Plant Histone HTB (H2B) Variants in Regulating Chromatin Structure and Function
Abstract
1. Introduction
2. Animal H2Bs
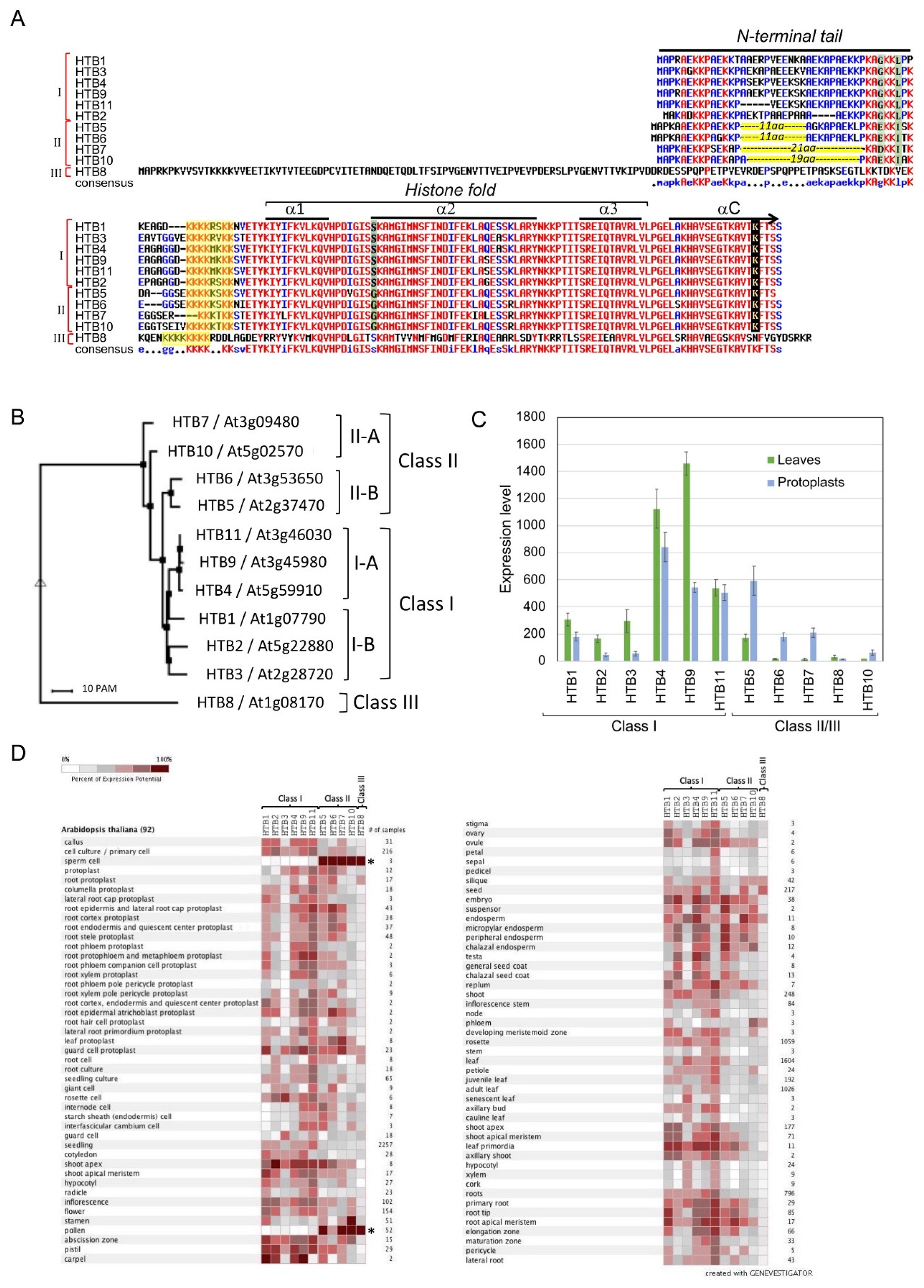
3. Plant H2Bs
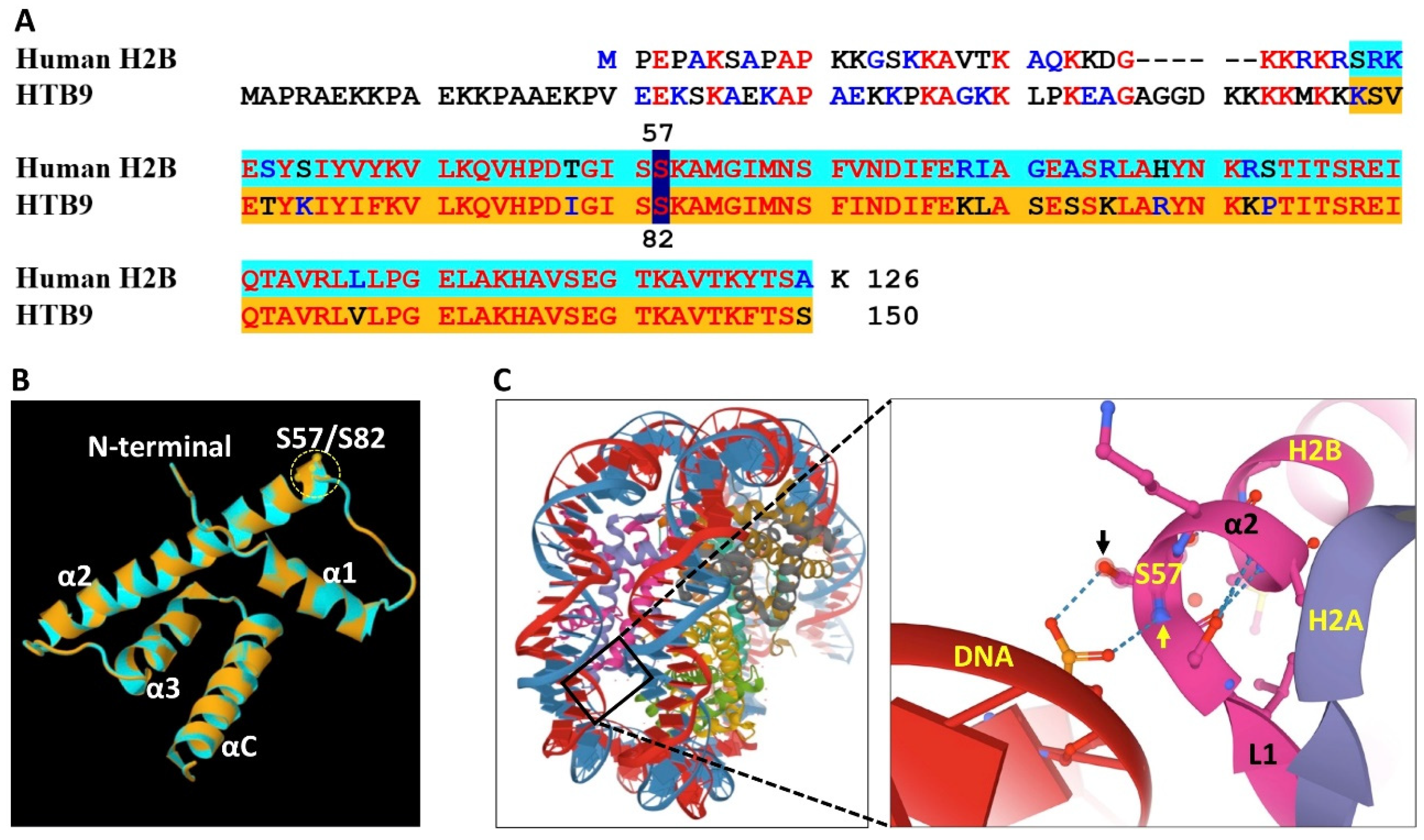
4. Amino Acid Substitutions and Functional Significance
5. H2B Variants Posttranslational Modifications
6. Expression of HTB Variants
7. Subnuclear Localization and Genomic Distribution of HTB Proteins
8. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Arents, G.; Moudrianakis, E.N. The histone fold: A ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 1995, 92, 11170–11174. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.V.; Desvoyes, B.; Gutierrez, C. Similar yet Critically Different: The distribution, dynamics and function of Histone Variants. J. Exp. Bot. 2020, 71, 5191–5204. [Google Scholar] [CrossRef]
- Steiner, F.A.; Henikoff, S. Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev. 2015, 31, 28–35. [Google Scholar] [CrossRef]
- Stroud, H.; Otero, S.; Desvoyes, B.; Ramírez-Parra, E.; Jacobsen, S.E.; Gutierrez, C. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 5370–5375. [Google Scholar] [CrossRef]
- Wollmann, H.; Stroud, H.; Yelagandula, R.; Tarutani, Y.; Jiang, D.; Jing, L.; Jamge, B.; Takeuchi, H.; Holec, S.; Nie, X.; et al. The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 2017, 18, 94. [Google Scholar] [CrossRef]
- Lei, B.; Berger, F. H2A variants in Arabidopsis: Versatile regulators of genome activity. Plant Commun. 2020, 1, 100015. [Google Scholar] [CrossRef]
- Osakabe, A.; Lorkovic, Z.J.; Kobayashi, W.; Tachiwana, H.; Yelagandula, R.; Kurumizaka, H.; Berger, F. Histone H2A variants confer specific properties to nucleosomes and impact on chromatin accessibility. Nucleic Acids Res. 2018, 46, 7675–7685. [Google Scholar] [CrossRef]
- Lorković, Z.J.; Park, C.; Goiser, M.; Jiang, D.; Kurzbauer, M.T.; Schlögelhofer, P.; Berger, F. Compartmentalization of DNA Damage Response between Heterochromatin and Euchromatin Is Mediated by Distinct H2A Histone Variants. Curr. Biol. 2017, 27, 1192–1199. [Google Scholar] [CrossRef]
- Roitinger, E.; Hofer, M.; Köcher, T.; Pichler, P.; Novatchkova, M.; Yang, J.; Schlögelhofer, P.; Mechtler, K. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteomics 2015, 14, 556–571. [Google Scholar] [CrossRef]
- Turinetto, V.; Giachino, C. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015, 43, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Lorković, Z.J.; Nishihama, R.; Ishizaki, K.; Axelsson, E.; Yelagandula, R.; Kohchi, T.; Berger, F. Diversification of histone H2A variants during plant evolution. Trends Plant Sci. 2015, 20, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. Histone variants--ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010, 11, 264–275. [Google Scholar] [CrossRef]
- Jiang, D.; Berger, F. Histone variants in plant transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 123–130. [Google Scholar] [CrossRef]
- Jiang, D.; Borg, M.; Lorković, Z.J.; Montgomery, S.A.; Osakabe, A.; Yelagandula, R.; Axelsson, E.; Berger, F. The evolution and functional divergence of the histone H2B family in plants. PLoS Genet. 2020, 16, e1008964. [Google Scholar] [CrossRef]
- Zambrano-Mila, M.S.; Aldaz-Villao, M.J.; Armando Casas-Mollano, J. Canonical histones and their variants in plants: Evolution and functions. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications; Alvarez-Venegas, R., De-la-Pena, C., Cassas-Molano, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 185–222. [Google Scholar]
- Susano Pinto, D.M.; Flaus, A. The human canonical core histone catalogue. BioRxiv 2019, 720235. [Google Scholar] [CrossRef]
- González-Romero, R.; Rivera-Casas, C.; Ausió, J.; Méndez, J.; Eirín-López, J.M. Birth-and-death long-term evolution promotes histone H2B variant diversification in the male germinal cell line. Mol. Biol. Evol. 2010, 27, 1802–1812. [Google Scholar] [CrossRef]
- Zalensky, A.O.; Siino, J.S.; Gineitis, A.A.; Zalenskaya, I.A.; Tomilin, N.V.; Yau, P.; Bradbury, E.M. Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J. Biol. Chem. 2002, 277, 43474–43480. [Google Scholar] [CrossRef]
- Govin, J.; Escoffier, E.; Rousseaux, S.; Kuhn, L.; Ferro, M.; Thévenon, J.; Catena, R.; Davidson, I.; Garin, J.; Khochbin, S.; et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 2007, 176, 283–294. [Google Scholar] [CrossRef]
- Montellier, E.; Boussouar, F.; Rousseaux, S.; Zhang, K.; Buchou, T.; Fenaille, F.; Shiota, H.; Debernardi, A.; Héry, P.; Curtet, S.; et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013, 27, 1680–1692. [Google Scholar] [CrossRef]
- Shinagawa, T.; Takagi, T.; Tsukamoto, D.; Tomaru, C.; Huynh, L.M.; Sivaraman, P.; Kumarevel, T.; Inoue, K.; Nakato, R.; Katou, Y.; et al. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 2014, 14, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Huynh, L.M.; Shinagawa, T.; Ishii, S. Two Histone Variants TH2A and TH2B Enhance Human Induced Pluripotent Stem Cell Generation. Stem Cells Dev. 2016, 25, 251–258. [Google Scholar] [CrossRef]
- Urahama, T.; Horikoshi, N.; Osakabe, A.; Tachiwana, H.; Kurumizaka, H. Structure of human nucleosome containing the testis-specific histone variant TSH2B. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Török, A.; Schiffer, P.H.; Schnitzler, C.E.; Ford, K.; Mullikin, J.C.; Baxevanis, A.D.; Bacic, A.; Frank, U.; Gornik, S.G. The cnidarian Hydractinia echinata employs canonical and highly adapted histones to pack its DNA. Epigenetics Chromatin 2016, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Dulac, C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. Elife 2012, 1, e00070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zang, C.; Rosenfeld, J.A.; Schones, D.E.; Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Peng, W.; Zhang, M.Q.; et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008, 40, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Chabouté, M.E.; Chaubet, N.; Gigot, C.; Philipps, G. Histones and histone genes in higher plants: Structure and genomic organization. Biochimie 1993, 75, 523–531. [Google Scholar] [CrossRef]
- Malik, H.S.; Henikoff, S. Phylogenomics of the nucleosome. Nat. Struct. Biol. 2003, 10, 882–891. [Google Scholar] [CrossRef]
- Hu, Y.; Lai, Y. Identification and expression analysis of rice histone genes. Plant Physiol. Biochem. 2015, 86, 55–65. [Google Scholar] [CrossRef]
- Bergmüller, E.; Gehrig, P.M.; Gruissem, W. Characterization of post-translational modifications of histone H2B-variants isolated from Arabidopsis thaliana. J. Proteome Res. 2007, 6, 3655–3668. [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Vipond, I.B.; Moon, B.J.; Halford, S.E. An isoleucine to leucine mutation that switches the cofactor requirement of the EcoRV restriction endonuclease from magnesium to manganese. Biochemistry 1996, 35, 1712–1721. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.Y.; Walsh, A.R.; Materne, E.C.; Hiltner, E.P.; Zielinski, B.; Miller, B.R., 3rd; Mawby, L.; Modeste, E.; Parish, C.A.; Barnes, W.M.; et al. A conservative isoleucine to leucine mutation causes major rearrangements and cold sensitivity in KlenTaq1 DNA polymerase. Biochemistry 2015, 54, 881–889. [Google Scholar] [CrossRef]
- Tachiwana, H.; Osakabe, A.; Shiga, T.; Miya, Y.; Kimura, H.; Kagawa, W.; Kurumizaka, H. Structures of human nucleosomes containing major histone H3 variants. Acta Cryst. 2011, D67, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Eberharter, A.; Becker, P.B. Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002, 3, 224–229. [Google Scholar] [CrossRef]
- Clayton, A.L.; Hazzalin, C.A.; Mahadevan, L.C. Enhanced histone acetylation and transcription: A dynamic perspective. Mol. Cell. 2006, 23, 289–296. [Google Scholar] [CrossRef]
- Shilatifard, A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006, 75, 243–269. [Google Scholar] [CrossRef]
- Fiorucci, A.S.; Bourbousse, C.; Concia, L.; Rougée, M.; Deton-Cabanillas, A.F.; Zabulon, G.; Layat, E.; Latrasse, D.; Kim, S.K.; Chaumont, N.; et al. Arabidopsis S2Lb links AtCOMPASS-like and SDG2 activity in H3K4me3 independently from histone H2B monoubiquitination. Genome Biol. 2019, 20, 100. [Google Scholar] [CrossRef]
- Bourbousse, C.; Ahmed, I.; Roudier, F.; Zabulon, G.; Blondet, E.; Balzergue, S.; Colot, V.; Bowler, C.; Barneche, F. Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 2012, 8, e1002825. [Google Scholar] [CrossRef]
- Himanen, K.; Woloszynska, M.; Boccardi, T.M.; De Groeve, S.; Nelissen, H.; Bruno, L.; Vuylsteke, M.; Van Lijsebettens, M. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 2012, 72, 249–260. [Google Scholar] [CrossRef]
- Sridhar, V.V.; Kapoor, A.; Zhang, K.; Zhu, J.; Zhou, T.; Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 2007, 447, 735–738. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Ji, Z.; Liu, Y.; Zhang, Q. BRHIS1 suppresses rice innate immunity through binding to monoubiquitinated H2A and H2B variants. EMBO Rep. 2015, 16, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Henry, K.W.; Wyce, A.; Lo, W.S.; Duggan, L.J.; Emre, N.C.; Kao, C.F.; Pillus, L.; Shilatifard, A.; Osley, M.A.; Berger, S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003, 17, 2648–2663. [Google Scholar] [CrossRef]
- Weake, V.M.; Lee, K.K.; Guelman, S.; Lin, C.H.; Seidel, C.; Abmayr, S.M.; Workman, J.L. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008, 27, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Pfab, A.; Bruckmann, A.; Nazet, J.; Merkl, R.; Grasser, K.D. The Adaptor Protein ENY2 Is a Component of the Deubiquitination Module of the Arabidopsis SAGA Transcriptional Co-activator Complex but not of the TREX-2 Complex. J. Mol. Biol. 2018, 430, 1479–1494. [Google Scholar] [CrossRef] [PubMed]
- Fleury, D.; Himanen, K.; Cnops, G.; Nelissen, H.; Boccar di, T.M.; Maere, S.; Beemster, G.T.; Neyt, P.; Anami, S.; Robles, P.; et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 2007, 19, 417–432. [Google Scholar] [CrossRef]
- Liu, Y.; Koornneef, M.; Soppe, W.J. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 2007, 19, 433–444. [Google Scholar] [CrossRef]
- Cao, Y.; Dai, Y.; Cui, S.; Ma, L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 2008, 20, 2586–2602. [Google Scholar] [CrossRef]
- Gu, X.; Jiang, D.; Wang, Y.; Bachmair, A.; He, Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009, 57, 522–533. [Google Scholar] [CrossRef]
- Xu, L.; Ménard, R.; Berr, A.; Fuchs, J.; Cognat, V.; Meyer, D.; Shen, W.H. The E2 ubiquitin- conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009, 57, 279–288. [Google Scholar] [CrossRef]
- Ménard, R.; Verdier, G.; Ors, M.; Erhardt, M.; Beisson, F.; Shen, W.H. Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Pei, B.L.; Zhang, L.F.; Li, Y.Z. Histone H2B monoubiquitination is involved in regulating the dynamics of microtubules during the defense response to Verticillium dahliae toxins in Arabidopsis. Plant Physiol. 2014, 164, 1857–1865. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, R.; Luo, H.; Foerster, A.M.; Abuqamar, S.; Du, H.N.; Briggs, S.D.; Mittelsten Scheid, O.; Mengiste, T. HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 2009, 21, 1000–1019. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Yang, D.L.; Shi, Z.; Dong, H.; Hua, J. Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol. 2014, 165, 309–318. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Cole, A.J.; Clifton-Bligh, R.; Marsh, D.J. Histone H2B monoubiquitination: Roles to play in human malignancy. Endocr. Relat. Cancer. 2015, 22, T19–T33. [Google Scholar] [CrossRef]
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012, 2, 26. [Google Scholar] [CrossRef]
- Robzyk, K.; Recht, J.; Osley, M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science 2000, 287, 501–504. [Google Scholar] [CrossRef]
- Nassrallah, A.; Rougée, M.; Bourbousse, C.; Drevensek, S.; Fonseca, S.; Iniesto, E.; Ait-Mohamed, O.; Deton-Cabanillas, A.-F.; Zabulon, G.; Ahmed, I.; et al. DET1-mediated degradation of a SAGA-like deubiquitination module controls H2Bub homeostasis. Elife 2018, 7, e37892. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Morozova, N.; Williams, L.; Libs, L.; Avivi, Y.; Grafi, G. Two phases of chromatin decondensation during dedifferentiation of plant cells: Distinction between competence for cell fate switch and a commitment for S phase. J. Biol. Chem. 2001, 276, 22772–22778. [Google Scholar] [CrossRef] [PubMed]
- Tessadori, F.; Chupeau, M.C.; Chupeau, Y.; Knip, M.; Germann, S.; van Driel, R.; Fransz, P.; Gaudin, V. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell Sci. 2007, 120, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Givaty-Rapp, Y.; Yadav, N.S.; Khan, A.; Grafi, G. S1-Type Endonuclease 2 in Dedifferentiating Arabidopsis Protoplasts: Translocation to the Nucleus in Senescing Protoplasts Is Associated with De-Glycosylation. PLoS ONE 2017, 12, e0170067. [Google Scholar] [CrossRef]
- Damri, M.; Granot, G.; Ben-Meir, H.; Avivi, Y.; Plaschkes, I.; Chalifa-Caspi, V.; Wolfson, M.; Fraifeld, V.; Grafi, G. Senescing cells share common features with dedifferentiating cells. Rejuvenation Res. 2009, 12, 435–443. [Google Scholar] [CrossRef]
- Hoffmann, R.D.; Palmgren, M.G. Epigenetic repression of male gametophyte-specific genes in the Arabidopsis sporophyte. Mol. Plant 2013, 6, 1176–1186. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Yang, N.; Wang, T. Proteomic analysis reveals the differential histone programs between male germline cells and vegetative cells in Lilium davidii. Plant J. 2016, 85, 660–674. [Google Scholar] [CrossRef]
- Borg, M.; Berger, F. Chromatin remodelling during male gametophyte development. Plant J. 2015, 83, 177–188. [Google Scholar] [CrossRef]
- Van Zanten, M.; Koini, M.A.; Geyer, R.; Liu, Y.; Brambilla, V.; Bartels, D.; Koornneef, M.; Fransz, P.; Soppe, W.J. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc. Natl. Acad. Sci. USA 2011, 108, 20219–20224. [Google Scholar] [CrossRef]
- Pesok, A. The Role of HTB5-a Histone H2B Variant-in Plant Development and Stress-Induced Dedifferentiation. Master’s Thesis, Ben Gurion University, Beersheba, Israel, 2014. [Google Scholar]
- Zemach, A.; Li, Y.; Wayburn, B.; Ben-Meir, H.; Kiss, V.; Avivi, Y.; Kalchenko, V.; Jacobsen, S.E.; Grafi, G. DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant Cell 2005, 17, 1549–1558. [Google Scholar] [CrossRef]
- Preuss, S.B.; Costa-Nunes, P.; Tucker, S.; Pontes, O.; Lawrence, R.J.; Mosher, R.; Kasschau, K.D.; Carrington, J.C.; Baulcombe, D.C.; Viegas, W.; et al. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell 2008, 32, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Di Padua-Mathieu, D.; Mura, C.V.; Frado, L.-L.Y.; Woodcock, C.L.; Stollar, B.D. Differing accessibility in chromatin of the antigenic sites of regions 1-58 and 63-125 of histone H2B. J. Cell Biol. 1981, 91, 135–141. [Google Scholar] [CrossRef]
- Sen Gupta, A.; Joshi, G.; Pawar, S.; Sengupta, K. Nucleolin modulates compartmentalization and dynamics of histone 2B-ECFP in the nucleolus. Nucleus 2018, 9, 350–367. [Google Scholar] [CrossRef] [PubMed]
- Musinova, Y.R.; Lisitsyna, O.M.; Golyshev, S.A.; Tuzhikov, A.I.; Polyakov, V.Y.; Sheval, E.V. Nucleolar localization/retention signal is responsible for transient accumulation of histone H2B in the nucleolus through electrostatic interactions. Biochim. Biophys. Acta 2011, 1813, 27–38. [Google Scholar] [CrossRef]
- Parra, M.A.; Kerr, D.; Fahy, D.; Pouchnik, D.J.; Wyrick, J.J. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol. Cell. Biol. 2006, 26, 3842–3852. [Google Scholar] [CrossRef]
- Brown, J.W.; Shaw, P.J.; Shaw, P.; Marshall, D.F. Arabidopsis nucleolar protein database (AtNoPDB). Nucleic Acids Res. 2005, 33, D633–D636. [Google Scholar] [CrossRef][Green Version]
- Lau, A.T.; Lee, S.Y.; Xu, Y.M.; Zheng, D.; Cho, Y.Y.; Zhu, F.; Kim, H.G.; Li, S.Q.; Zhang, Z.; Bode, A.M.; et al. Phosphorylation of histone H2B serine 32 is linked to cell transformation. J. Biol. Chem. 2011, 286, 26628–26637. [Google Scholar] [CrossRef]
- Tessarz, P.; Santos-Rosa, H.; Robson, S.C.; Sylvestersen, K.B.; Nelson, C.J.; Nielsen, M.L.; Kouzarides, T. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 2014, 505, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Loza-Muller, L.; Rodríguez-Corona, U.; Sobol, M.; Rodríguez-Zapata, L.C.; Hozak, P.; Castano, E. Fibrillarin methylates H2A in RNA polymerase I trans-active promoters in Brassica oleracea. Front. Plant Sci. 2015, 6, 976. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khadka, J.; Pesok, A.; Grafi, G. Plant Histone HTB (H2B) Variants in Regulating Chromatin Structure and Function. Plants 2020, 9, 1435. https://doi.org/10.3390/plants9111435
Khadka J, Pesok A, Grafi G. Plant Histone HTB (H2B) Variants in Regulating Chromatin Structure and Function. Plants. 2020; 9(11):1435. https://doi.org/10.3390/plants9111435
Chicago/Turabian StyleKhadka, Janardan, Anat Pesok, and Gideon Grafi. 2020. "Plant Histone HTB (H2B) Variants in Regulating Chromatin Structure and Function" Plants 9, no. 11: 1435. https://doi.org/10.3390/plants9111435
APA StyleKhadka, J., Pesok, A., & Grafi, G. (2020). Plant Histone HTB (H2B) Variants in Regulating Chromatin Structure and Function. Plants, 9(11), 1435. https://doi.org/10.3390/plants9111435






