An Improved Oil Palm Genome Assembly as a Valuable Resource for Crop Improvement and Comparative Genomics in the Arecoideae Subfamily
Abstract
1. Introduction
2. Results
2.1. Construction of Multiple High-Density Oil Palm Linkage Maps
2.2. Improvement of the Oil Palm Physical Genome Assembly by Integration of Multiple Oil Palm Linkage Maps
2.3. Comparative Genomic Analysis of Three Palm Species
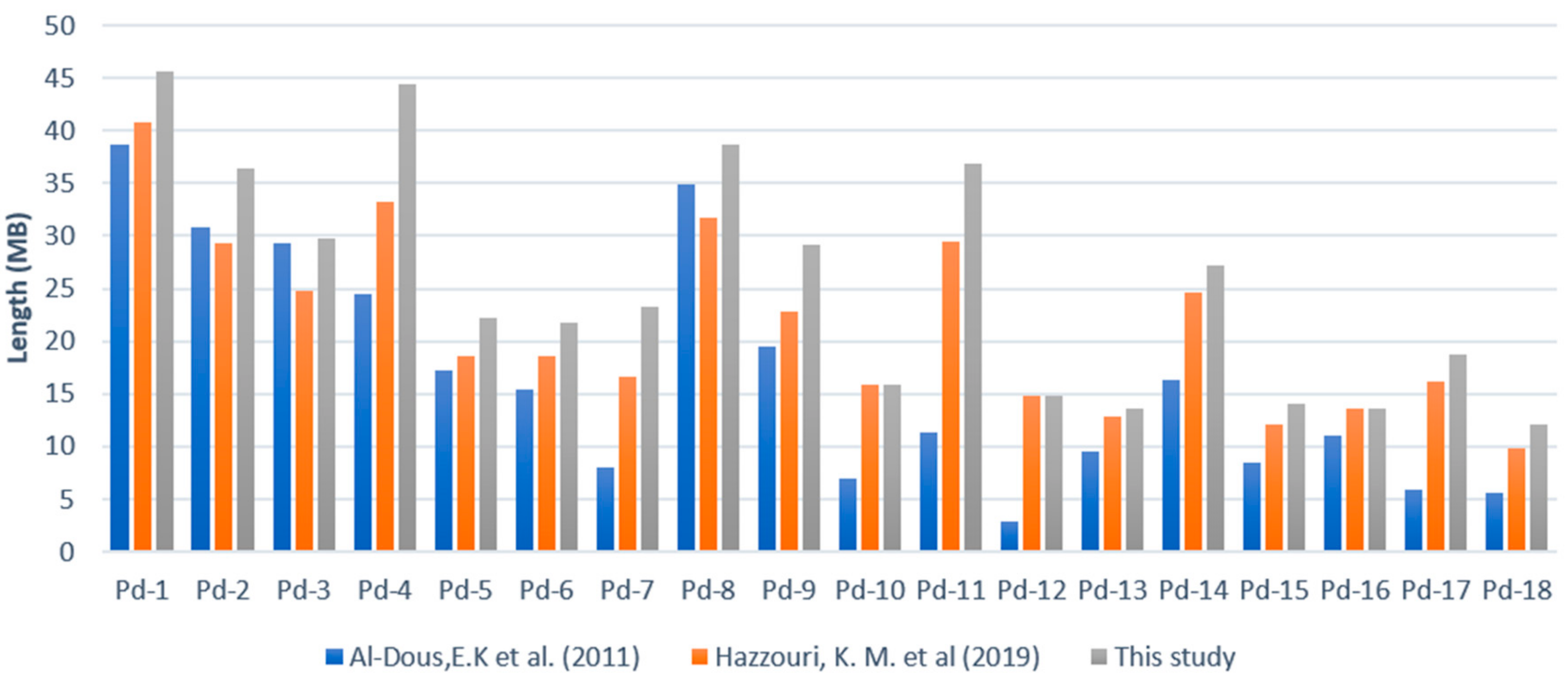
2.4. Oil-Palm-Guided Improvement of the Date Palm Genome
3. Discussion
4. Materials and Methods
4.1. Mapping Populations and DNA Preparations
4.2. SNPs Identification and Genotyping
4.3. Construction of Linkage Maps
4.4. Anchoring of Scaffolds to Linkage Groups
4.5. Genome and Gene Comparisons
4.6. Reconstruction of Date Palm Pseudomolecules
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Hardon, J.J. Interspecific hybrids in the genus Elaeis II. vegetative growth and yield of F1 hybrids E. guineensis x E. oleifera. Euphytica 1969, 18, 380–388. [Google Scholar] [CrossRef]
- Basiron, Y.; Balu, N.; Chandramohan, D. Palm oil: The driving force of world oils and fats economy. Oil Palm Ind. Econ. J. 2004, 4, 1–10. [Google Scholar]
- Prades, A.; Salum, U.N.; Pioch, D. New era for the coconut sector. What prospects for research? OCL Oilseeds Fats Crop. Lipids 2016, 23. [Google Scholar] [CrossRef]
- LLP, A.A. Coconut Products Market by Type Application and Form: Global Opportunity Analysis and Industry Forecast, 2019–2026. 2019. Available online: https://www.globenewswire.com/news-release/2020/02/21/1988574/0/en/Coconut-Products-Market-Study-2019-2026-World-Market-Projected-to-Cross-31-Billion-by-2026.html (accessed on 22 January 2020).
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef] [PubMed]
- Munier, P. Le palmier-dattier. In Munier 1973 Palmier; Maisonneuve&Larose: Paris, France, 1973; Volume 24. [Google Scholar]
- Al-Yahyai, R.; Manickavasagan, A. Dates: Production, Processing, Food, and Medicinal Values; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar] [CrossRef]
- Shahbandeh, M. Global Date Palm Industry Value 2014–2023. Available online: https://www.statista.com/statistics/960213/date-palm-market-value-worldwide/ (accessed on 22 January 2020).
- Mayes, S.; Jack, P.L.; Corley, R.H.; Marshall, D.F. Construction of a RFLP genetic linkage map for oil palm (Elaeis guineensis Jacq.). Genome 1997, 40, 116–122. [Google Scholar] [CrossRef]
- Batugal, P.; Bourdeix, R.; Baudouin, L. Coconut breeding. In Breeding Plantation Tree Crops: Tropical Species; Springer: Berlin, Germany, 2009; pp. 327–375. ISBN 9780387712031. [Google Scholar]
- Batugal, P.; Bourdeix, R. Conventional coconut breeding. Coconut Genet. Resour. 2005, 251. [Google Scholar]
- Hadrami, I.E.; Hadrami, A. El; Hadrami, A. El Breeding date palm. In Breeding Plantation Tree Crops: Tropical Species; Springer: New York, NY, USA, 2009; pp. 191–216. ISBN 9780387712031. [Google Scholar]
- Hardon, J.J. Oil palm breeding, Introduction. Oil Palm Breed. Introd. 1976, 98–107. [Google Scholar]
- Rance, K.A.; Mayes, S.; Price, Z.; Jack, P.L.; Corley, R.H.V. Quantitative trait loci for yield components in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 2001, 103, 1302–1310. [Google Scholar] [CrossRef]
- Moretzsohn, M.C.; Nunes, C.D.M.; Ferreira, M.E.; Grattapaglia, D. RAPD linkage mapping of the shell thickness locus in oil palm (Elaeis guineensis jacq.). Theor. Appl. Genet. 2000, 100, 63–70. [Google Scholar] [CrossRef]
- Ting, N.C.; Jansen, J.; Mayes, S.; Massawe, F.; Sambanthamurthi, R.; Ooi, L.C.L.; Chin, C.W.; Arulandoo, X.; Seng, T.Y.; Alwee, S.S.R.S.; et al. High density SNP and SSR-based genetic maps of two independent oil palm hybrids. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.T.; Wong, W.C.; Wong, C.K.; Soh, A.C.; Kilian, A.; Low, E.T.L.; Massawe, F.; Mayes, S. High density SNP and DArT-based genetic linkage maps of two closely related oil palm populations. J. Appl. Genet. 2018, 59, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Billotte, N.; Marseillac, N.; Risterucci, A.M.; Adon, B.; Brottier, P.; Baurens, F.C.; Singh, R.; Herrán, A.; Asmady, H.; Billot, C.; et al. Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 2005, 110, 754–765. [Google Scholar] [CrossRef]
- Seng, T.Y.; Saad, S.H.; Chin, C.W.; Ting, N.C.; Singh, R.S.H.; Zaman, F.; Tan, S.G.; Alwee, S.S.R. Genetic linkage map of a high yielding FELDA Deli×Yangambi oil palm cross. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Xia, J.H.; Zou, Z.; Ye, J.; Rahmadsyah; Alfiko, Y.; Jin, J.; Lieando, J.V.; Purnamasari, M.I.; Lim, C.H.; et al. A consensus linkage map of oil palm and a major QTL for stem height. Sci. Rep. 2015, 5, 8232. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ong-Abdullah, M.; Low, E.-T.L.; Manaf, M.A.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.; Chan, K.-L.; Halim, M.A.; et al. Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef]
- Teh, C.K.; Ong, A.L.; Kwong, Q.B.; Apparow, S.; Chew, F.T.; Mayes, S.; Mohamed, M.; Appleton, D.; Kulaveerasingam, H. Genome-wide association study identifies three key loci for high mesocarp oil content in perennial crop oil palm. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Kwong, Q.B.; Teh, C.K.; Ong, A.L.; Heng, H.Y.; Lee, H.L.; Mohamed, M.; Low, J.Z.B.; Apparow, S.; Chew, F.T.; Mayes, S.; et al. Development and Validation of a High-Density SNP Genotyping Array for African Oil Palm. Mol. Plant 2016, 9, 1132–1141. [Google Scholar] [CrossRef]
- Hazzouri, K.M.; Gros-Balthazard, M.; Flowers, J.M.; Copetti, D.; Lemansour, A.; Lebrun, M.; Masmoudi, K.; Ferrand, S.; Dhar, M.I.; Fresquez, Z.A.; et al. Genome-wide association mapping of date palm fruit traits. Nat. Commun. 2019, 10, 4680. [Google Scholar] [CrossRef]
- Xiao, Y.; Xu, P.; Fan, H.; Baudouin, L.; Xia, W.; Bocs, S.; Xu, J.; Li, Q.; Guo, A.; Zhou, L.; et al. The genome draft of coconut (Cocos nucifera). Gigascience 2017, 6. [Google Scholar] [CrossRef]
- Rival, A.; Beule, T.; Barre, P.; Hamon, S.; Duval, Y.; Noirot, M. Comparative flow cytometric estimation of nuclear DNA content in oil palm (Elaeis guineensis jacq) tissue cultures and seed-derived plants. Plant Cell Rep. 1997, 16, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, J.; Leitch, I.J. The Plant DNA C-values database (release 7.1): An updated online repository of plant genome size data for comparative studies. New Phytol. 2020, 226, 301–305. [Google Scholar] [CrossRef]
- Ong, A.L.; Teh, C.K.; Kwong, Q.B.; Tangaya, P.; Appleton, D.R.; Massawe, F.; Mayes, S. Linkage-based genome assembly improvement of oil palm (Elaeis guineensis). Sci. Rep. 2019, 9, 6619. [Google Scholar] [CrossRef]
- Kianian, P.M.A.; Wang, M.; Simons, K.; Ghavami, F.; He, Y.; Dukowic-Schulze, S.; Sundararajan, A.; Sun, Q.; Pillardy, J.; Mudge, J.; et al. High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 2018, 9, 2370. [Google Scholar] [CrossRef]
- He, Z.; Zhang, Z.; Guo, W.; Zhang, Y.; Zhou, R.; Shi, S. De Novo Assembly of Coding Sequences of the Mangrove Palm (Nypa fruticans) Using RNA-Seq and Discovery of Whole-Genome Duplications in the Ancestor of Palms. PLoS ONE 2015, 10, e0145385. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Low, E.T.L.; Ooi, L.C.L.; Ong-Abdullah, M.; Ting, N.C.; Nagappan, J.; Nookiah, R.; Amiruddin, M.D.; Rosli, R.; Manaf, M.A.A.; et al. The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 2013, 500, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.F.; Mathew, L.S.; Ahmed, I.; Al-Azwani, I.K.; Krueger, R.; Rivera-Nuñez, D.; Mohamoud, Y.A.; Clark, A.G.; Suhre, K.; Malek, J.A. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun. 2018, 9, 3969. [Google Scholar] [CrossRef] [PubMed]
- Al-Dous, E.K.; George, B.; Al-Mahmoud, M.E.; Al-Jaber, M.Y.; Wang, H.; Salameh, Y.M.; Al-Azwani, E.K.; Chaluvadi, S.; Pontaroli, A.C.; Debarry, J.; et al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat. Biotechnol. 2011, 29, 521. [Google Scholar] [CrossRef] [PubMed]
- Mathew, L.S.; Spannagl, M.; Al-Malki, A.; George, B.; Torres, M.F.; Al-Dous, E.K.; Al-Azwani, E.K.; Hussein, E.; Mathew, S.; Mayer, K.F.X.; et al. A first genetic map of date palm (Phoenix dactylifera) reveals long-range genome structure conservation in the palms. BMC Genom. 2014, 15, 285. [Google Scholar] [CrossRef]
- Pamin, K. A Hundred and Fifty Years of Oil Palm Development in Indonesia from the Bogor Botanical Garden to the Industry; Medan IOPRI: Medan Maimun, Kota Medan, Indonesia, 1998. [Google Scholar]
- Rajanaidu, N.; Kushairi, A.; Rafii, M.; Mohd, D.A.; Maizura, I.; Jalani, B.S. Oil palm breeding and genetic resources. Adv. Oil Palm Res. 2000, 1, 171–237. [Google Scholar]
- Shen, C.; Li, X.; Zhang, R.; Lin, Z. Genome-wide recombination rate variation in a recombination map of cotton. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Phillips, R. Cell Biology by the Numbers; Garland Science: New York, NY, USA, 2016; pp. 334–338. [Google Scholar] [CrossRef]
- Sidhu, D.; Gill, K.S. Distribution of genes and recombination in wheat and other eukaryotes. Plant Cell. Tissue Organ Cult. 2005, 79, 257–270. [Google Scholar] [CrossRef]
- Jordan, K.W.; Wang, S.; He, F.; Chao, S.; Lun, Y.; Paux, E.; Sourdille, P.; Sherman, J.; Akhunova, A.; Blake, N.K.; et al. The genetic architecture of genome-wide recombination rate variation in allopolyploid wheat revealed by nested association mapping. Plant J. 2018, 95, 1039–1054. [Google Scholar] [CrossRef]
- Teh, C.K.; Muaz, S.D.; Tangaya, P.; Fong, P.Y.; Ong, A.L.; Mayes, S.; Chew, F.T.; Kulaveerasingam, H.; Appleton, D. Characterizing haploinsufficiency of SHELL gene to improve fruit form prediction in introgressive hybrids of oil palm. Sci. Rep. 2017, 7, 3118. [Google Scholar] [CrossRef] [PubMed]
- Corley, R.H.V.; Tinker, P.B. The Oil Palm: Fifth Edition; John Wiley&Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Hardon, J.J.; Corley, R.H.V.; Lee, C.H. Breeding and Selecting the Oil Palm. In Improving Vegetatively Propagated Crops; Academic Press: London, UK, 1987. [Google Scholar]
- Bellow, S.A.; Agunsoye, J.O.; Adebisi, J.A.; Kolawole, F.O.; Hassan, S.B. Physical properties of coconut shell nanoparticles. Kathmandu Univ. J. Sci. Eng. Technol. 2018, 12, 63–79. [Google Scholar] [CrossRef]
- Budi, E.; Umiatin, U.; Nasbey, H.; Bintoro, R.A.; Wulandari, F.; Erlina, E. Adsorption and Pore of Physical-Chemical Activated Coconut Shell Charcoal Carbon. In IOP Conference Series: Materials Science and Engineering; Management Science and Engineering: Stanford, CA, USA, 2018. [Google Scholar]
- Mohd Iqbaldin, M.N.; Khudzir, I.; Mohd Azlan, M.I.; Zaidi, A.G.; Surani, B.; Zubri, Z. Properties of coconut shell activated carbon. J. Trop. For. Sci. 2013, 25, 497–503. [Google Scholar]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Corley, R.H.V. Oil palm physiology-a review. In Proceedings of the International Oil Palm Conference, Kuala Lumpur, Malaysia, 16–18 November 1973. [Google Scholar]
- Couvreur, T.L.P.; Forest, F.; Baker, W.J. Origin and global diversification patterns of tropical rain forests: Inferences from a complete genus-level phylogeny of palms. BMC Biol. 2011, 9, 44. [Google Scholar] [CrossRef]
- Barrett, C.F.; Bacon, C.D.; Antonelli, A.; Cano, Á.; Hofmann, T. An introduction to plant phylogenomics with a focus on palms. Bot. J. Linn. Soc. 2016, 182, 234–255. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Rastas, P.; Paulin, L.; Hanski, I.; Lehtonen, R.; Auvinen, P.; Brudno, M. Lep-MAP: Fast and accurate linkage map construction for large SNP datasets. Bioinformatics 2013, 29, 3128–3134. [Google Scholar] [CrossRef] [PubMed]
- Rastas, P. Lep-MAP3: Robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 2017, 33, 3726–3732. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, X.; Miao, C.; Zhang, J.; Ming, R.; Schnable, J.C.; Schnable, P.S.; Lyons, E.; Lu, J. ALLMAPS: Robust scaffold ordering based on multiple maps. Genome Biol. 2015, 16, 3. [Google Scholar] [CrossRef]
- Al-Mssallem, I.S.; Hu, S.; Zhang, X.; Lin, Q.; Liu, W.; Tan, J.; Yu, X.; Liu, J.; Pan, L.; Zhang, T.; et al. Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 2013, 4, 2274. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Clustalw, U.; To, C.; Multiple, D.O. ClustalW and ClustalX. Options 2003, 1–22. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Rambaut, A. Fig Tree v. 1.4.4. 2018. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 12 June 2020).
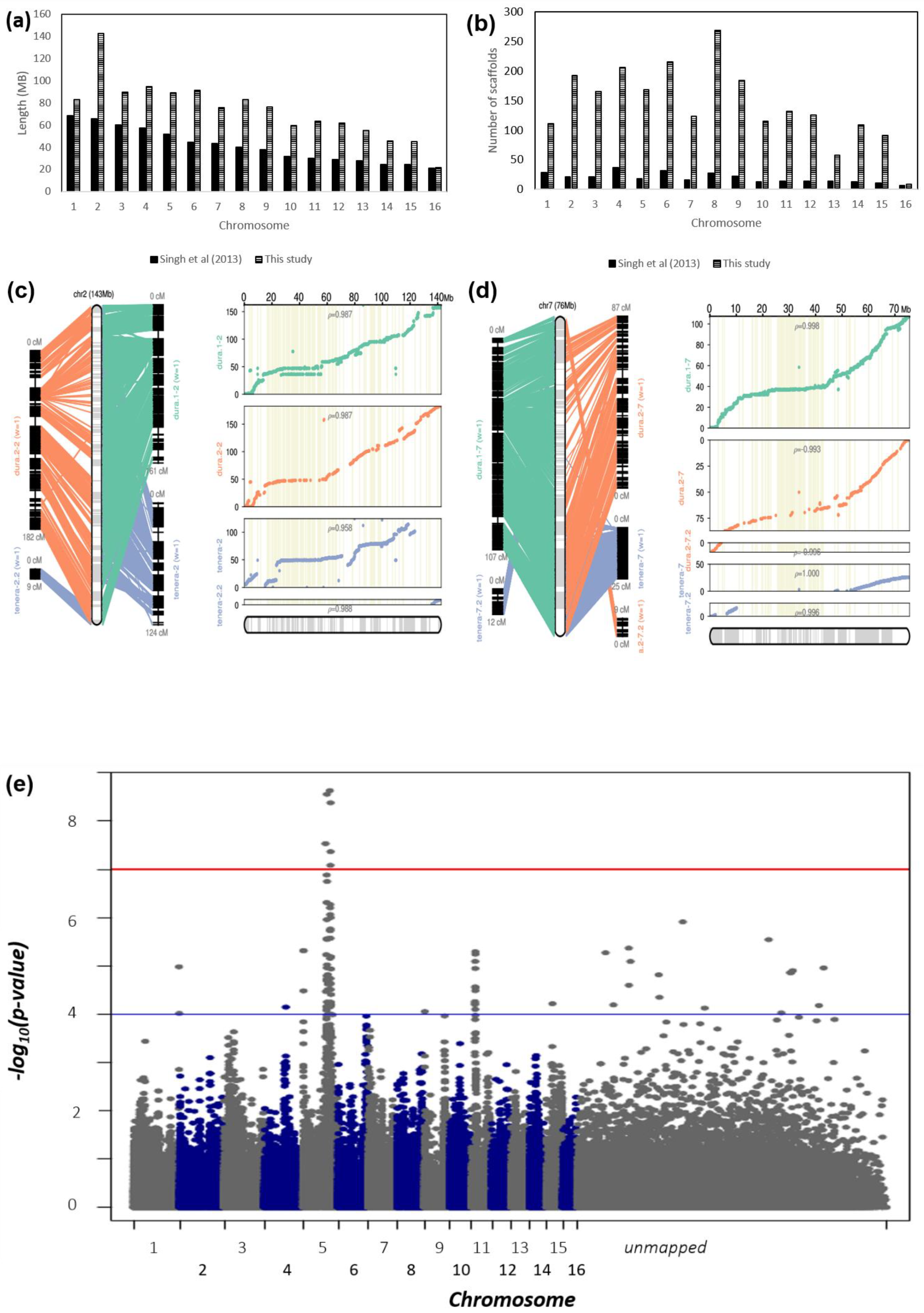
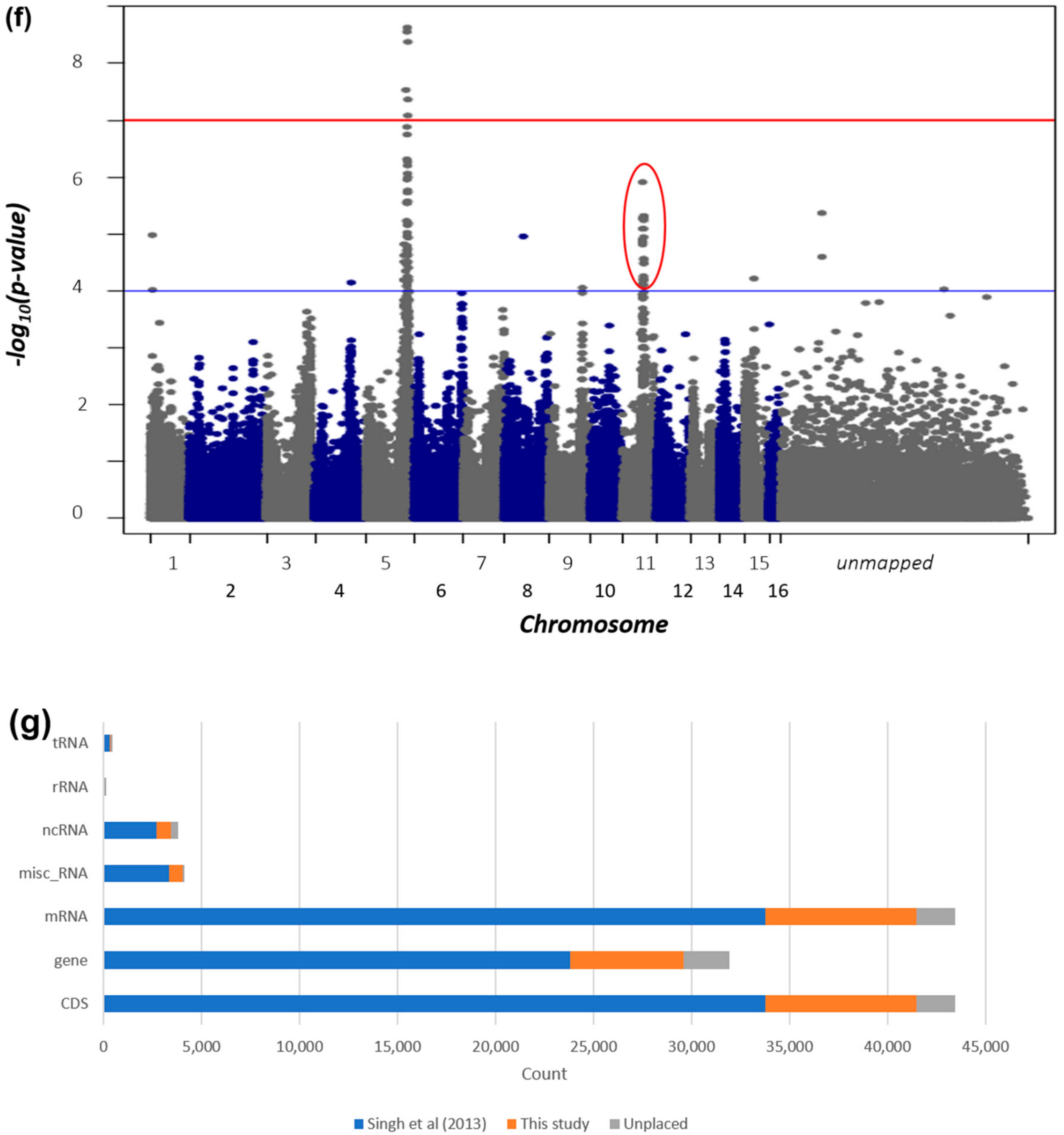
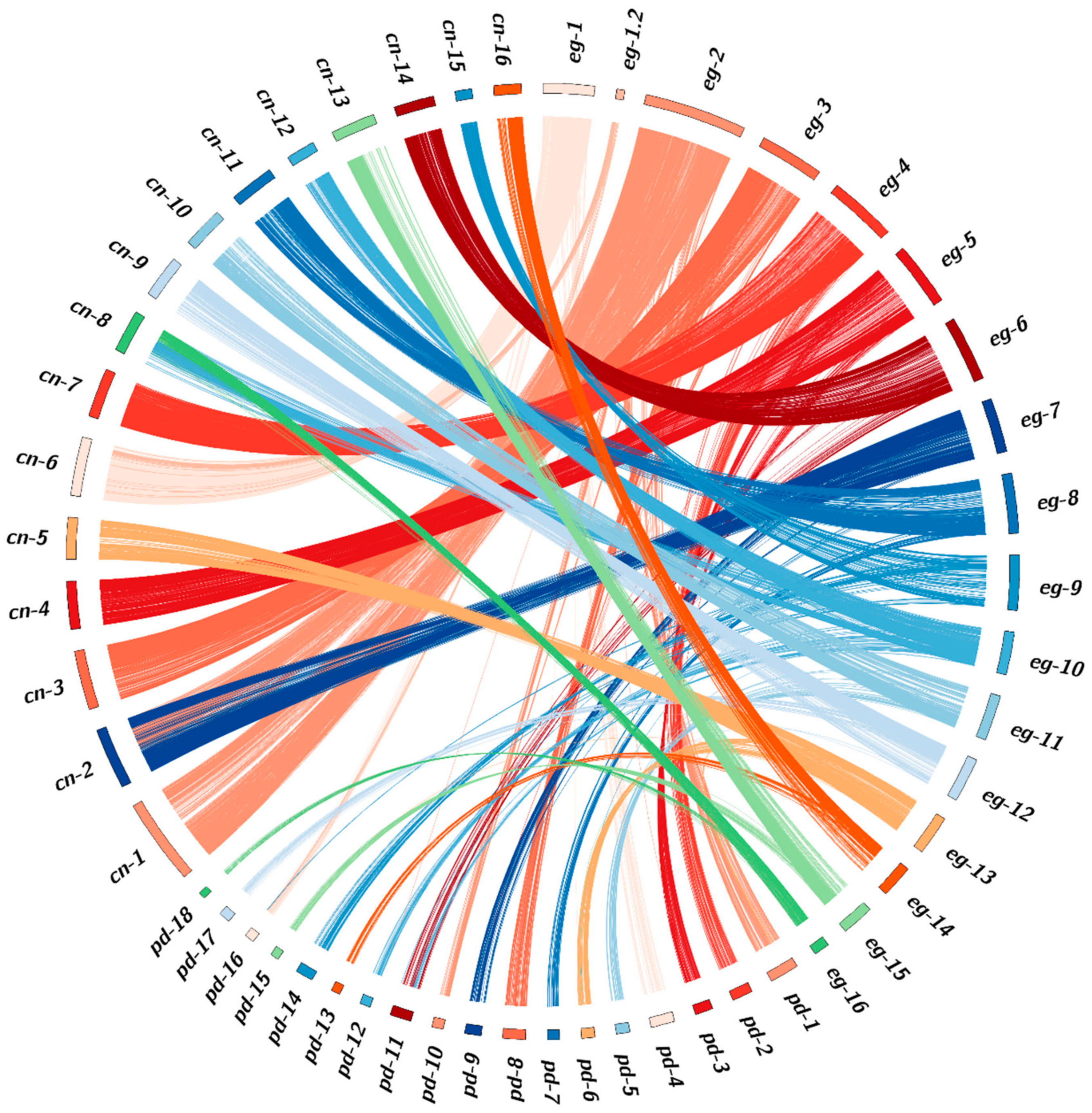
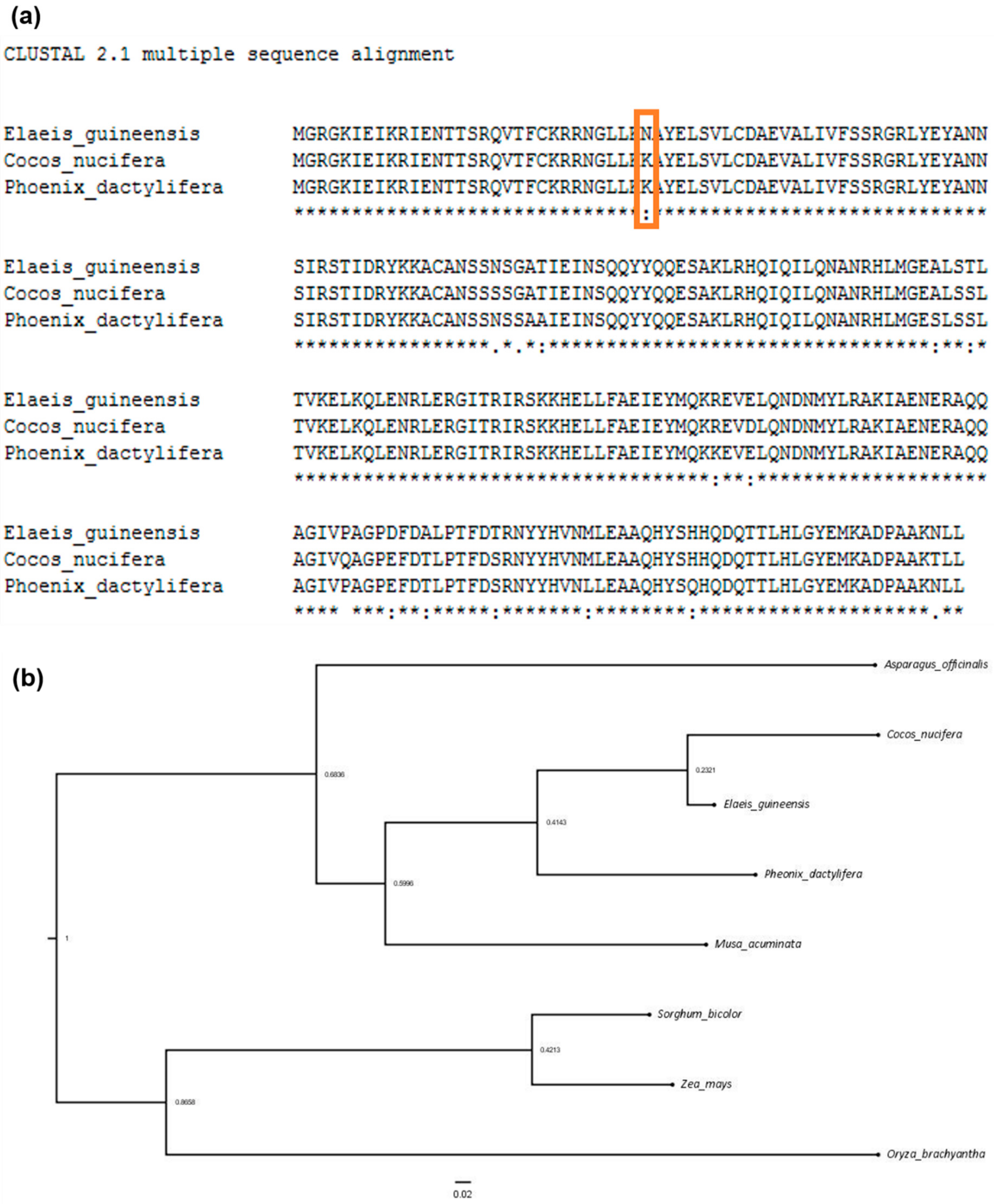
| Population | Number of Linked Markers | Number of Linkage Groups (LGs) | Length of Linkage Map (cM) | Marker Interval (cM) | Genome-Wide Recombination Rate of P5-Build (cM/Mb) | Genome-Wide Recombination Rate of PMv6 (cM/Mb) |
|---|---|---|---|---|---|---|
| Deli dura × AVROS pisifera * | 27,890 | 19 | 1151.70 | 0.04 | 1.75 | 0.98 |
| Deli dura × Nigerian dura | 32,650 | 17 | 1646.95 | 0.05 | 2.50 | 1.40 |
| Johore Labis dura | 6920 | 24 | 1268.26 | 0.18 | 1.93 | 1.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong, A.-L.; Teh, C.-K.; Mayes, S.; Massawe, F.; Appleton, D.R.; Kulaveerasingam, H. An Improved Oil Palm Genome Assembly as a Valuable Resource for Crop Improvement and Comparative Genomics in the Arecoideae Subfamily. Plants 2020, 9, 1476. https://doi.org/10.3390/plants9111476
Ong A-L, Teh C-K, Mayes S, Massawe F, Appleton DR, Kulaveerasingam H. An Improved Oil Palm Genome Assembly as a Valuable Resource for Crop Improvement and Comparative Genomics in the Arecoideae Subfamily. Plants. 2020; 9(11):1476. https://doi.org/10.3390/plants9111476
Chicago/Turabian StyleOng, Ai-Ling, Chee-Keng Teh, Sean Mayes, Festo Massawe, David Ross Appleton, and Harikrishna Kulaveerasingam. 2020. "An Improved Oil Palm Genome Assembly as a Valuable Resource for Crop Improvement and Comparative Genomics in the Arecoideae Subfamily" Plants 9, no. 11: 1476. https://doi.org/10.3390/plants9111476
APA StyleOng, A.-L., Teh, C.-K., Mayes, S., Massawe, F., Appleton, D. R., & Kulaveerasingam, H. (2020). An Improved Oil Palm Genome Assembly as a Valuable Resource for Crop Improvement and Comparative Genomics in the Arecoideae Subfamily. Plants, 9(11), 1476. https://doi.org/10.3390/plants9111476






