.; Denessiouk, K.; Laanemets, K.; et al. The receptor-like pseudokinase GHR1 is required for stomatal closure. Plant Cell 2018, 30, 2813–2837. [Google Scholar] [CrossRef]
- Wang, S.; Lei, C.; Wang, J.; Ma, J.; Tang, S.; Wang, C.; Zhao, K.; Tian, P.; Zhang, H.; Qi, C.; et al. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 2017, 68, 899–913. [Google Scholar] [CrossRef]
- Tang, C.; Deng, L.; Chang, D.; Chen, S.; Wang, X.; Kang, Z. TaADF3, an Actin-Depolymerizing factor, negatively modulates wheat resistance against Puccinia striiformis. Front. Plant Sci. 2016, 6, 1214. [Google Scholar] [CrossRef]
- Moreno, J.I.; Martín, R.; Castresana, C. Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 2005, 41, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Resjö, S.; Zahid, M.A.; Burra, D.D.; Lenman, M.; Levander, F.; Andreasson, E. Proteomics of PTI and two ETI immune reactions in potato leaves. Int. J. Mol. Sci. 2019, 20, 4726. [Google Scholar] [CrossRef]
- Boller, T.; He, S.Y. Innate Immunity in Plants: An Arms Race Between Pattern Recognition Receptors in Plants and Effectors in Microbial Pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef]
- Jelenska, J.; Van Hal, J.A.; Greenberg, J.T. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Natl. Acad. Sci. USA 2010, 107, 13177–13182. [Google Scholar] [CrossRef]
- Kallamadi, P.R.; Dandu, K.; Kirti, P.B.; Rao, C.M.; Thakur, S.S.; Mulpuri, S. An Insight into Powdery Mildew–Infected, Susceptible, Resistant, and Immune Sunflower Genotypes. Proteomics 2018, 18, 1700418. [Google Scholar] [CrossRef]
- Sirover, M.A. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: Biochemical mechanisms and regulatory control. Biochim. Biophys. Acta Gen. Subj. 2011, 1810, 741–751. [Google Scholar] [CrossRef]
- Meng, X.; Song, T.; Fan, H.; Yu, Y.; Cui, N.; Zhao, J.; Meng, K. A comparative cell wall proteomic analysis of cucumber leaves under Sphaerotheca fuliginea stress. Acta Physiol. Plant. 2016, 38, 260. [Google Scholar] [CrossRef]
- Roshan, P.; Kulshreshtha, A.; Hallan, V. Identification of host cellular targets of AC4 and AV2 proteins of tomato leaf curl palampur virus and their sub-cellular localization studies. VirusDisease 2017, 28, 390–400. [Google Scholar] [CrossRef]
- Wang, R.Y.-L.; Nagy, P.D. Tomato bushy stunt virus Co-Opts the RNA-Binding Function of a Host Metabolic Enzyme for Viral Genomic RNA Synthesis. Cell Host Microbe 2008, 3, 178–187. [Google Scholar] [CrossRef]
- Prasanth, K.R.; Huang, Y.-W.; Liou, M.-R.; Wang, R.Y.-L.; Hu, C.-C.; Tsai, C.-H.; Meng, M.; Lin, N.-S.; Hsu, Y.-H. Glyceraldehyde 3-Phosphate Dehydrogenase Negatively Regulates the Replication of Bamboo Mosaic Virus and Its Associated Satellite RNA. J. Virol. 2011, 85, 8829–8840. [Google Scholar] [CrossRef]
- Li, X.; Bi, X.; An, M.; Xia, Z.; Wu, Y. iTRAQ-Based Proteomic Analysis of Watermelon Fruits in Response to Cucumber green mottle mosaic virus Infection. Int. J. Mol. Sci. 2020, 21, 2541. [Google Scholar] [CrossRef]
- Liu, B.; Qian, S.B. Translational reprogramming in cellular stress response. WIREs RNA 2014, 5, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Liu, Y.; Wu, Y.; Xie, Q. The sHSP22 heat shock protein requires the ABI1 protein phosphatase to modulate polar auxin transport and downstream responses. Plant Physiol. 2018, 176, 2406–2425. [Google Scholar] [CrossRef]
- Haider, M.S.; Zhang, C.; Kurjogi, M.M.; Pervaiz, T.; Zheng, T.; Zhang, C.; Lide, C.; Shangguan, L.; Fang, J. Insights into grapevine defense response against drought as revealed by biochemical, physiological and RNA-Seq analysis. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Zhu, C.; Li, X.; Zheng, J. Transcriptome profiling using Illumina- and SMRT-based RNA-seq of hot pepper for in-depth understanding of genes involved in CMV infection. Gene 2018, 666, 123–133. [Google Scholar] [CrossRef]
- Bilgin, D.D.; Zavala, J.A.; Zhu, J.; Clough, S.J.; Ort, D.R.; Delucia, E.H. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 2010, 33, 1597–1613. [Google Scholar] [CrossRef]
- Biochemistry and Molecular Biology of Plants, 2nd ed.; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; Wiley: Hoboken, NJ, USA, 2015; ISBN 978-0-470-71421-8. [Google Scholar]
- Yu, H.-D.; Yang, X.-F.; Chen, S.-T.; Wang, Y.-T.; Li, J.-K.; Shen, Q.; Liu, X.-L.; Guo, F.-Q. Downregulation of Chloroplast RPS1 Negatively Modulates Nuclear Heat-Responsive Expression of HsfA2 and Its Target Genes in Arabidopsis. PLoS Genet. 2012, 8, e1002669. [Google Scholar] [CrossRef]
- Wu, L.; Han, Z.; Wang, S.; Wang, X.; Sun, A.; Zu, X.; Chen, Y. Comparative proteomic analysis of the plant-virus interaction in resistant and susceptible ecotypes of maize infected with sugarcane mosaic virus. J. Proteom. 2013, 89, 124–140. [Google Scholar] [CrossRef]
- Samaniego, R.; Jeong, S.Y.; Meier, I.; Díaz De La Espina, S.M. Dual location of MAR-binding, filament-like protein 1 in Arabidopsis, tobacco, and tomato. Planta 2006, 223, 1201–1206. [Google Scholar] [CrossRef]
- Chen, F.; Li, Q.; He, Z. Proteomic analysis of rice plasma membrane-associated proteins in response to chitooligosaccharide elicitors. J. Integr. Plant Biol. 2007, 49, 863–870. [Google Scholar] [CrossRef]
- Van Aubel, G.; Buonatesta, R.; Cutsem, P. Van COS-OGA, a new oligosaccharidic elicitor that induces protection against a wide range of plant pathogens. IOBC-WPRS Bull. 2013, 89, 403–407. [Google Scholar]
- van Aubel, G.; Buonatesta, R.; Van Cutsem, P. COS-OGA: A novel oligosaccharidic elicitor that protects grapes and cucumbers against powdery mildew. Crop Prot. 2014, 65, 129–137. [Google Scholar] [CrossRef]
- Su, T.; Li, W.; Wang, P.; Ma, C. Dynamics of peroxisome homeostasis and its role in stress response and signaling in plants. Front. Plant Sci. 2019, 10, 705. [Google Scholar] [CrossRef]
- Hao, J.; Wu, W.; Wang, Y.; Yang, Z.; Liu, Y.; Lv, Y.; Zhai, Y.; Yang, J.; Liang, Z.; Huang, K.; et al. Arabidopsis thaliana defense response to the ochratoxin A-producing strain (Aspergillus ochraceus 3.4412). Plant Cell Rep. 2015, 34, 705–719. [Google Scholar] [CrossRef]
- Agostini, R.B.; Postigo, A.; Rius, S.P.; Rech, G.E.; Campos-Bermudez, V.A.; Vargas, W.A. Long-Lasting Primed State in Maize Plants: Salicylic Acid and Steroid Signaling Pathways as Key Players in the Early Activation of Immune Responses in Silks. Mol. Plant-Microbe Interact. 2019, 32, 95–106. [Google Scholar] [CrossRef]
- Monteiro, S.; Barakat, M.; Piçarra-Pereira, M.A.; Teixeira, A.R.; Ferreira, R.B. Osmotin and Thaumatin from Grape: A Putative General Defense Mechanism Against Pathogenic Fungi. Phytopathology 2003, 93, 1505–1512. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Yang, Y.; Li, T.; Lu, W.; Du, Y.; Qiang, X.; Wen, Q.; Shan, W. A Phytophthora capsici RXLR Effector Targets and Inhibits a Plant PPIase to Suppress Endoplasmic Reticulum-Mediated Immunity. Mol. Plant 2018, 11, 1067–1083. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, Y.; Bi, C.; Kang, Z. Quantitative proteomics reveals the defense response of wheat against Puccinia striiformis f. sp. tritici. Sci. Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef]
- Godoy, A.V.; Lazzaro, A.S.; Casalongué, C.A.; San Segundo, B. Expression of a Solanum tuberosum cyclophilin gene is regulated by fungal infection and abiotic stress conditions. Plant Sci. 2000, 152, 123–134. [Google Scholar] [CrossRef]
- Kong, H.Y.; Lee, S.C.; Hwang, B.K. Expression of pepper cyclophilin gene is differentially regulated during the pathogen infection and abiotic stress conditions. Physiol. Mol. Plant Pathol. 2001, 59, 189–199. [Google Scholar] [CrossRef]
- Park, S.C.; Jung, R.L.; Shin, S.O.; Ji, H.J.; Young, M.L.; Son, H.; Park, Y.; Sang, Y.L.; Hahm, K.S. Purification and characterization of an antifungal protein, C-FKBP, from Chinese cabbage. J. Agric. Food Chem. 2007, 55, 5277–5281. [Google Scholar] [CrossRef]
- Park, S.-W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.R.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef]
- Kang, M.; Lee, S.; Abdelmageed, H.; Reichert, A.; Lee, H.K.; Fokar, M.; Mysore, K.S.; Allen, R.D. Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 2017, 40, 702–716. [Google Scholar] [CrossRef]
- Petriccione, M.; Salzano, A.M.; Di Cecco, I.; Scaloni, A.; Scortichini, M. Proteomic analysis of the Actinidia deliciosa leaf apoplast during biotrophic colonization by Pseudomonas syringae pv. actinidiae. J. Proteom. 2014, 101, 43–62. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Wang, H.; Bao, Y.; Zhang, W. Examination of the leaf proteome during flooding stress and the induction of programmed cell death in maize. Proteome Sci. 2014, 12, 33. [Google Scholar] [CrossRef]
- Moura, H.F.N.; Vasconcelos, I.M.; Souza, C.E.A.; Silva, F.D.A.; Moreno, F.B.M.B.; Lobo, M.D.P.; Monteiro-Moreira, A.C.O.; Moura, A.A.; Costa, J.H.; Oliveira, J.T.A. Proteomics changes during the incompatible interaction between cowpea and Colletotrichum gloeosporioides (Penz.) Penz and Sacc. Plant Sci. 2014, 217–218, 158–175. [Google Scholar] [CrossRef]
- Welter, L.J.; Göktürk-Baydar, N.; Akkurt, M.; Maul, E.; Eibach, R.; Töpfer, R.; Zyprian, E.M. Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol. Breed. 2007, 20, 359–374. [Google Scholar] [CrossRef]
- Kortekamp, A. Expression analysis of defence-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiol. Biochem. 2006, 44, 58–67. [Google Scholar] [CrossRef]
- Sebastiana, M.; Figueiredo, A.; Monteiro, F.; Martins, J.; Franco, C.; Coelho, A.V.; Vaz, F.; Simões, T.; Penque, D.; Pais, M.S.; et al. A possible approach for gel-based proteomic studies in recalcitrant woody plants. Springerplus 2013, 2, 210. [Google Scholar] [CrossRef]
- Llombart, V.; García-Berrocoso, T.; Bech-Serra, J.J.; Simats, A.; Bustamante, A.; Giralt, D.; Reverter-Branchat, G.; Canals, F.; Hernández-Guillamon, M.; Montaner, J. Characterization of secretomes from a human blood brain barrier endothelial cells in-vitro model after ischemia by stable isotope labeling with aminoacids in cell culture (SILAC). J. Proteom. 2016, 133, 100–112. [Google Scholar] [CrossRef]
- Zhang, X.; Smits, A.H.; Van Tilburg, G.B.A.; Ovaa, H.; Huber, W.; Vermeulen, M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 2018, 13, 530–550. [Google Scholar] [CrossRef]
- Huber, W.; von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef]
- Winter, D.J. Rentrez: An R package for the NCBI eUtils API. R J. 2017, 9, 520. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.40.0. Bioconductor. Available online: http://www.bioconductor.org/packages/release/bioc/html/topGO.html (accessed on 4 November 2020).
| Protein Name | Protein Code (NCBI Database) | log2 (FC) | |
|---|---|---|---|
| 6 hpi | 12 hpi | ||
| PREDICTED: 50S ribosomal protein L3, chloroplastic (Vitis vinifera) | XP_002271466.1 | 2.4 | −2.3 |
| PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 (Vitis vinifera) | XP_003634163.1 | 3.4 | −1.6 |
| PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 1 (Vitis vinifera) | XP_003632619.1 | 1.3 | −2.2 |
| PREDICTED: major allergen Pru ar 1 (Vitis vinifera) | XP_002273790.2 | 3.0 | −1.5 |
| serine/threonine kinase-like (Vitis vinifera) | NP_001268124.1 | 1.5 | −2.1 |
| Protein Name | Protein Code (NCBI Database) | log2 (FC) | |
|---|---|---|---|
| 6 hpi | 12 hpi | ||
| PREDICTED: aminomethyltransferase, mitochondrial (Vitis vinifera) | XP_002272701.1 | −1.7 | 5.4 |
| PREDICTED: elongation factor 1-alpha (Vitis vinifera) | XP_002277159.1 | −3.1 | 2.6 |
| PREDICTED: actin-depolymerizing factor 2 (Vitis vinifera) | XP_002284292.1 | −4.2 | 2.9 |
| PREDICTED: serine hydroxymethyltransferase, mitochondrial (Vitis vinifera) | XP_010646402.1 | −4.3 | 2.3 |
| PREDICTED: heat shock 70 kDa protein, mitochondrial isoform X1 (Vitis vinifera) | XP_002263457.1 | −2.1 | 2.1 |
| PREDICTED: glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic (Vitis vinifera) | XP_002273754.1 | −0.9 | 2.2 |
| Protein Name | Protein Code (NCBI Database) | log2 (FC) | |
|---|---|---|---|
| 6 hpi | 12 hpi | ||
| PREDICTED: 40S ribosomal protein S14 (Vitis vinifera) | XP_002274381.1 | −1.7 | −3.3 |
| PREDICTED: 22.0 kDa class IV heat shock protein (Vitis vinifera) | XP_002263376.1 | −3.2 | −2.8 |
| PREDICTED: H/ACA ribonucleoprotein complex subunit 1 (Vitis vinifera) | XP_002277849.1 | −2.1 | −4.0 |
| photosystem II CP43 chlorophyll apoprotein (chloroplast) (Vitis vinifera) | ABE47530.1 | −2.8 | −4.1 |
| PREDICTED: thiol protease aleurain-like isoform X1 (Vitis vinifera) | XP_002278624.1 | −6.6 | −1.4 |
| Protein Name | Protein Code (NCBI Database) | log2 (FC) | |
|---|---|---|---|
| 6 hpi | 12 hpi | ||
| PREDICTED: 50S ribosomal protein L1, chloroplastic (Vitis vinifera) | XP_002274498.1 | 2.0 | 1.3 |
| PREDICTED: 30S ribosomal protein S1, chloroplastic (Vitis vinifera) | XP_002280604.1 | 1.7 | 2.4 |
| PREDICTED: MAR-binding filament-like protein 1-1 isoform X1 (Vitis vinifera) | XP_002284745.2 | 3.9 | 2.2 |
| PREDICTED: peroxisome biogenesis protein 19-2 (Vitis vinifera) | XP_002269360.1 | 2.3 | 3.0 |
| PREDICTED: protein P21 (Vitis vinifera) | XP_002283030.1 | 2.3 | 3.0 |
| PREDICTED: peptidyl-prolyl cis-trans isomerase (Vitis vinifera) | XP_002273421.2 | 2.1 | 3.4 |
| PREDICTED: ubiquitin receptor RAD23c (Vitis vinifera) | XP_002283656.1 | 2.8 | 2.4 |
| PREDICTED: fruit protein pKIWI502 (Vitis vinifera) | XP_002283966.1 | 2.0 | 3.9 |
| PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At3g48420 (Vitis vinifera) | XP_002277650.1 | 2.7 | 6.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
B. Santos, R.; Nascimento, R.; V. Coelho, A.; Figueiredo, A. Grapevine–Downy Mildew Rendezvous: Proteome Analysis of the First Hours of an Incompatible Interaction. Plants 2020, 9, 1498. https://doi.org/10.3390/plants9111498
B. Santos R, Nascimento R, V. Coelho A, Figueiredo A. Grapevine–Downy Mildew Rendezvous: Proteome Analysis of the First Hours of an Incompatible Interaction. Plants. 2020; 9(11):1498. https://doi.org/10.3390/plants9111498
Chicago/Turabian StyleB. Santos, Rita, Rui Nascimento, Ana V. Coelho, and Andreia Figueiredo. 2020. "Grapevine–Downy Mildew Rendezvous: Proteome Analysis of the First Hours of an Incompatible Interaction" Plants 9, no. 11: 1498. https://doi.org/10.3390/plants9111498
APA StyleB. Santos, R., Nascimento, R., V. Coelho, A., & Figueiredo, A. (2020). Grapevine–Downy Mildew Rendezvous: Proteome Analysis of the First Hours of an Incompatible Interaction. Plants, 9(11), 1498. https://doi.org/10.3390/plants9111498