Insights into the Superoxide Dismutase Gene Family and Its Roles in Dendrobium catenatum under Abiotic Stresses
Abstract
1. Introduction
2. Results
2.1. Identification of SOD Gene Family Members in Dendrobium catenatum
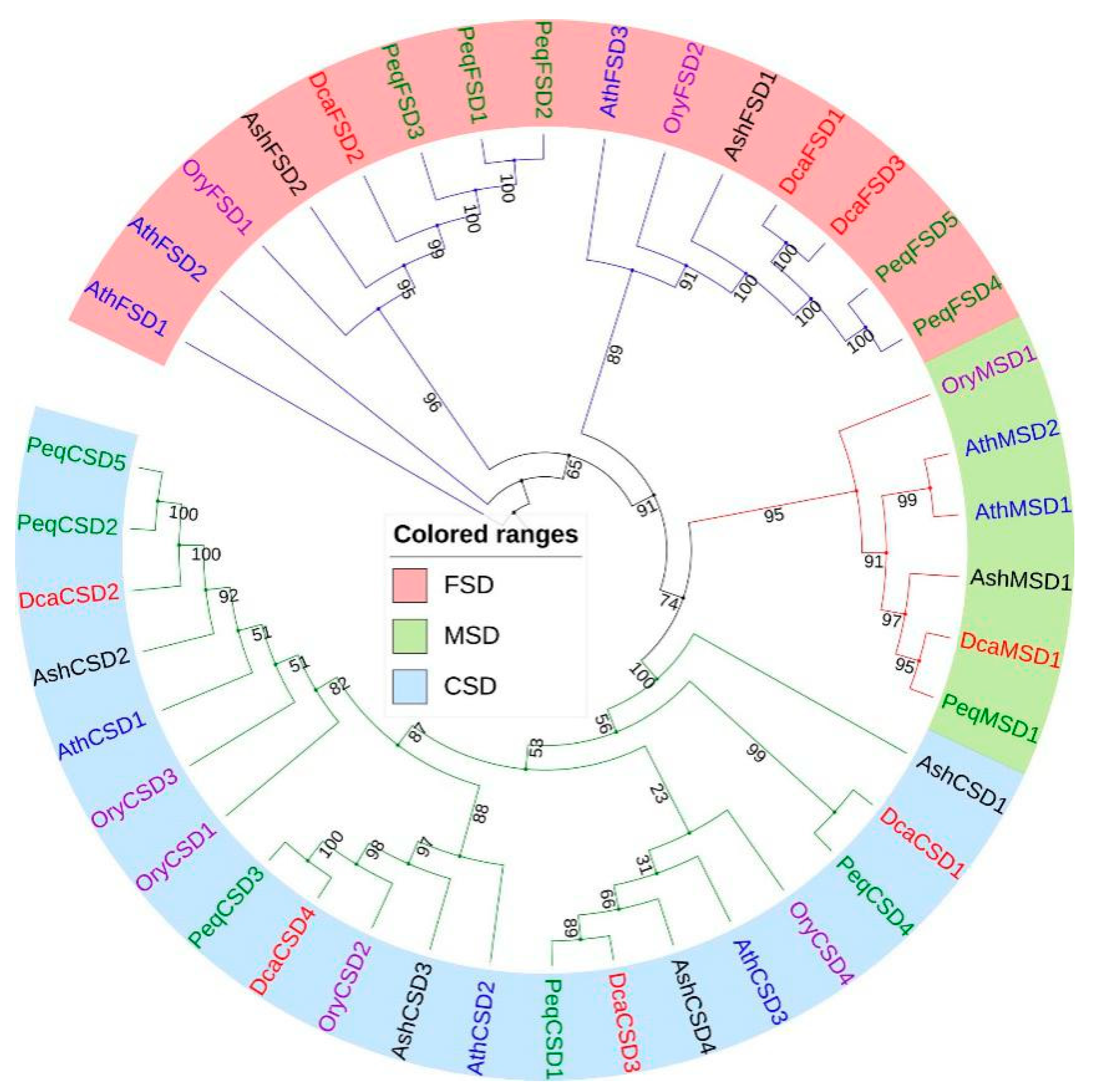
2.2. Phylogenetic Analysis of DcaSOD Proteins
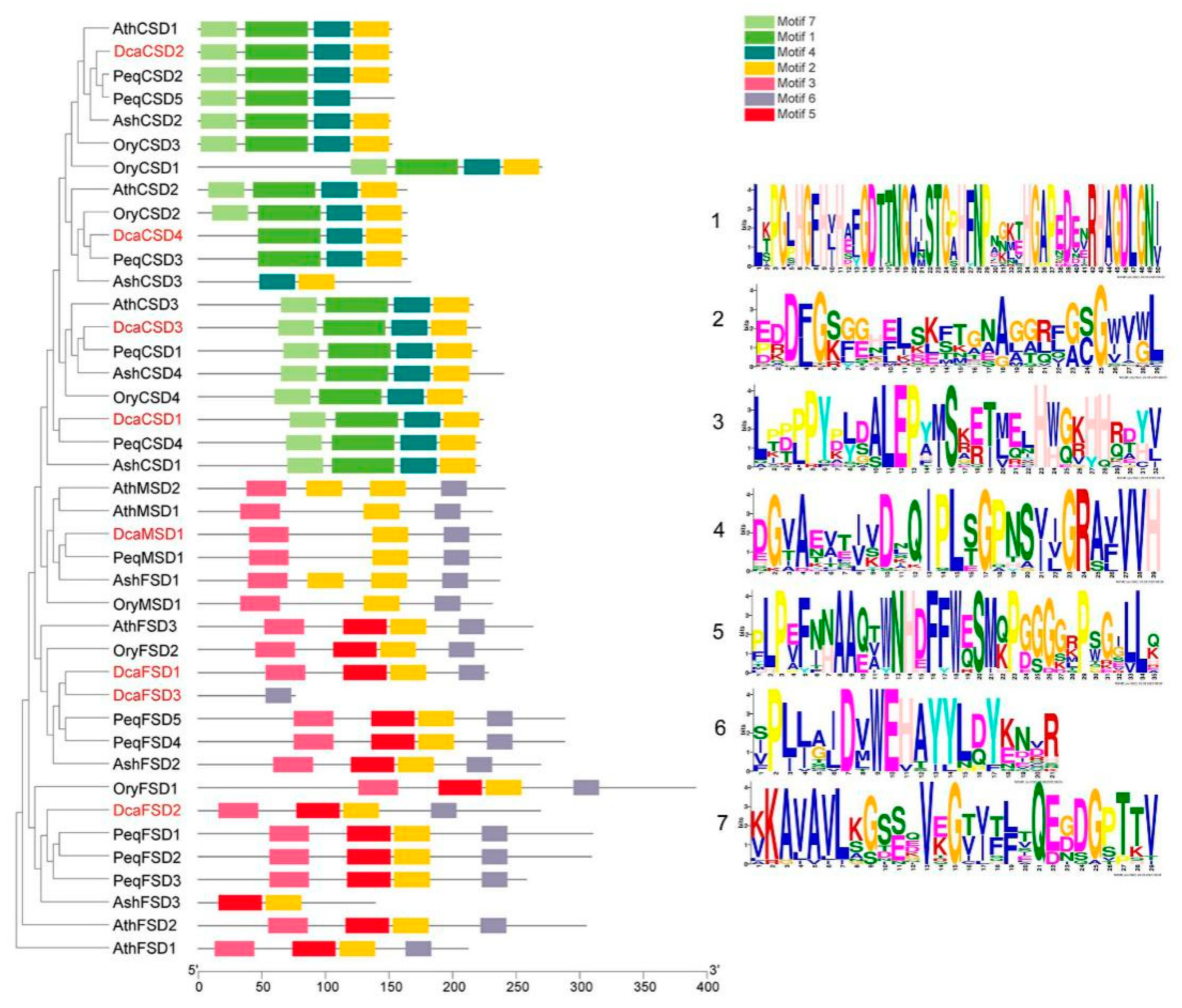
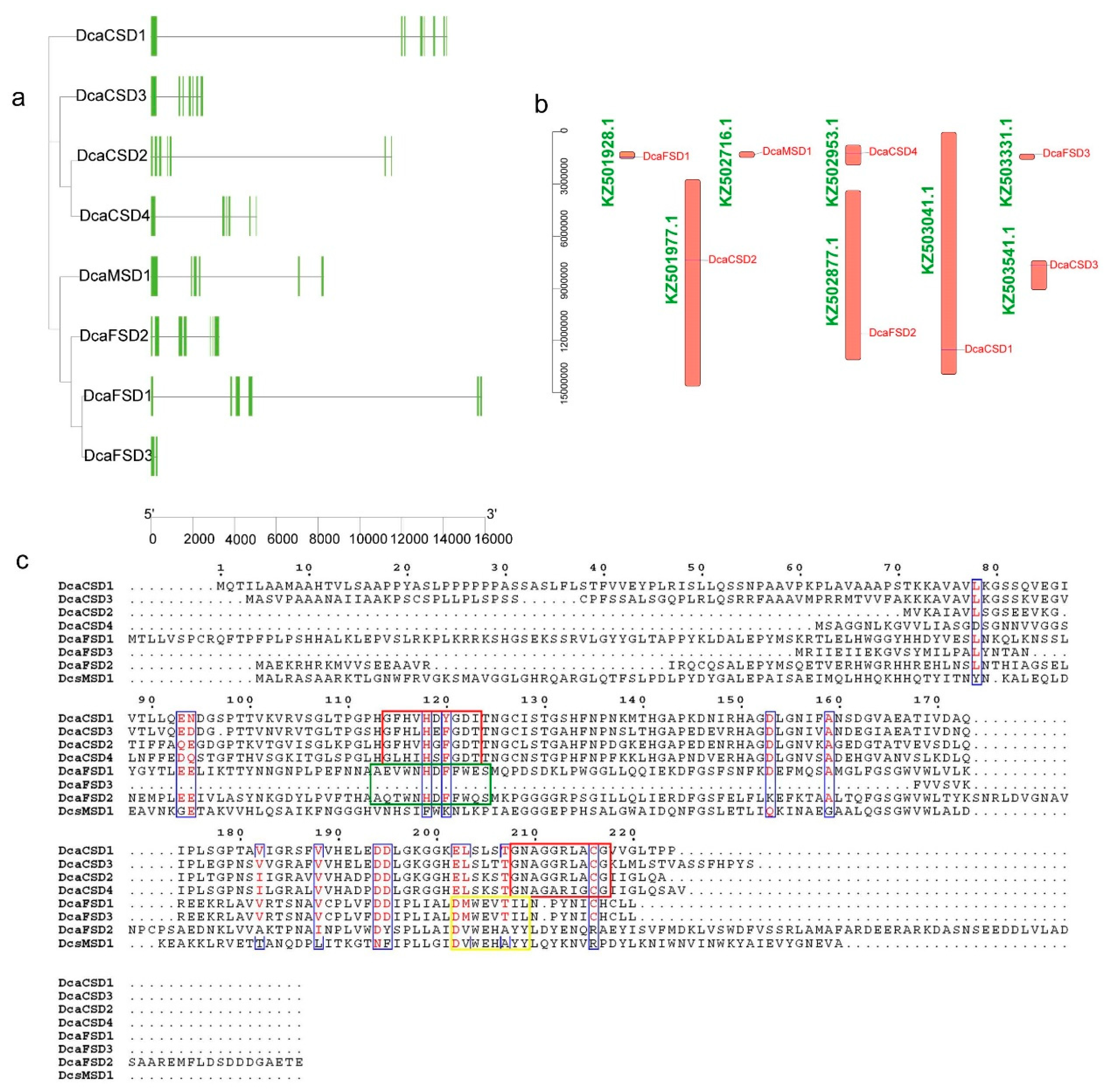
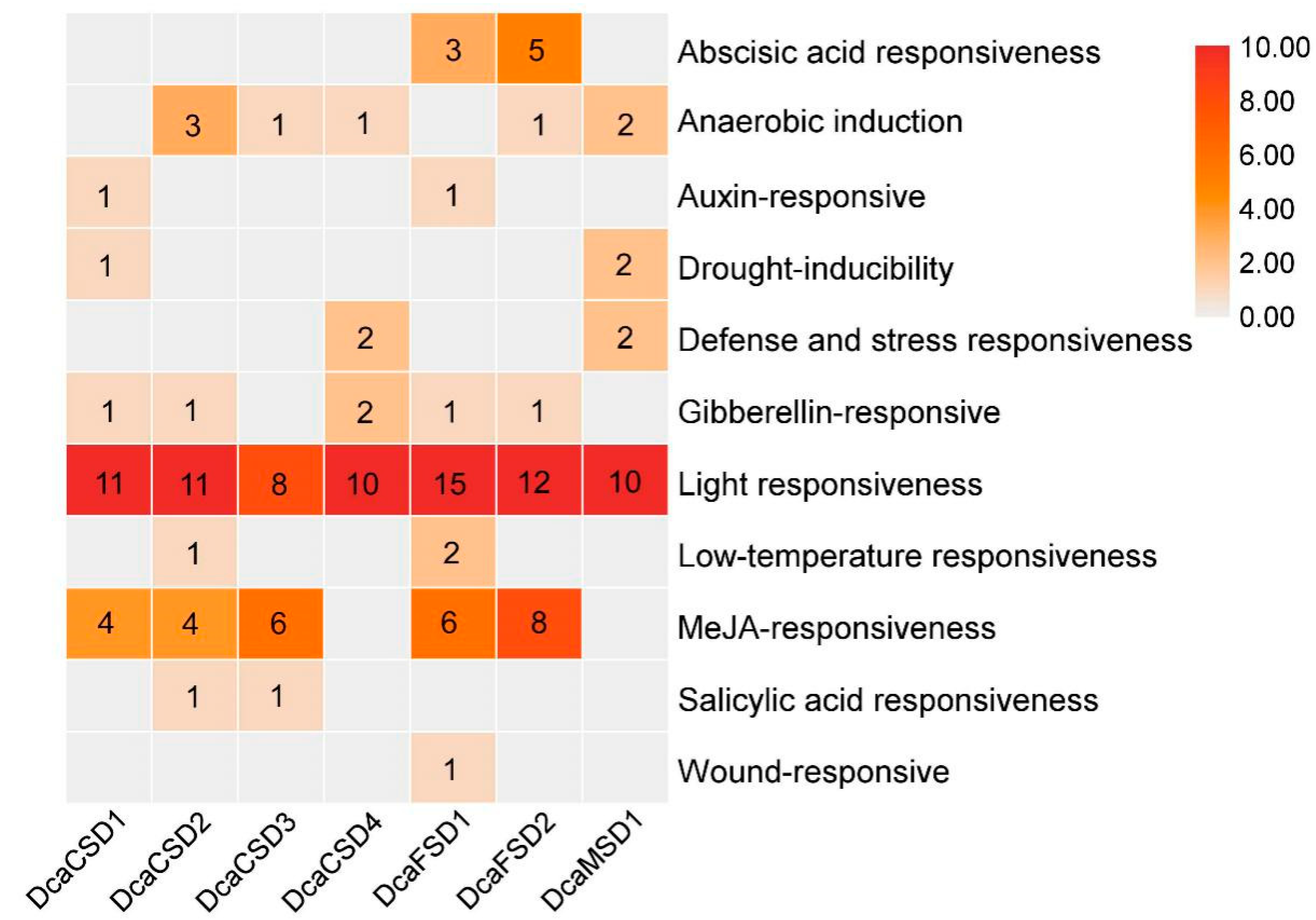
2.3. Conserved Motifs, Gene Structures, Distribution, and cis-Elements Analysis
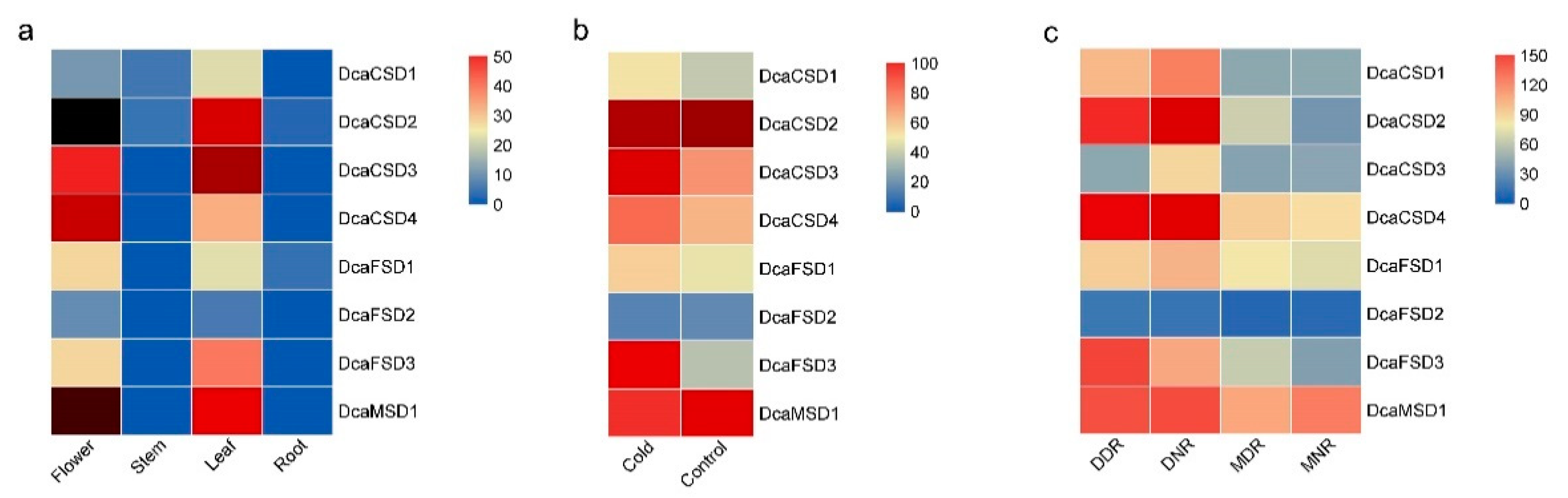
2.4. Distinct Expression Profiles of DcaSOD Genes in Different Tissues, Cold and Drought Responses
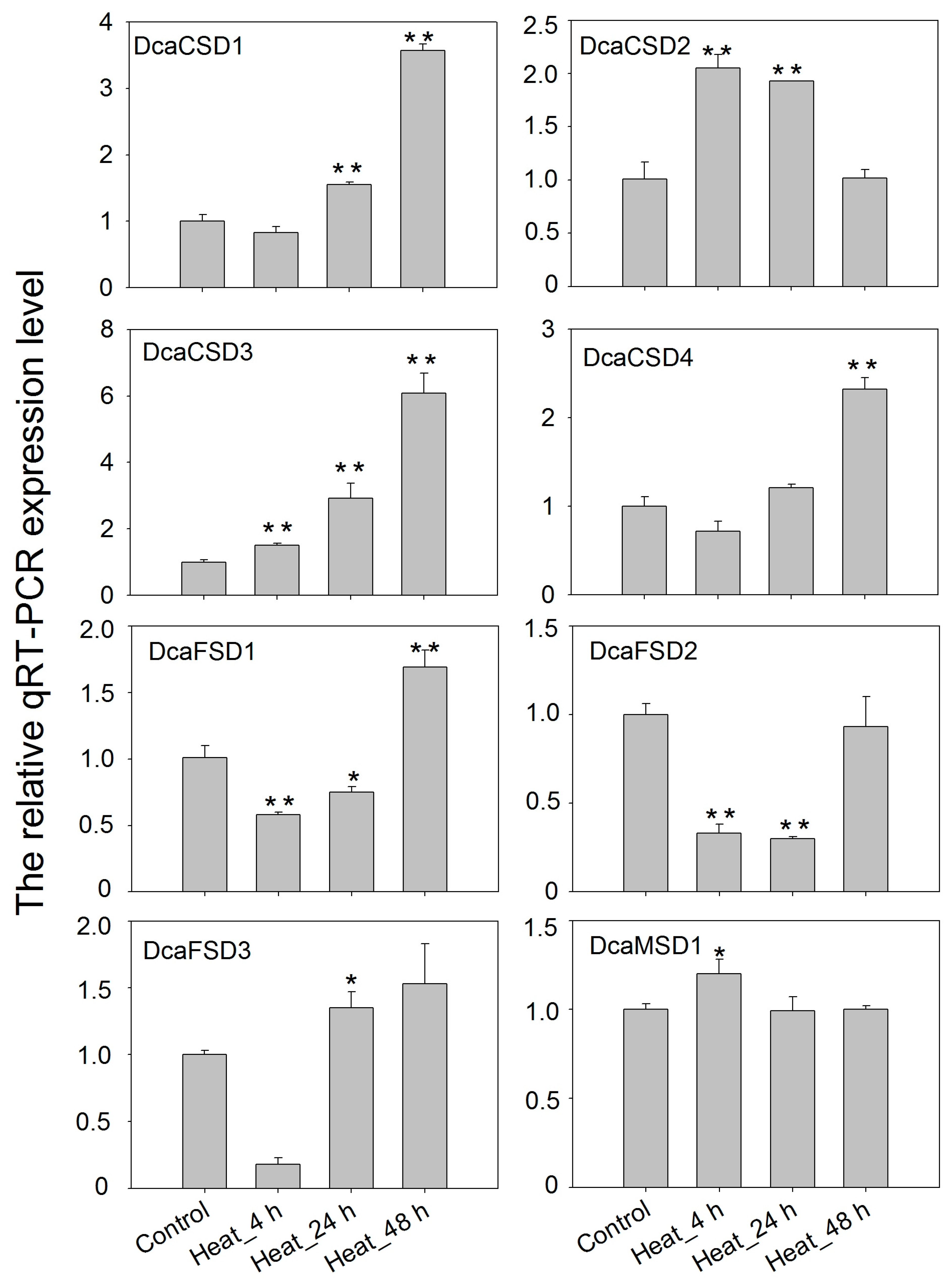
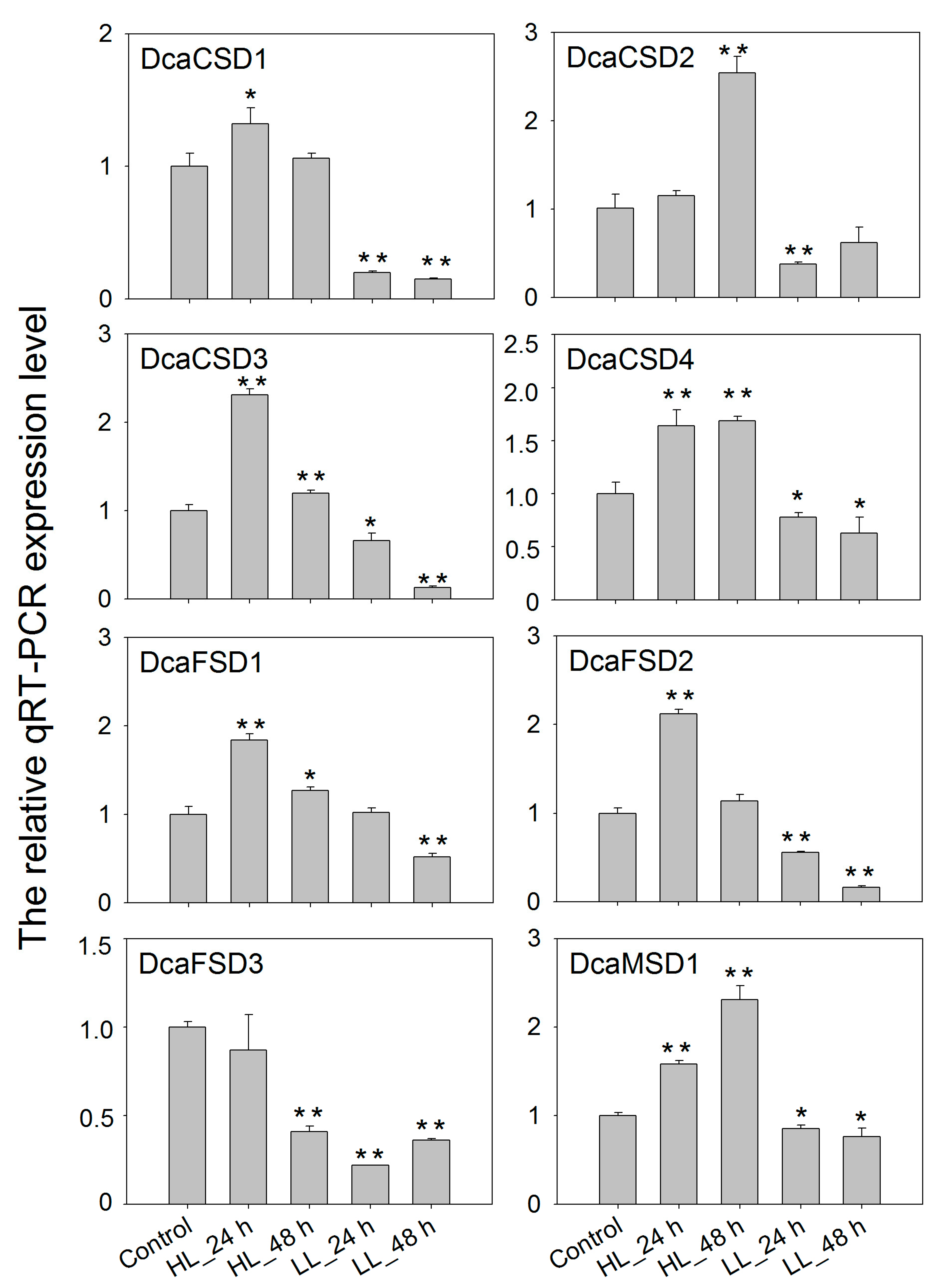
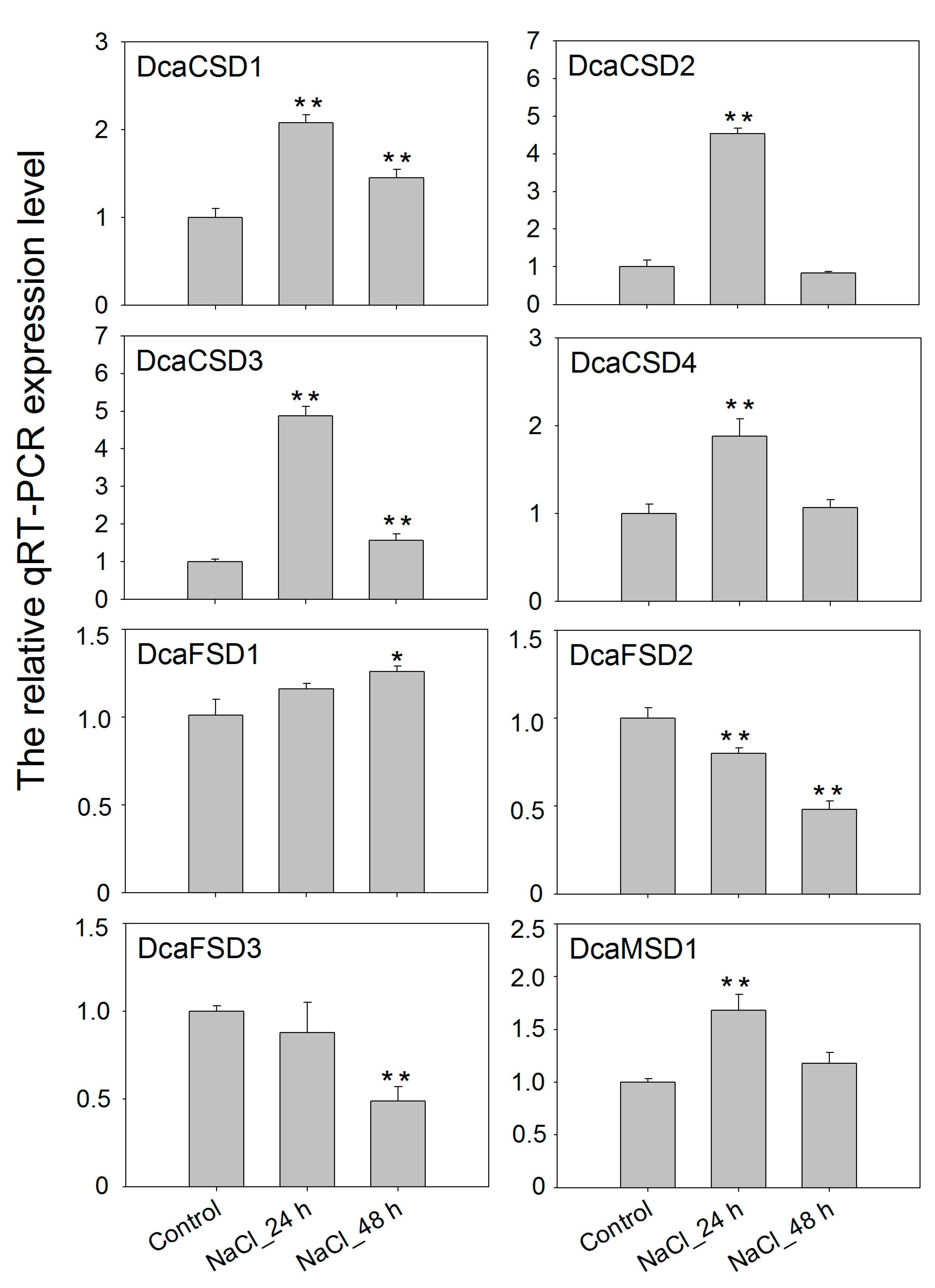
2.5. qRT-PCR Verified the Expression of DcaSOD Genes in Response to Heat, Light and NaCl Treatments
3. Discussion
4. Materials and Methods
4.1. Identification and Sequence Analysis of DcaSOD Genes in D. catenatum
4.2. Phylogenetic Analysis
4.3. Conserved Motifs, Gene Structures, Locations, Multiple Sequence Alignments and Cis-elements Analyses of DcaSOD Genes
4.4. Transcriptome Analysis
4.5. Plant Material, Growth Conditions and Treatments
4.6. Real-Time PCR Experiment
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Atwood, J. The size of Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana 1986, 9, 171–186. [Google Scholar]
- Su, W.H.; Zhang, G.F. The photosynthesis pathway in leaves of Dendrobium catenatum. Chin. J. Plant Ecol. 2003, 27, 631–637. [Google Scholar]
- Zhang, Z.; Dongxian, H.; Rongfu, G. Concomitant CAM and C3 photosynthetic pathways in Dendrobium catenatum plants. J. Am. Soc. Hortic. Sci. 2014, 139, 290–298. [Google Scholar] [CrossRef]
- Zhang, S.B.; Dai, Y.; Hao, G.Y.; Li, J.W.; Fu, X.W.; Zhang, J.L. Differentiation of water-related traits in terrestrial and epiphytic Cymbidium species. Front. Plant Sci. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of Dendrobium officinale illuminates the biology of the important traditional chinese orchid herb. Mol. Plant 2015, 8, 922–934. [Google Scholar] [CrossRef]
- Zhang, G.-Q.; Xu, Q.; Bian, C.; Tsai, W.-C.; Yeh, C.-M.; Liu, K.-W.; Yoshida, K.; Zhang, L.-S.; Chang, S.-B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef]
- Hsieh, Y.S.; Chien, C.; Liao, S.K.; Liao, S.F.; Hung, W.T.; Yang, W.B.; Lin, C.C.; Cheng, T.J.; Chang, C.C.; Fang, J.M. Structure and bioactivity of the polysaccharides in medicinal plant Dendrobium huoshanense. Bioorganic Med. Chem. 2008, 16, 6054–6068. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Sze, S.C.W.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef]
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive oxygen species in plants: Their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar]
- Gupta, S.; Dong, Y.; Dijkwel, P.P.; Mueller-Roeber, B.; Gechev, T.S. Genome-wide analysis of ROS antioxidant genes in resurrection species suggest an involvement of distinct ROS detoxification systems during desiccation. Int. J. Mol. Sci. 2019, 20, 3101. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.; Chen, J.; Yuan, R.; Deng, F.; Shen, F. Gene expression characteristics and regulation mechanisms of superoxide dismutase and its physiological roles in plants under stress. Biochemistry 2016, 81, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.; Perl-Treves, R.; Galili, S.; Aviv, D.; Shalgi, E.; Malkin, S.; Galun, E. Enhanced oxidative-stress defense in transgenic potato expressing tomato Cu, Zn superoxide dismutases. Theor. Appl. Genet. 1993, 85, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Pilon, M.; Ravet, K.; Tapken, W. The biogenesis and physiological function of chloroplast superoxide dismutases. BBA Bioenerg. 2011, 1807, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Asensio, A.C.; Gil-Monreal, M.; Pires, L.; Gogorcena, Y.; Aparicio-Tejo, P.M.; Moran, J.F. Two Fe-superoxide dismutase families respond differently to stress and senescence in legumes. J. Plant Physiol. 2012, 169, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom. 2015, 16, 823. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD gene family in tomato: Identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Wu, H.; Jiang, L.; Liu, S. Genome-wide identification and transcriptional expression analysis of cucumber superoxide dismutase (SOD) family in response to various abiotic stress. Int. J. Genom. 2017, 2017, 7243973. [Google Scholar]
- Wang, W.; Zhang, X.; Deng, F.; Yuan, R.; Shen, F. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genom. 2017, 18, 376. [Google Scholar] [CrossRef]
- Jiang, W.Q.; Yang, L.; He, Y.Q.; Zhang, H.T.; Li, W.; Chen, H.G.; Ma, D.F.; Yin, J.L. Genome-wide identification and transcriptional expression analysis of superoxide dismutase (SOD) family in wheat (Triticum aestivum). PeerJ 2019, 7, e8062. [Google Scholar] [CrossRef]
- Han, L.M.; Hua, W.P.; Cao, X.Y.; Yan, J.A.; Chen, C.; Wang, Z.Z. Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Lai, Z.X. Superoxide dismutase multigene family in longan somatic embryos: A comparison of CuZn-SOD, Fe-SOD, and Mn-SOD gene structure, splicing, phylogeny, and expression. Mol. Breed. 2013, 32, 595–615. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Liu, K.W.; Li, Z.; Lohaus, R.; Hsiao, Y.Y.; Niu, S.-C.; Wang, J.-Y.; Lin, Y.-C.; Xu, Q.; Chen, L.-J.; et al. The Apostasia genome and the evolution of orchids. Nature 2017, 549, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Song, H.; Zhang, B.; Lu, Q.; Liu, Z.; Zhang, S.; Guo, R.; Wang, C.; Zhao, Z.; Liu, J. Genome-wide identification, characterization, and expression analysis of superoxide dismutase (SOD) genes in foxtail millet (Setaria italica L.). 3 Biotech 2018, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Chen, J.; Li, R.; Shang, S.; Tang, X. Genome-wide analysis of the superoxide dismutase (SOD) gene family in Zostera marina and expression profile analysis under temperature stress. PeerJ 2020, 8, 9063. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.S.; Heinen, J.L.; Holaday, A.S.; Burke, J.J.; Allen, R.D. Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 1993, 90, 1629–1633. [Google Scholar] [CrossRef] [PubMed]
- McKersie, B.D.; Chen, Y.; de Beus, M.; Bowley, S.R.; Bowler, C.; Inze, D.; D’Halluin, K.; Botterman, J. Superoxide dismutase enhances tolerance of freezing stress in transgenic alfalfa (Medicago sativa L.). Plant Physiol. 1993, 103, 1155–1163. [Google Scholar] [CrossRef]
- Jain, R.; Chandra, A.; Venugopalan, V.K.; Solomon, S. Physiological changes and expression of SOD and P5CS genes in response to water deficit in Sugarcane. Sugar Tech 2015, 17, 276–282. [Google Scholar] [CrossRef]
- Milla, M.A.R.; Maurer, A.; Huete, A.R.; Gustafson, J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003, 36, 602–615. [Google Scholar] [CrossRef]
- Hwang, Y.; Lee, H.; Lee, Y.S.; Cho, H.T. Cell wall-associated ROOT HAIR SPECIFIC 10, a proline-rich receptor-like kinase, is a negative modulator of Arabidopsis root hair growth. J. Exp. Bot. 2016, 67, 2007–2022. [Google Scholar] [CrossRef]
- Pillet, J.; Yu, H.W.; Chambers, A.H.; Whitaker, V.M.; Folta, K.M. Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 2015, 66, 4455–4467. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Fu, L.; Tan, D.; Sun, X.; Zhang, J. An integrative transcriptomic and genomic analysis reveals novel insights into the hub genes and regulatory networks associated with rubber synthesis in H. brasiliensis. Ind. Crop. Prod. 2020, 153, 112562. [Google Scholar] [CrossRef]
- Lu, T.; Meng, Z.; Zhang, G.; Qi, M.; Sun, Z.; Liu, Y.; Li, T. Sub-high temperature and high light intensity induced irreversible inhibition on photosynthesis system of tomato plant (Solanum lycopersicum L.). Front. Plant Sci. 2017, 8, 365. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, R.; Ślesak, I.; Orzechowska, A.; Kruk, J. Physiological and biochemical responses to high light and temperature stress in plants. Environ. Exp. Bot. 2017, 139, 165–177. [Google Scholar] [CrossRef]
- Wang, Y.; Ying, Y.; Chen, J.; Wang, X. Transgenic Arabidopsis overexpressing MnSOD enhanced salt-tolerance. Plant Sci. 2004, 4, 671–677. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools-an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wan, X.; Zou, L.H.; Zheng, B.Q.; Tian, Y.Q.; Wang, Y. Transcriptomic profiling for prolonged drought in Dendrobium catenatum. Sci. Data 2018, 5, 180233. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.Y.; Huang, H.; Tong, Y.; Xia, E.H.; Gao, L.Z. Transcriptome analysis identifies candidate genes related to triacylglycerol biosynthesis, flower coloration and flowering time control in Camellia reticulata (Theaceae), a well-known ornamental and oil-producing plant. Front. Plant Sci. 2016, 7, 163. [Google Scholar] [CrossRef] [PubMed]
| No. | Gene Name | Gene ID | Functional Annotations | Position | Protein Length (aa) | MW | pI | Predicted Subcellular Localization | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | DcaCSD1 | Dca002609 | Cu/ZnSOD | KZ503041.1:12,499,497–2,513,682 | 224 | 22,761.91 | 6.43 | Chloroplast | 0.086 |
| 2 | DcaCSD2 | Dca003218 | Cu/ZnSOD | KZ501977.1:4,636,681–4,648,211 | 152 | 15,312.98 | 5.34 | Cytoplasmic | −0.208 |
| 3 | DcaCSD3 | Dca012338 | Cu/ZnSOD | KZ503541.1:64,465–66,952 | 222 | 22,926.95 | 6.29 | Cytoplasmic | −0.008 |
| 4 | DcaCSD4 | Dca016361 | Cu/ZnSOD | KZ502953.1:468,317–473,374 | 164 | 16,547.33 | 6.16 | Cytoplasmic | −0.151 |
| 5 | DcaFSD1 | Dca023874 | FeSOD | KZ501928.1:290,962–306,815 | 228 | 26,325.25 | 6.71 | Chloroplast | −0.344 |
| 6 | DcaFSD2 | Dca004481 | FeSOD | KZ502877.1:8,227,560–8,230,822 | 269 | 30,783.39 | 4.91 | Chloroplast | −0.488 |
| 7 | DcaFSD3 | Dca024864 | FeSOD | KZ503331.1:79–387 | 76 | 26,554.21 | 5.66 | None | 0.603 |
| 8 | DcaMSD1 | Dca024548 | MnSOD | KZ502716.1:147,498–155,759 | 238 | 26,554.21 | 8.61 | Mitochondrial | −0.343 |
| Gene ID | Correlation Coefficient | p-Value | Description |
|---|---|---|---|
| DcaCSD1 | |||
| DN40231_c3_g1_i3 | 0.999 | 0.0004 | MADS box protein DOMADS2 |
| DN32316_c5_g1_i3 | 0.999 | 0.0005 | HVA22-like protein e |
| DN28998_c0_g1_i2 | −0.999 | 0.0007 | Cytochrome P450 90B2 |
| DN33200_c0_g6_i4 | 0.999 | 0.0008 | / |
| DN37544_c1_g1_i1 | 0.999 | 0.0012 | CDT1-like protein a, chloroplastic |
| DN31187_c4_g1_i2 | 0.999 | 0.001 | Sucrose synthase |
| DN36361_c1_g5_i2 | −0.997 | 0.0026 | Trimethyltridecatetraene synthase |
| DN33357_c2_g2_i1 | 0.996 | 0.0038 | Putative glutathione peroxidase 7, chloroplastic |
| DN24425_c0_g1_i2 | 0.996 | 0.0039 | Dermcidin |
| DN32586_c2_g4_i1 | 0.996 | 0.0041 | Aldehyde dehydrogenase family 2 member C4 |
| DN36550_c0_g2_i4 | 0.996 | 0.0041 | DEAD-box ATP-dependent RNA helicase 50 |
| DN29711_c0_g1_i2 | 0.996 | 0.0044 | Keratin, type I cytoskeletal 14 |
| DN78496_c0_g1_i1 | 0.996 | 0.0044 | Apolipoprotein E |
| DN39306_c2_g1_i5 | 0.995 | 0.0048 | Keratin, type I cytoskeletal 10 |
| DN29035_c0_g1_i1 | 0.995 | 0.0049 | Probable histone H2A.2 |
| DcaCSD2 | |||
| DN29768_c0_g1_i3 | −0.995 | 0.0046 | Proline-rich receptor-like protein kinase PERK13 |
| DN33141_c1_g5_i1 | −0.995 | 0.0049 | / |
| DN33417_c1_g1_i2 | −0.999 | 0.0013 | Zinc-finger homeodomain protein 2 |
| DN34829_c2_g1_i11 | −0.997 | 0.0027 | Receptor-like serine/threonine-protein kinase SD1-7 |
| DN36012_c3_g1_i1 | −0.999 | 0.0008 | WAT1-related protein At5g64700 |
| DN36361_c1_g1_i2 | −0.996 | 0.0037 | Trimethyltridecatetraene synthase |
| DN36986_c4_g1_i2 | 0.997 | 0.0033 | Starch branching enzyme I |
| DN37712_c8_g3_i2 | −0.999 | 0.0003 | 3-O-acetylpapaveroxine carboxylesterase CXE1 |
| DN38190_c0_g2_i6 | −0.997 | 0.0027 | Ricin B-like lectin R40C1 |
| DN38200_c5_g1_i1 | −0.995 | 0.0049 | / |
| DN38969_c0_g3_i6 | −0.998 | 0.0020 | RNA-directed DNA polymerase homolog |
| DN40032_c2_g2_i2 | −0.999 | 0.0008 | Transposon Ty3-I Gag-Pol polyprotein |
| DN40530_c1_g4_i2 | 0.998 | 0.0025 | Probable alpha-mannosidase At5g13980 |
| DN40586_c9_g5_i1 | 0.997 | 0.0030 | Homeobox-leucine zipper protein HOX32 |
| DN40622_c1_g3_i1 | −0.998 | 0.0018 | Protein ASPARTIC PROTEASE IN GUARD CELL 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Wang, H.; Tong, Y.; Wang, Y. Insights into the Superoxide Dismutase Gene Family and Its Roles in Dendrobium catenatum under Abiotic Stresses. Plants 2020, 9, 1452. https://doi.org/10.3390/plants9111452
Huang H, Wang H, Tong Y, Wang Y. Insights into the Superoxide Dismutase Gene Family and Its Roles in Dendrobium catenatum under Abiotic Stresses. Plants. 2020; 9(11):1452. https://doi.org/10.3390/plants9111452
Chicago/Turabian StyleHuang, Hui, Hui Wang, Yan Tong, and Yuhua Wang. 2020. "Insights into the Superoxide Dismutase Gene Family and Its Roles in Dendrobium catenatum under Abiotic Stresses" Plants 9, no. 11: 1452. https://doi.org/10.3390/plants9111452
APA StyleHuang, H., Wang, H., Tong, Y., & Wang, Y. (2020). Insights into the Superoxide Dismutase Gene Family and Its Roles in Dendrobium catenatum under Abiotic Stresses. Plants, 9(11), 1452. https://doi.org/10.3390/plants9111452





