Linkage Map Development by EST-SSR Markers and QTL Analysis for Inflorescence and Leaf Traits in Chrysanthemum (Chrysanthemum morifolium Ramat.)
Abstract
1. Introduction
2. Results
2.1. EST-SSR Marker Segregation
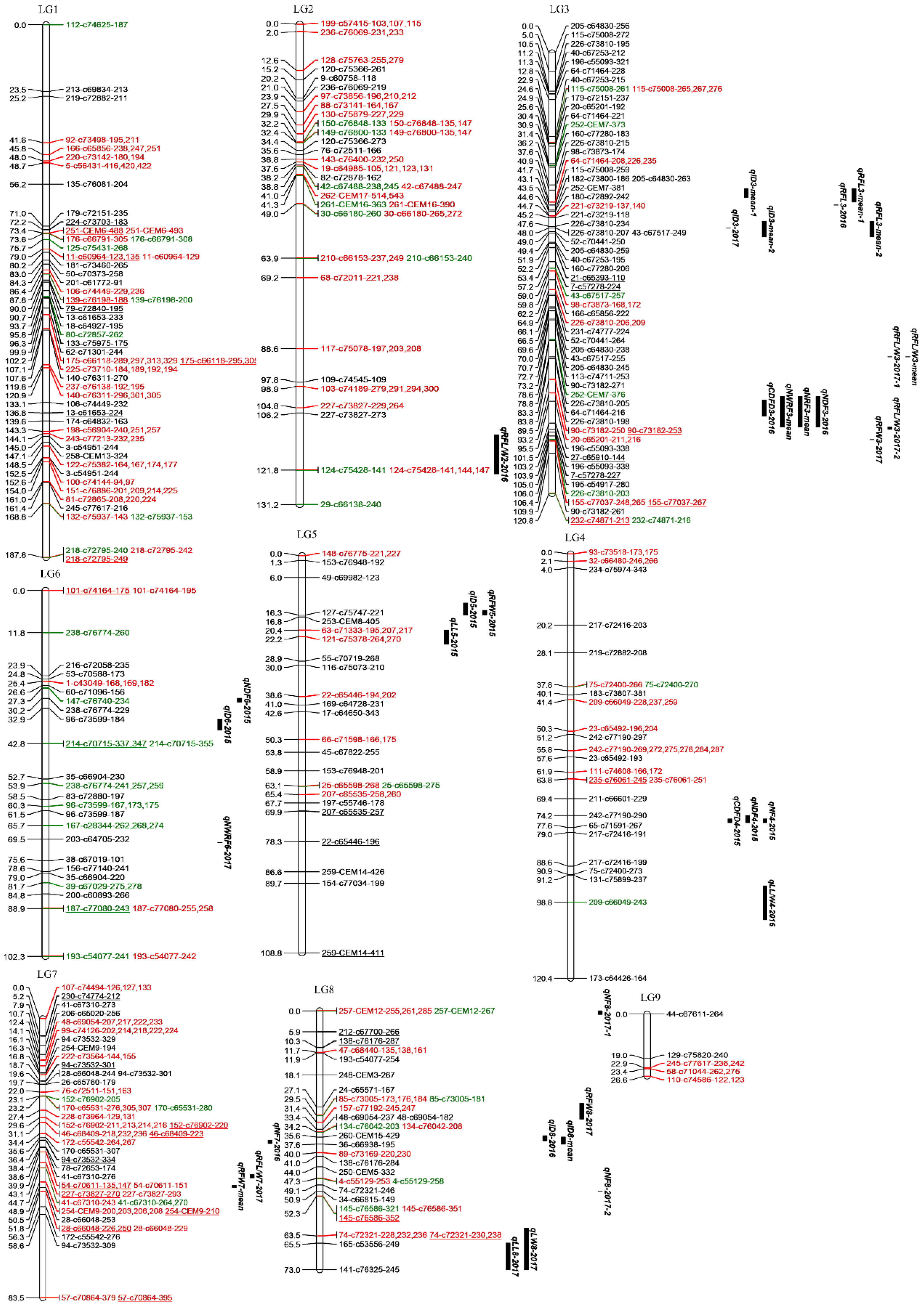
2.2. Linkage Map Construction
2.3. Phenotypic Evaluation
2.4. QTL Identification
2.4.1. Inflorescence Traits
2.4.2. Leaf Traits
3. Discussion
3.1. EST-SSR Markers
3.2. Genetic Map Construction
3.3. Phenotypic Characterization
3.4. QTL Mapping
4. Materials and Method
4.1. Plant Materials and DNA Extraction
4.2. Genotyping of Mapping Population
4.3. Marker Scoring and Marker Segregation Type
4.4. Linkage Map Construction
4.5. Phenotyping and Statistical Analysis
4.6. QTL Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, Y.; Jiang, Z.; Guo, J.; Yang, X.; Xu, N.; Chen, Z.; Hao, J.; Li, J.; Pang, J.; Shen, C.; et al. Transcriptomic analyses of Chrysanthemum morifolium Ramat. under UV-B radiation treatment reveal variations in the metabolisms associated with bioactive components. Ind. Crop. Prod. 2018, 124, 475–486. [Google Scholar] [CrossRef]
- Pharmacopoeia of People’s Republic of China. The State of Pharmacopoeia Commission of People’s Republic of China; Chemical Industry Press: Beijing, China, 2010; Volume 1. [Google Scholar]
- Anderson, N.O.; Ascher, P.D. Fertility changes in inbred families of self-incompatible chrysanthemums (Dendranthema grandiflora). J. Am. Soc. Hortic. Sci. 2000, 125, 619–625. [Google Scholar] [CrossRef]
- Song, A.; Song, C.; Liu, Y.; Dong, G.; Dong, B.; Zhao, H.; Sun, W.; Ramakrishnan, S.; Wang, S.; Wang, Y.; et al. The Chrysanthemum nankingense Genome Provides Insights into the Evolution and Diversification of Chrysanthemum Flowers and Medicinal Traits. Mol. Plant. 2018, 11, 1482–1491. [Google Scholar] [CrossRef]
- Hirakawa, H.; Sumitomo, K.; Hisamatsu, T.; Nagano, S.; Shirasawa, K.; Higuchi, Y.; Kusaba, M.; Koshioka, M.; Nakano, Y.; Yagi, M.; et al. De novo whole-genome assembly in Chrysanthemum seticuspe, a model species of Chrysanthemums, and its application to genetic and gene discovery analysis. DNA Res. 2019, 26, 195–203. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, S.; Chen, F.; Fang, W.; Li, F. A preliminary genetic linkage map of chrysanthemum (Chrysanthemum morifolium) cultivars using RAPD, ISSR and AFLP markers. Sci. Hortic. 2010, 125, 422–428. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, S.; Chen, F.; Fang, W.; Deng, Y.; Chang, Q.; Liu, P. Genetic analysis and associated SRAP markers for flowering traits of chrysanthemum (Chrysanthemum morifolium). Euphytica 2011, 177, 15–24. [Google Scholar] [CrossRef]
- Klie, M.; Schie, S.; Linde, M.; Debener, T. The type of ploidy of chrysanthemum is not black or white: A comparison of a molecular approach to published cytological methods. Front. Plant. Sci. 2014, 5, 479. [Google Scholar] [CrossRef]
- Park, S.K.; Arens, P.; Esselink, D.; Lim, J.H.; Shin, H.K. Analysis of inheritance mode in chrysanthemum using EST-derived SSR markers. Sci. Hortic. 2015, 192, 80–88. [Google Scholar] [CrossRef]
- Van Geest, G.; Voorrips, R.E.; Esselink, D.; Post, A.; Visser, R.G.; Arens, P. Conclusive evidence for hexasomic inheritance in chrysanthemum based on analysis of a 183 k SNP array. BMC Genomics 2017, 18, 585. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Young, N.D.; Bonierbale, M.W. RFLP Mapping in Plant Breeding: New Tools for an Old Science. Biotechnology 1989, 7, 257–264. [Google Scholar] [CrossRef]
- Sargent, D.J.; Passey, T.; Šurbanovski, N.; Lopez Girona, E.; Kuchta, P.; Davik, J.; Harrison, R.; Passey, A.; Whitehouse, A.B.; Simpson, D.W. A microsatellite linkage map for the cultivated strawberry (Fragaria × ananassa) suggests extensive regions of homozygosity in the genome that may have resulted from breeding and selection. Theor. Appl. Genet. 2012, 124, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Yu, X.; Jie, Q.; Li, H.; Li, H.; Hu, J.; Zhai, H.; He, S.; Liu, Q. A genetic linkage map based on AFLP and SSR markers and mapping of QTL for dry-matter content in sweetpotato. Mol. Breed. 2013, 32, 807–820. [Google Scholar] [CrossRef]
- Ukoskit, K.; Posudsavang, G.; Pongsiripat, N.; Chatwachirawong, P.; Klomsa-ard, P.; Poomipant, P.; Tragoonrung, S. Detection and validation of EST-SSR markers associated with sugar-related traits in sugarcane using linkage and association mapping. Genomics 2019, 111, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Chen, S.; Chen, F.; Fang, W.; Chen, Y.; Li, F. SRAP-based mapping and QTL detection for inflorescence-related traits in chrysanthemum (Dendranthema morifolium). Mol. Breed. 2011, 27, 11–23. [Google Scholar] [CrossRef]
- Zhang, F.; Jiang, J.; Chen, S.; Chen, F.; Fang, W. Mapping single-locus and epistatic quantitative trait loci for plant architectural traits in chrysanthemum. Mol. Breed. 2012, 30, 1027–1036. [Google Scholar] [CrossRef]
- Zhang, F.; Jiang, J.; Chen, S.; Chen, F.; Fang, W. Detection of quantitative trait loci for leaf traits in chrysanthemum. J. Hortic. Sci. Biotechnol. 2012, 87, 613–618. [Google Scholar] [CrossRef]
- Van Geest, G.; Bourke, P.M.; Voorrips, R.E.; Marasek-Ciolakowska, A.; Liao, Y.; Post, A.; van Meeteren, U.; Visser, R.G.F.; Maliepaard, C.; Arens, P. An ultra-dense integrated linkage map for hexaploid chrysanthemum enables multi-allelic QTL analysis. Theor. Appl. Genet. 2017, 130, 2527–2541. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Yang, X.; Zhang, F.; Wu, S.; Xiong, S.; Shi, L.; Guan, Z.; Fang, W.; Chen, F. Dynamic and epistatic QTL mapping reveals the complex genetic architecture of waterlogging tolerance in chrysanthemum. Planta 2018, 247, 899–924. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, S.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F. Genetic mapping of quantitative trait loci underlying flowering time in chrysanthemum (Chrysanthemum morifolium). PLoS ONE 2013, 8, e83023. [Google Scholar] [CrossRef][Green Version]
- Song, X.; Xu, Y.; Gao, K.; Fan, G.; Zhang, F.; Deng, C.; Dai, S.; Huang, H.; Xin, H.; Li, Y. High-density genetic map construction and identification of loci controlling flower-type traits in Chrysanthemum (Chrysanthemum × morifolium Ramat.). Hortic. Res. 2020, 7, 108. [Google Scholar] [CrossRef]
- Truco, M.J.; Antonise, R.; Lavelle, D.; Ochoa, O.; Kozik, A.; Witsenboer, H.; Fort, S.B.; Jeuken, M.J.W.; Kesseli, R.V.; Lindhout, P.; et al. A high-density, integrated genetic linkage map of lettuce (Lactuca spp.). Theor. Appl. Genet. 2007, 115, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Yagi, M.; Kimura, T.; Yamamoto, T.; Isobe, S.; Tabata, S.; Onozaki, T. QTL analysis for resistance to bacterial wilt (Burkholderia caryophylli) in carnation (Dianthus caryophyllus) using an SSR-based genetic linkage map. Mol. Breed. 2012, 30, 495–509. [Google Scholar] [CrossRef]
- Tan, L.; Wang, L.; Xu, L.; Wu, L.; Peng, M.; Zhang, C.; Wei, K.; Bai, P.; Li, H.; Cheng, H.; et al. SSR-based genetic mapping and QTL analysis for timing of spring bud flush, young shoot color, and mature leaf size in tea plant (Camellia sinensis). Tree Genet. Genomes 2016, 12. [Google Scholar] [CrossRef]
- Van Geest, G. Disentangling hexaploid genetics: Towards DNA-informed breeding for postharvest performance in chrysanthemum. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2017. [Google Scholar]
- Bourke, P.M.; Voorrips, R.E.; Visser, R.G.F.; Maliepaard, C. Tools for genetic studies in experimental populations of polyploids. Front. Plant. Sci. 2018, 9, 513. [Google Scholar] [CrossRef]
- Zhang, Y.; Sledge, M.K.; Bouton, J.H. Genome mapping of white clover (Trifolium repens L.) and comparative analysis within the Trifolieae using cross-species SSR markers. Theor. Appl. Genet. 2007, 114, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Luo, L.; Pan, H.; Guo, X.; Wan, H.; Zhang, Q. Filling gaps with construction of a genetic linkage map in tetraploid roses. Front. Plant. Sci. 2015, 5, 796. [Google Scholar] [CrossRef]
- Julier, B.; Flajoulot, S.; Barre, P.; Cardinet, G.; Santoni, S.; Huguet, T.; Huyghe, C. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant. Biol. 2003, 3, 9. [Google Scholar] [CrossRef][Green Version]
- Li, M.; Yuyama, N.; Hirata, M.; Wang, Y.; Han, J.; Cai, H. An integrated SSR based linkage map for Zoysia matrella L. and Z. japonica Steud. Mol. Breed. 2010, 26, 467–476. [Google Scholar] [CrossRef]
- Kesseli, R.V.; Michelmore, I.P.A.R. Analysis of a detailed genetic linkage map of Lactuca sativa (Lettuce) constructed from RFLP and RAPD markers. Genetics 1994, 136, 1435–1446. [Google Scholar]
- Song, X.; Zhao, X.; Fan, G.; Gao, K.; Dai, S.; Zhang, M.; Ma, C.; Wu, X. Genetic analysis of the corolla tube merged degree and the relative number of ray florets in chrysanthemum (Chrysanthemum × morifolium Ramat.). Sci. Hortic. 2018, 242, 214–224. [Google Scholar] [CrossRef]
- DeVicente, M.C.; Tanksley, S.D. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 1993, 134, 585–596. [Google Scholar] [PubMed]
- Koide, Y.; Sakaguchi, S.; Uchiyama, T.; Ota, Y.; Tezuka, A.; Nagano, A.J.; Ishiguro, S.; Takamure, I.; Kishima, Y. Genetic properties responsible for the transgressive segregation of days to heading in rice. G3 2019, 9, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; He, K.; Chang, L.; Cui, T.; Zhao, Z.; Xue, J.; Liu, J. QTL mapping and genetic analysis for maize kernel size and weight in multi-environments. Euphytica 2018, 214, 119. [Google Scholar] [CrossRef]
- Xin, F.; Zhu, T.; Wei, S.; Han, Y.; Zhao, Y.; Zhang, D.; Ma, L.; Ding, Q. QTL mapping of kernel traits and validation of a major QTL for kernel length-width ratio using SNP and bulked segregant analysis in wheat. Sci. Rep. 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, Z.H.; Doebley, J.F. Genetic dissection of a genomic region with pleiotropic effects on domestication traits in maize reveals multiple linked QTL. Genetics 2014, 198, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Gao, Y.; Gao, Y.; Wu, Z.; Liu, H.; Zhang, Q. Characterization and development of EST-SSR markers from transcriptome sequences of chrysanthemum (Chrysanthemum × morifolium Ramat.). Hortscience 2019, 54, 772–778. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Q.X.; Sun, M.; Pan, H.T.; Kong, Z.X. Development of expressed sequence tag-simple sequence repeat markers for Chrysanthemum morifolium and closely related species. Genet. Mol. Res. 2015, 14, 7578–7586. [Google Scholar] [CrossRef]
- Sun, L.; Yang, W.; Zhang, Q.; Cheng, T.; Pan, H.; Xu, Z.; Zhang, J.; Chen, C. Genome-wide characterization and linkage mapping of simple sequence repeats in mei (Prunus mume Sieb. et Zucc.). PLoS ONE 2013, 8, e59562. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Sederoff, R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: Mapping strategy and RAPD markers. Genetics 1994, 137, 1121–1137. [Google Scholar]
- Van Ooijen, J.W. JoinMap ® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma B.V.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Cervantes-Flores, J.C.; Yencho, G.C.; Kriegner, A.; Pecota, K.V.; Faulk, M.A.; Mwanga, R.O.M.; Sosinski, B.R. Development of a genetic linkage map and identification of homologous linkage groups in sweetpotato using multiple-dose AFLP markers. Mol. Breed. 2008, 21, 511–532. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. MapQTL ® 6, Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma B.V.: Wageningen, The Netherlands, 2009. [Google Scholar]

| Marker Types | Autohexaploid (Hexasomic) | Allohexaploid (Disomic) | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of Alleles Present in the Paternal Parent | No. of Alleles Present in the Maternal Parent | No. of Alleles Present in Both Parents | Total | No. of Alleles Present in the Paternal Parent | No. of Alleles Present in the Maternal Parent | No. of Alleles Present in Both Parents | Total | |
| Monomorphic | 3 | 2 | 89 | 94 | 3 | 2 | 89 | 94 |
| Polymorphic | ||||||||
| Simplex × nulliplex | 183 | 179 | - | 362 | 183 | 179 | - | 362 |
| Duplex × nulliplex | 50 | 37 | - | 87 | 28 | 28 | - | 56 |
| Triplex × nulliplex | 23 | 15 | - | 38 | 28 | 21 | - | 49 |
| Simplex × simplex | - | - | 94 | 94 | - | - | 94 | 94 |
| Duplex × simplex | 79 | 79 | - | - | 45 | 45 | ||
| Unidentified dose | 0 | 0 | 158 | 158 | 8 | 6 | 150 | 164 |
| Distorted at α < 0.01 | 26 | 38 | 24 | 88 | 35 | 35 | 66 | 136 |
| Total | 285 | 271 | 444 | 1000 | 285 | 271 | 444 | 1000 |
| Linkage Group | Number of SSR Marker Alleles | Duplex Markers | Number of Markers Exhibiting Segregation Distortion (p < 0.01) | Map Length (cM) | Map Density (loci/cM) | Average Distance Between Markers (cM) | Largest Gap Between Markers (cM) |
|---|---|---|---|---|---|---|---|
| LG1 | 45 | 3 | 2 | 187.80 | 0.24 | 4.17 | 23.49 |
| LG2 | 29 | 1 | 0 | 131.22 | 0.22 | 4.52 | 19.44 |
| LG3 | 58 | 4 | 2 | 120.79 | 0.48 | 2.08 | 10.92 |
| LG4 | 23 | 1 | 0 | 120.39 | 0.19 | 5.23 | 21.64 |
| LG5 | 23 | 0 | 3 | 108.81 | 0.21 | 4.73 | 19.15 |
| LG6 | 24 | 1 | 0 | 102.32 | 0.23 | 4.26 | 13.43 |
| LG7 | 33 | 1 | 2 | 83.48 | 0.40 | 2.53 | 24.92 |
| LG8 | 24 | 0 | 1 | 73.04 | 0.33 | 3.04 | 11.16 |
| LG9 | 5 | 0 | 0 | 26.61 | 0.19 | 5.32 | 18.96 |
| Total | 264 | 11 | 10 | 954.46 | 0.28 | 3.62 | 24.92 |
| Trait | Trait Code | Year | Parent Mean ± SD | Significant Mean Difference Among Parental Values (t-Test) | F1 Population Mean ± SD | Range | Skewness | Kurtosis | Heritability | |
|---|---|---|---|---|---|---|---|---|---|---|
| F | M | |||||||||
| Inflorescence diameter | ID | 2015 | 44.52 ± 2.2 | 45.79 ± 2.84 | n.s | 40.57 ± 7.95 | 16.68–60.7 | −0.32 | 0.23 | 0.81 |
| 2016 | 63.26 ± 1.42 | 47.62 ± 5.16 | Yes, p < 0.001 | 45.86 ± 7.42 | 6.20–63.27 | −1.04 | 4.59 | |||
| 2017 | 59.98 ± 2.11 | 41.09 ± 2.94 | Yes, p < 0.01 | 45.01 ± 8.58 | 20.06–65.86 | −0.01 | −0.45 | |||
| Central disc flower diameter | CDFD | 2015 | 10 ± 0.75 | 3.03 ± 0.80 | Yes, p < 0.01 | 8.80 ± 1.69 | 3.95–12.05 | −0.39 | −0.18 | 0.90 |
| 2016 | 10.16 ± 0.84 | 3.32 ± 0.46 | Yes, p < 0.001 | 8.58 ± 1.99 | 2.92–14.33 | −0.46 | 0.61 | |||
| 2017 | 12.79 ± 0.49 | 3.5 ± 0.28 | Yes, p < 0.001 | 8.72 ± 1.71 | 4.11–12.95 | −0.15 | −0.07 | |||
| Number of whorls of ray florets | NWRF | 2015 | 1 ± 0 | 9.67 ± 1.53 | Yes, p < 0.05 | 2.83 ± 1.44 | 0–8.5 | 1.20 | 2.43 | 0.93 |
| 2016 | 1 ± 0 | 8.5 ± 0.50 | Yes, p < 0.05 | 3.28 ± 1.52 | 1–12.67 | 1.95 | 7.98 | |||
| 2017 | 1 ± 0 | 8.67 ± 0.58 | Yes, p < 0.05 | 3.32 ± 1.44 | 1–8.33 | 0.95 | 0.63 | |||
| Number of ray florets | NRF | 2015 | 14.33 ± 0.58 | 209.67 ± 43.11 | Yes, p < 0.01 | 60.01 ± 31.60 | 13–173 | 1.31 | 1.45 | 0.95 |
| 2016 | 13.33 ± 1.15 | 239 ± 14.11 | Yes, p < 0.001 | 70.71 ± 38.5 | 17.67–245.67 | 1.52 | 3.34 | |||
| 2017 | 16 ± 2.00 | 297.33 ± 25.17 | Yes, p < 0.001 | 76.51 ± 40.34 | 21–213.33 | 1.02 | 0.72 | |||
| Number of disc florets | NDF | 2015 | 118 ± 14.73 | 16.67 ± 3.06 | Yes, p < 0.01 | 131.28 ± 54.40 | 12–274.33 | 0.41 | −0.08 | 0.88 |
| 2016 | 92.33 ± 10.50 | 17.67 ± 6.03 | Yes, p < 0.001 | 121.56 ± 49.96 | 16–292 | 0.43 | 0.32 | |||
| 2017 | 226 ± 6.00 | 29 ± 6.56 | Yes, p < 0.001 | 144.53 ± 46.03 | 41.33–265.33 | 0.27 | −0.24 | |||
| Number of florets | NF | 2015 | 132.33 ± 15.28 | 226.33 ± 40.80 | n.s | 191.67 ± 55.74 | 53.00–355 | 0.46 | 0.16 | 0.85 |
| 2016 | 105.67 ± 11.50 | 256.67 ± 20.13 | Yes, p < 0.001 | 192.41 ± 57.35 | 95.67–396.33 | 0.66 | 0.39 | |||
| 2017 | 242 ± 8.00 | 326.33 ± 30.11 | Yes, p < 0.01 | 221.57 ± 42.22 | 122.67–421.5 | 0.82 | 2.23 | |||
| Ray floret length | RFL | 2015 | 26.47 ± 2.02 | 21.18 ± 1.54 | Yes, p < 0.05 | 21.86 ± 4.23 | 13.17–33.9 | −0.05 | 0.02 | 0.83 |
| 2016 | 34.77 ± 0.62 | 21.63 ± 3.55 | Yes, p < 0.05 | 23.56 ± 3.54 | 10.51–32.54 | −0.22 | 1.00 | |||
| 2017 | 32.7 ± 0.25 | 21.52 ± 0.83 | Yes, p < 0.05 | 23.15 ± 4.48 | 11.12–35.79 | 0.16 | 0.06 | |||
| Ray floret width | RFW | 2015 | 7 ± 0.58 | 5.40 ± 0.43 | Yes, p < 0.05 | 5.39 ± 0.99 | 2.9–8.61 | −0.01 | 0.76 | 0.86 |
| 2016 | 8.14 ± 1.12 | 4.75 ± 0.30 | Yes, p < 0.05 | 5.97 ± 1.32 | 3.2–15.42 | 2.81 | 16.57 | |||
| 2017 | 7.97 ± 0.30 | 4.91 ± 0.05 | Yes, p < 0.05 | 5.90 ± 1.17 | 2.36–9.06 | −0.24 | 0.25 | |||
| Ray floret length/width | RFL/W | 2015 | 3.78 ± 0.08 | 3.93 ± 0.37 | n.s | 4.12 ± 0.67 | 2.98–7.52 | 1.30 | 4.90 | 0.90 |
| 2016 | 4.32 ± 0.55 | 4.54 ± 0.50 | n.s | 4.06 ± 0.62 | 2.71–15.42 | 0.46 | −0.04 | |||
| 2017 | 4.11 ± 0.13 | 4.39 ± 0.20 | n.s | 4.04 ± 0.87 | 1.88–9.50 | 1.69 | 7.92 | |||
| Leaf length | LL | 2015 | 44.05 ± 0.58 | 60.07 ± 7.49 | Yes, p < 0.05 | 71.94 ± 15.95 | 39.95–111.21 | 0.45 | −0.15 | 0.78 |
| 2016 | 46.15 ± 4.83 | 58.04 ± 3.59 | Yes, p < 0.05 | 58.93 ± 14.13 | 22.47–110.24 | 0.75 | 1.13 | |||
| 2017 | 45.91 ± 3.13 | 51.43 ± 4.74 | n.s | 54.25 ± 15.31 | 23.82–110.59 | 1.04 | 1.80 | |||
| Leaf width | LW | 2015 | 26.11 ± 0.45 | 36.69 ± 8.43 | n.s | 40.48 ± 9.26 | 24.1–63.8 | 0.51 | −0.25 | 0.76 |
| 2016 | 30.28 ± 3.34 | 33.52 ± 3.30 | n.s | 35.18 ± 8.35 | 6.22–61.47 | 0.33 | 0.63 | |||
| 2017 | 28.48 ± 3.34 | 30.43 ± 2.64 | n.s | 31.82 ± 9.73 | 15.77–77 | 1.54 | 4.24 | |||
| Leaf length/width | LL/W | 2015 | 1.69 ± 0.05 | 1.66 ± 0.17 | n.s | 1.80 ± 0.20 | 1.32–2.23 | 0.00 | −0.31 | 0.82 |
| 2016 | 1.53 ± 0.04 | 1.74 ± 0.15 | n.s | 1.70 ± 8.35 | 1.24–3.84 | 4.09 | 32.89 | |||
| 2017 | 1.62 ± 0.14 | 1.69 ± 0.07 | n.s | 1.73 ± 0.20 | 1.31–2.37 | 0.33 | −0.01 | |||
| Traits | QTL | Year | Linkage Group | Marker Interval | QTL Position (cM) | Max LOD Value | Contributions (%) |
|---|---|---|---|---|---|---|---|
| Inflorescence diameter | qID5-2015 | 2015 | 5 | 127-c75747-221 | 13.968 | 3.69 | 14.7 |
| qID6-2015 | 2015 | 6 | 96-c73599-184 | 37.884 | 3.87 | 15.3 | |
| qID8-2016 | 2016 | 8 | 260-CEM15-429 | 35.602 | 4.41 | 12.4 | |
| qID3-2017 | 2017 | 3 | 226-c73810-207; 43-c67517-249 | 48.031 | 3.81 | 9.3 | |
| qID3-mean-1 | Mean | 3 | 98-c73873-174 | 37.579 | 4.51 | 10.5 | |
qID3-mean-2 | Mean | 3 | 226-c73810-234; 226-c73810-207; 43-c67517-249; 52-c70441-250; 205-c64830-259 | 49.403 | 5.45 | 12.6 | |
| qID8-mean | Mean | 8 | 260-CEM15-429; 36-c66938-195 | 36.602 | 4.48 | 9.1 | |
| Central disc flower diameter | qCDFD4-2015 | 2015 | 4 | 242-c77190-290 | 75.158 | 3.6 | 14.2 |
| qCDFD3-2016 | 2016 | 3 | 196-c55093-348 | 95.174 | 4.28 | 11.9 | |
| Number of whorls of ray florets | qNWRF6-2017 | 2017 | 6 | 203-c64705-232 | 70.453 | 3.71 | 9.1 |
| qNWRF3-mean | Mean | 3 | 196-c55093-348; 27-c65910-144 | 99.468 | 4.04 | 9.5 | |
| Number of ray florets | qNRF3-mean | Mean | 3 | 196-c55093-348; 27-c65910-144 | 67.734 | 3.19 | 6.8 |
| Number of disc florets | qNDF4-2015 | 2015 | 4 | 242-c77190-290 | 76.158 | 4.92 | 18.9 |
| qNDF6-2015 | 2015 | 6 | 238-c76774-229 | 30.209 | 3.69 | 14.5 | |
| qNDF3-2016 | 2016 | 3 | 196-c55093-348; 27-c65910-144 | 102.478 | 4.45 | 12.4 | |
| Number of florets | qNF4-2015 | 2015 | 4 | 242-c77190-290 | 76.158 | 3.79 | 14.9 |
| qNF7-2016 | 2016 | 7 | 94-c73532-334 | 36.357 | 4.03 | 11.4 | |
| qNF8-2017-1 | 2017 | 8 | 257-CEM12-285 | 0 | 3.99 | 9.8 | |
| qNF8-2017-2 | 2017 | 8 | 34-c66815-149 | 50.854 | 3.63 | 8.9 | |
| Ray floret length | qRFL3-2016 | 2016 | 3 | 115-c75008-259 | 41.748 | 3.74 | 10.7 |
| qRFL8-mean | Mean | 8 | 260-CEM15-429 | 36.602 | 3.97 | 9.3 | |
| qRFL3-mean-1 | Mean | 3 | 98-c73873-174; 64-c71464-208 | 37.579 | 4.29 | 10 | |
| qRFL3-mean-2 | Mean | 3 | 226-c73810-234; 226-c73810-207; 43-c67517-249; 52-c70441-250; 205-c64830-259 | 47.975 48.031 | 5.14 5.14 | 11.9 11.9 | |
| Ray floret width | qRFW5-2015 | 2015 | 5 | 127-c75747-221 | 16.261 | 3.69 | 14.9 |
| qRFW3-2017 | 2017 | 3 | 226-c73810-203 | 106.019 | 3.98 | 9.7 | |
| qRFW8-2017 | 2017 | 8 | 24-c65571-167; 85-c73005 | 29.128 | 4.07 | 9.9 | |
| qRFW7-mean | Mean | 7 | 28-c66048-253 | 50.477 | 3.67 | 8.6 | |
| Ray floret length/width | qRFL/W2-2016 | 2016 | 2 | 124-c75428-144 | 117.158 | 4.4 | 12.4 |
| qRFL/W3-2017-1 | 2017 | 3 | 64-c71464-216 | 83.312 | 3.77 | 9.2 | |
| qRFL/W3-2017-2 | 2017 | 3 | 196-c55093-338 | 102.478 | 3.91 | 9.6 | |
| qRFL/W7-2017 | 2017 | 7 | 254-CEM9-208 | 47.708 | 3.82 | 9.4 | |
| qRFL/W3-mean | Mean | 3 | 64-c71464-216 | 83.312 | 3.65 | 8.6 | |
| Leaf length | qLL5-2015 | 2015 | 5 | 63-c71333-217; 121-c75378-264 | 20.383 | 3.68 | 14.8 |
| qLL8-2017 | 2017 | 8 | 165-c53556-249; 141-c76325-245 | 73.038 | 4.65 | 11.3 | |
| Leaf width | qLW8-2017 | 2017 | 8 | 165-c53556-249; 141-c76325-245 | 73.038 | 5 | 12.1 |
| Leaf length/width | qLL/W4-2016 | 2016 | 4 | 209-c66049-243 | 97.241 | 5.67 | 15.7 |
| Marker Dosage | Hypothesis I | Hypothesis II |
|---|---|---|
| Autohexaploid (Hexasomic) | Allohexaploid (Disomic) | |
| Simplex × nulliplex | 1:1 | 1:1 |
| Duplex × nulliplex | 4:1 | 3:1 |
| 1:0 | ||
| Triplex × nulliplex | 19:1 | 7:1 |
| 1:0 | ||
| Simplex × simplex | 3:1 | 3:1 |
| Duplex × simplex | 9:1 | 7:1 |
| Triplex × simplex | 39:1 | 15:1 |
| Duplex × duplex | 24:1 | 15:1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, M.; Gao, Y.; Wu, Z.; Zhang, Q. Linkage Map Development by EST-SSR Markers and QTL Analysis for Inflorescence and Leaf Traits in Chrysanthemum (Chrysanthemum morifolium Ramat.). Plants 2020, 9, 1342. https://doi.org/10.3390/plants9101342
Fan M, Gao Y, Wu Z, Zhang Q. Linkage Map Development by EST-SSR Markers and QTL Analysis for Inflorescence and Leaf Traits in Chrysanthemum (Chrysanthemum morifolium Ramat.). Plants. 2020; 9(10):1342. https://doi.org/10.3390/plants9101342
Chicago/Turabian StyleFan, Min, Yike Gao, Zhiping Wu, and Qixiang Zhang. 2020. "Linkage Map Development by EST-SSR Markers and QTL Analysis for Inflorescence and Leaf Traits in Chrysanthemum (Chrysanthemum morifolium Ramat.)" Plants 9, no. 10: 1342. https://doi.org/10.3390/plants9101342
APA StyleFan, M., Gao, Y., Wu, Z., & Zhang, Q. (2020). Linkage Map Development by EST-SSR Markers and QTL Analysis for Inflorescence and Leaf Traits in Chrysanthemum (Chrysanthemum morifolium Ramat.). Plants, 9(10), 1342. https://doi.org/10.3390/plants9101342




