Peptide Extracts from Seven Medicinal Plants Discovered to Inhibit Oomycete Phytophthora infestans, a Causative Agent of Potato Late Blight Disease
Abstract
1. Introduction
2. Results
2.1. Screening of Plant Peptide Extracts to Suppress Phytophthora infestans In Vitro
2.2. The Antifungal Activity of Plant Peptide Extracts toward P. infestans by Inoculation of Potato Tuber Discs
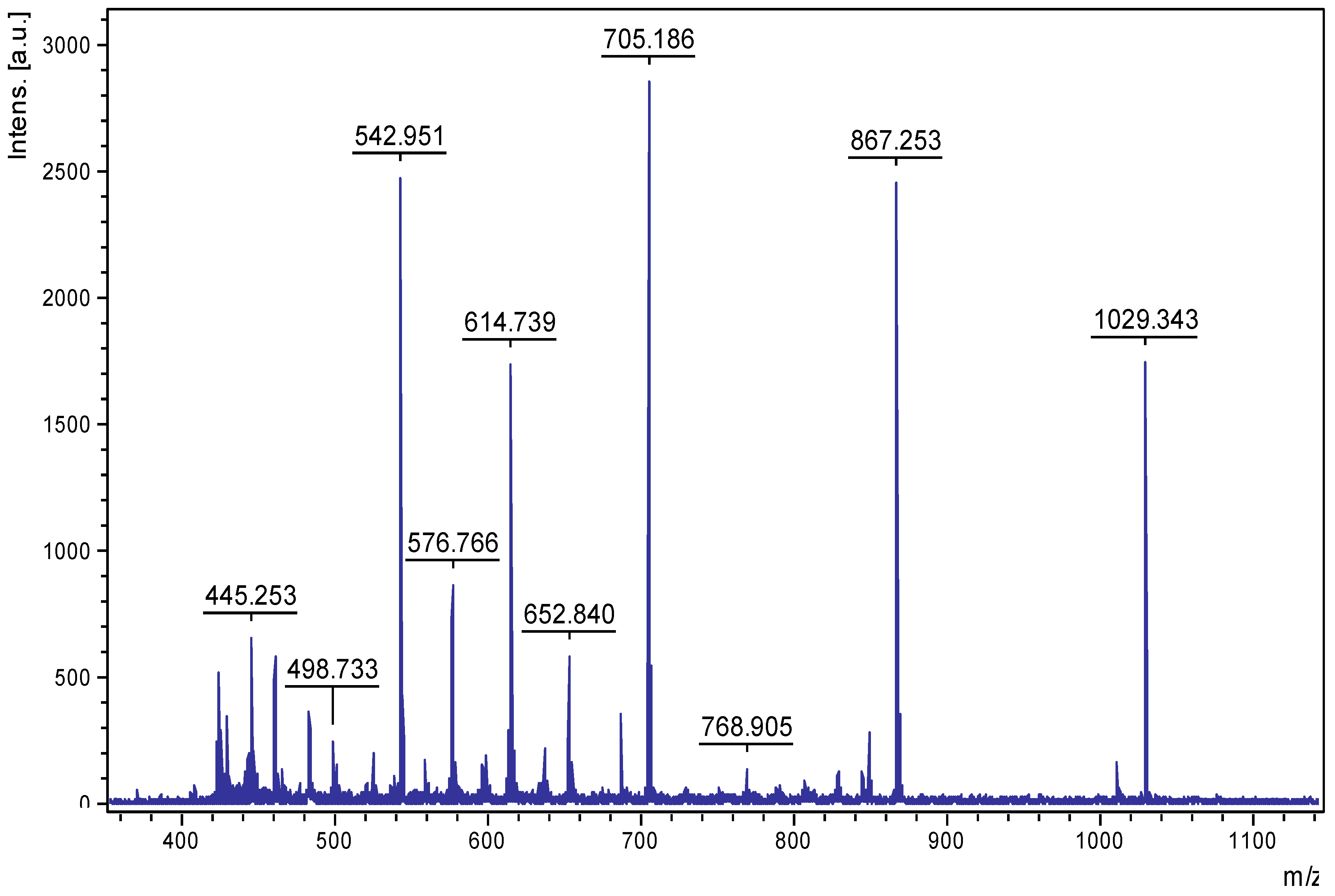
2.3. Initial Structural Characterization of PE-Eqi
2.4. HPLC Analysis of PE-Eqi and Structure Determination of Individual Compounds
3. Discussion
4. Materials and Methods
4.1. Medicinal Plants
4.2. Oomycete Origin and Cultivation Conditions
4.3. Peptide Extracts
4.4. Amino Acid Analysis
4.5. MALDI TOF Mass Spectrometry
4.6. Ion-Exchange HPLC
4.7. Reversed-Phase HPLC
4.8. Edman Sequencing
4.9. Antifungal Assays In Vitro
4.10. Antifungal Assay by Inoculation of Potato Tuber Discs
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MALDI-TOF MS | matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| Asn | asparagine |
| Asp | aspartate |
| Gln | glutamine |
| Glu | glutamate |
| HPLC | high performance liquid chromatography |
| IC50 | inhibition concentration caused half-effect |
| ICmin | inhibition concentration caused sub-effect (15%); |
| NCBI | National Center of Biotechnology Information, USA |
| BLASTP | basic local alignment search tool protein; |
| MQ | deionized water |
| EDTA | ethylenediaminetetraacetic acid |
| PBS | phosphate-buffered saline |
| NaCl | sodium chloride |
| UV | ultraviolet |
| RP-HPLC | reversed-phase high performance liquid chromatography; |
| MeCN | acetonitrile |
| PTH | phenylthiohydantoin |
References
- Gisi, U.A.L. Advances in Downy Mildew Research; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002. [Google Scholar]
- Leesutthiphonchai, W.; Vu, A.L.; Ah-Fong, A.M.V.; Judelson, H.S. How Does Phytophthora infestans Evade Control Efforts? Modern Insight into the Late Blight Disease. Phytopathology 2018, 108, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Jiao, F. Effectors of Phytophthora pathogens are powerful weapons for manipulating host immunity. Planta 2019, 250, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Stephan, D.; Eckhard, K. Screening of plant extracts, rnicro-organisms and commercial preparations for biocontrol of Phytophthora infestans on detached potato leaves. IOBC WPRS Bull. 2006, 25, 391–394. [Google Scholar]
- Fravel, D.R.; Connick, W.J.; Lewis, J.A. Formulation of Microorganisms to Control Plant Diseases. In Formulation of Microbial Biopesticides; Springer Nature: Dordrecht, The Netherlands, 1998; pp. 187–202. [Google Scholar]
- Stephan, D.; Schmitt, A.; Martins Carvalho, S.; Seddon, B.; Koch, E. Evaluation of biocontrol preparations and plant extracts for the control of Phytophthora infestans on potato leaves. Eur. J. Plant Pathol. 2005, 112, 235–246. [Google Scholar] [CrossRef]
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef]
- Fry, W.E. Phytophthora infestans: New Tools (and Old Ones) Lead to New Understanding and Precision Management. Annu. Rev. Phytopathol. 2016, 54, 529–547. [Google Scholar] [CrossRef]
- Quimby, P.C.; King, L.R.; Grey, W.E. Biological control as a means of enhancing the sustainability of crop/land management systems. Agric. Ecosyst. Environ. 2002, 88, 147–152. [Google Scholar] [CrossRef]
- Andreu, A.B.; Guevara, M.G.; Wolski, E.A.; Daleo, G.R.; Caldiz, D.O. Enhancement of natural disease resistance in potatoes by chemicals. Pest Manag. Sci. 2006, 62, 162–170. [Google Scholar] [CrossRef]
- Chen, F.; Zhou, Q.; Qin, C.; Li, Y.; Zhan, J. Low evolutionary risk of iprovalicarb resistance in Phytophthora infestans. Pestic. Biochem. Physiol. 2018, 152, 76–83. [Google Scholar] [CrossRef]
- Jindal, K.K.; Singh, H.; Meeta, M. Biological control of Phytophthora infestans on potato. Indian J. Plant Pathol. 1988, 6, 59–62. [Google Scholar]
- Yan, Z.; Reddy, M.S.; Ryu, C.M.; McInroy, J.A.; Wilson, M.; Kloepper, J.W. Induced systemic protection against tomato late blight elicited by plant growth-promoting rhizobacteria. Phytopathology 2002, 92, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Lamsal, K.; Kim, S.W.; Kim, Y.S.; Lee, Y.S. Biocontrol of late blight and plant growth promotion in tomato using rhizobacterial isolates. J. Microbiol. Biotechnol. 2013, 23, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Tomar, S.; Singh, B.P.; Lal, M.; Khan, M.A.; Hussain, T.; Sharma, S.; Kaushik, S.K.; Kumar, S. Screening of novel microorganisms for biosurfactant and biocontrol activity against Phytophthora infestans. J. Environ. Biol. 2014, 35, 893–899. [Google Scholar] [PubMed]
- Cray, J.A.; Connor, M.C.; Stevenson, A.; Houghton, J.D.R.; Rangel, D.E.N.; Cooke, L.R.; Hallsworth, J.E. Biocontrol agents promote growth of potato pathogens, depending on environmental conditions. Microb. Biotechnol. 2016, 9, 330–354. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.; von Dahlen, J.K.; Schnake, A.; Ginschel, S.; Schulz, B.; Rose, L.E. Broad-spectrum inhibition of Phytophthora infestans by fungal endophytes. FEMS Microbiol. Ecol. 2018, 94, 1–15. [Google Scholar] [CrossRef]
- Ali, M.; Kim, B.; Belfield, K.D.; Norman, D.; Brennan, M.; Ali, G.S. Inhibition of Phytophthora parasitica and P. capsici by silver nanoparticles synthesized using aqueous extract of Artemisia absinthium. Phytopathology 2015, 105, 1183–1190. [Google Scholar] [CrossRef]
- Divya, K.; Smitha, V.; Jisha, M.S. Antifungal, antioxidant and cytotoxic activities of chitosan nanoparticles and its use as an edible coating on vegetables. Int. J. Biol. Macromol. 2018, 114, 572–577. [Google Scholar] [CrossRef]
- Ghorbani, R.; Wilcockson, S.; Leifert, C. Alternative treatments for late blight control in organic potato: Antagonistic micro-organisms and compost extracts for activity against Phytophthora infestans. Potato Res. 2005, 48, 181–189. [Google Scholar] [CrossRef]
- Wianowska, D.; Garbaczewska, S.; Cieniecka-Roslonkiewicz, A.; Dawidowicz, A.L.; Jankowska, A. Comparison of antifungal activity of extracts from different Juglans regia cultivars and juglone. Microb. Pathog. 2016, 100, 263–267. [Google Scholar] [CrossRef]
- Cayuela, M.L.; Millner, P.D.; Meyer, S.L.F.; Roig, A. Potential of olive mill waste and compost as biobased pesticides against weeds, fungi, and nematodes. Sci. Total Environ. 2008, 399, 11–18. [Google Scholar] [CrossRef]
- Kang, S.W.; Kim, S.W. New antifungal activity of penicillic acid against Phytophthora species. Biotechnol. Lett. 2004, 26, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Son, S.W.; Kim, H.Y.; Choi, G.J.; Lim, H.K.; Jang, K.S.; Lee, S.O.; Lee, S.; Sung, N.D.; Kim, J.C. Bikaverin and fusaric acid from Fusarium oxysporum show antioomycete activity against Phytophthora infestans. J. Appl. Microbiol. 2008, 104, 692–698. [Google Scholar] [CrossRef]
- Wang, G.; Liu, Z.; Lin, R.; Li, E.; Mao, Z.; Ling, J.; Yang, Y.; Yin, W.B.; Xie, B. Biosynthesis of Antibiotic Leucinostatins in Bio-control Fungus Purpureocillium lilacinum and Their Inhibition on Phytophthora Revealed by Genome Mining. PLoS Pathog. 2016, 12, e1005685. [Google Scholar] [CrossRef]
- Dahlin, P.; Müller, M.C.; Ekengren, S.; McKee, L.S.; Bulone, V. The impact of steroidal glycoalkaloids on the physiology of Phytophthora infestans, the causative agent of potato late blight. Mol. Plant Microbe Interact. 2017, 30, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Portz, D.; Koch, E.; Slusarenko, A.J. Effects of garlic (Allium sativum) juice containing allicin on Phytophthora infestans and downy mildew of cucumber caused by Pseudoperonospora cubensis. Eur. J. Plant Pathol. 2008, 122, 197–206. [Google Scholar] [CrossRef]
- Kim, J.C.; Choi, G.J.; Lee, S.W.; Kim, J.S.; Chung, K.Y.; Cho, K.Y. Screening extracts of Achyranthes japonica and Rumex crispus for activity against various plant pathogenic fungi and control of powdery mildew. Pest Manag. Sci. 2004, 60, 803–808. [Google Scholar] [CrossRef]
- Manter, D.K.; Kelsey, R.G.; Karchesy, J.J. Antimicrobial activity of extractable conifer heartwood compounds toward Phytophthora ramorum. J. Chem. Ecol. 2007, 33, 2133–2147. [Google Scholar] [CrossRef]
- Halama, P.; Van Haluwin, C. Antifungal activity of lichen extracts and lichenic acids. BioControl 2004, 49, 95–107. [Google Scholar] [CrossRef]
- Blaeser, P.; Steiner, U.; Lyr, H.; Russel, P.E.; Dehne, H.W.; Sisler, H.D. Antifungal activity of plant extracts against potato late blight (Phytophthora infestans). In Modern fungicides and antifungal compounds II, Proceedings of the 12th International Reinhardsbrunn Symposium, Friedrichroda, Thuringia, Germany, 24–29 May 1998; Intercept: New York, NY, USA, 1999; pp. 491–499. [Google Scholar]
- Pretorius, J.C.; Zietsman, P.C.; Eksteen, D. Fungitoxic properties of selected South African plant species against plant pathogens of economic importance in agriculture. Ann. Appl. Biol. 2002, 141, 117–124. [Google Scholar] [CrossRef]
- Okemo, P.O.; Bais, H.P.; Vivanco, J.M. In vitro activities of Maesa lanceolata extracts against fungal plant pathogens. Fitoterapia 2003, 74, 312–316. [Google Scholar] [CrossRef]
- Dales, M.J. A review of plant material used for controlling insect pests of stored products. Nat. Resour. Inst. Bull. 1996, 65, 1–91. [Google Scholar]
- Chaadaeva, A.V.; Tepkeeva, I.I.; Moiseeva, E.V.; Svirshchevskaya, E.V.; Demshkin, V.P. Antitumor activity of the plant remedy peptide extract pe-pm in a new mouse t-lymphoma/leukemia model. Biomeditsinskaya Khimiya 2009, 55, 81–88. [Google Scholar]
- Tepkeeva, I.I.; Moiseeva, E.V.; Chaadaeva, A.V.; Zhavoronkova, E.V.; Kessler, Y.V.; Semushina, S.G.; Demushkin, V.P. Evaluation of antitumor activity of peptide extracts from medicinal plants on the model of transplanted breast cancer in CBRB-Rb(8.17)1Iem mice. Bull. Exp. Biol. Med. 2008, 145, 464–466. [Google Scholar] [CrossRef] [PubMed]
- Rakotonindraina, T.; Chauvin, J.É.; Pellé, R.; Faivre, R.; Chatot, C.; Savary, S.; Aubertot, J.N. Modeling of yield losses caused by potato late blight on eight cultivars with different levels of resistance to phytophthora infestans. Plant Dis. 2012, 96, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.H.; Jang, J.Y.; Choi, G.J.; Choi, Y.H.; Jang, K.S.; Nguyen, V.T.; Min, B.S.; Le Dang, Q.; Kim, J.C. Antifungal activity of sterols and dipsacus saponins isolated from Dipsacus asper roots against phytopathogenic fungi. Pestic. Biochem. Physiol. 2017, 141, 103–108. [Google Scholar] [CrossRef]
- Yoon, M.Y.; Choi, G.J.; Choi, Y.H.; Jang, K.S.; Park, M.S.; Cha, B.; Kim, J.C. Effect of polyacetylenic acids from Prunella vulgaris on various plant pathogens. Lett. Appl. Microbiol. 2010, 51, 511–517. [Google Scholar] [CrossRef]
- Schuster, C.; Konstantinidou-Doltsinis, S.; Schmitt, A. Glycyrrhiza glabra extract protects plants against important phytopathogenic fungi. Commun. Agric. Appl. Biol. Sci. 2010, 75, 531–540. [Google Scholar]
- Turóczi, B.; Bakonyi, J.; Szabó, K.A.; Bálint, J.; Máthé, I.; Lányi, S.; Balog, A. In vitro and in vivo effect of poplar bud extracts on Phytophthora infestans: A new effective biological method in potato late blight control. Plants 2020, 9, 217. [Google Scholar] [CrossRef]
- Gao, L.; Bradeen, J.M. Contrasting Potato Foliage and Tuber Defense Mechanisms against the Late Blight Pathogen Phytophthora infestans. PLoS ONE 2016, 11, e0159969. [Google Scholar] [CrossRef]
- Arafa, R.A.; Rakha, M.T.; Soliman, N.E.K.; Moussa, O.M.; Kamel, S.M.; Shirasawa, K. Rapid identification of candidate genes for resistance to tomato late blight disease using next-generation sequencing technologies. PLoS ONE 2017, 12, e0189951. [Google Scholar] [CrossRef]
- Ha, J.H.; Jang, H.A.; Moon, K.B.; Baek, K.H.; Choi, G.J.; Choi, D.; Cho, H.S.; Kwon, S.Y.; Jeon, J.H.; Oh, S.K.; et al. Nicotiana benthamiana Matrix Metalloprotease 1 (NMMP1) gene confers disease resistance to Phytophthora infestans in tobacco and potato plants. J. Plant Physiol. 2017, 218, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, B.; Kalsi, H.S.; Godbole, P.; Malankar, N.; Thiagarayaselvam, A.; Siddappa, S.; Thulasiram, H.V.; Chakrabarti, S.K.; Banerjee, A.K. MiRNA160 is associated with local defense and systemic acquired resistance against Phytophthora infestans infection in potato. J. Exp. Bot. 2018, 69, 2023–2036. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Farooqi, M.S.; Mishra, D.C.; Kumar, S.; Rai, A.; Chaturvedi, K.K.; Lal, S.B.; Sharma, A. Prediction of miRNA and Identification of their Relationship Network Related to Late Blight Disease of Potato. MicroRNA 2017, 7, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Niderman, T.; Genetet, I.; Bruyere, T.; Gees, R.; Stintzi, A.; Legrand, M.; Fritig, B.; Mosinger, E. Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995, 108, 17–27. [Google Scholar] [CrossRef]
- Hoegen, E.; Strömberg, A.; Pihlgren, U.; Kombrink, E. Primary structure and tissue-specific expression of the pathogenesis-related protein PR-1b in potato. Mol. Plant Pathol. 2002, 3, 329–345. [Google Scholar] [CrossRef]
- Thomas, C.; Mabon, R.; Andrivon, D.; Val, F. The effectiveness of induced defense responses in a susceptible potato genotype depends on the growth rate of Phytophthora infestans. Mol. Plant Microbe Interact. 2019, 32, 76–85. [Google Scholar] [CrossRef]
- Belbahri, L.; Calmin, G.; Mauch, F.; Andersson, J.O. Evolution of the cutinase gene family: Evidence for lateral gene transfer of a candidate Phytophthora virulence factor. Gene 2008, 408, 1–8. [Google Scholar] [CrossRef]
- Zhang, Y.; Fletcher, K.; Han, R.; Michelmore, R.; Yang, R. Genome-wide analysis of cyclophilin proteins in 21 oomycetes. Pathogens 2020, 9, 24. [Google Scholar] [CrossRef]
- Meijer, H.J.G.; Schoina, C.; Wang, S.; Bouwmeester, K.; Hua, C.; Govers, F. Phytophthora infestans small phospholipase D-like proteins elicit plant cell death and promote virulence. Mol. Plant Pathol. 2019, 20, 180–193. [Google Scholar] [CrossRef]
- Gvozdeva, E.L.; Ievleva, E.V.; Gerasimova, N.G.; Ozeretskovskaya, O.L.; Valueva, T.A. Exoproteinases of the oomycete Phytophthora infestans. Appl. Biochem. Microbiol. 2004, 40, 165–169. [Google Scholar] [CrossRef]
- Boutemy, L.S.; King, S.R.F.; Win, J.; Hughes, R.K.; Clarke, T.A.; Blumenschein, T.M.A.; Kamoun, S.; Banfield, M.J. Structures of Phytophthora RXLR effector proteins: A conserved but adaptable fold underpins functional diversity. J. Biol. Chem. 2011, 286, 35834–35842. [Google Scholar] [CrossRef] [PubMed]
- Wawra, S.; Trusch, F.; Matena, A.; Apostolakis, K.; Linne, U.; Zhukov, I.; Stanek, J.; Koźmiński, W.; Davidson, I.; Secombes, C.J.; et al. The RxLR motif of the host targeting effector AVR3a of Phytophthora infestans is cleaved before secretion. Plant Cell 2017, 29, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Ŝtajner, D.; Popović, B.M.; Ĉanadanović-Brunet, J.; Anaĉkov, G. Exploring Equisetum arvense L., Equisetum ramosissimum L. and Equisetum telmateia L. as sources of natural antioxidants. Phyther. Res. 2009, 23, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Francescato, L.N.; Debenedetti, S.L.; Schwanz, T.G.; Bassani, V.L.; Henriques, A.T. Identification of phenolic compounds in Equisetum giganteum by LC-ESI-MS/MS and a new approach to total flavonoid quantification. Talanta 2013, 105, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Patova, O.A.; Smirnov, V.V.; Golovchenko, V.V.; Vityazev, F.V.; Shashkov, A.S.; Popov, S.V. Structural, rheological and antioxidant properties of pectins from Equisetum arvense L. and Equisetum sylvaticum L. Carbohydr. Polym. 2019, 209, 239–249. [Google Scholar] [CrossRef]
- Hase, T.; Wada, K.; Matsubara, H. Horsetail (Equisetum arvense) ferredoxins I and II: Amino acid sequences and gene duplication. J. Biochem. 1977, 82, 277–286. [Google Scholar] [CrossRef]
- Brumme, S.; Kruft, V.; Schmitz, U.K.; Braun, H.P. New insights into the co-evolution of cytochrome c reductase and the mitochondrial processing peptidase. J. Biol. Chem. 1998, 273, 13143–13149. [Google Scholar] [CrossRef]
- Al Mohammed, H.I.; Paray, B.A.; Rather, I.A. Anticancer activity of EA1 extracted from Equisetum arvense. Pak. J. Pharm. Sci. 2017, 30, 1947–1950. [Google Scholar]
- Do Monte, F.H.M.; Dos Santos, J.G.; Russi, M.; Bispo Lanziotti, V.M.N.; Moreira Leal, L.K.A.; De Andrade Cunha, G.M. Antinociceptive and anti-inflammatory properties of the hydroalcoholic extract of stems from Equisetum arvense L. in mice. Pharmacol. Res. 2004, 49, 239–243. [Google Scholar] [CrossRef]
- Garcia, D.; Ramos, A.J.; Sanchis, V.; Marín, S. Equisetum arvense hydro-alcoholic extract: Phenolic composition and antifungal and antimycotoxigenic effect against Aspergillus flavus and Fusarium verticillioides in stored maize. J. Sci. Food Agric. 2013, 93, 2248–2253. [Google Scholar] [CrossRef]
- Wojnicz, D.; Kucharska, A.Z.; Sokół-Łętowska, A.; Kicia, M.; Tichaczek-Goska, D. Medicinal plants extracts affect virulence factors expression and biofilm formation by the uropathogenic Escherichia coli. Urol. Res. 2012, 40, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, C.; Rémus-Borel, W.; Vivancos, J.; Labbé, C.; Belzile, F.; Bélanger, R.R. Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J. 2012, 72, 320–330. [Google Scholar] [CrossRef]
- Inamine, S.; Onaga, S.; Ohnuma, T.; Fukamizo, T.; Taira, T. Purification, cDNA cloning, and characterization of LysM-containing plant chitinase from horsetail (Equisetum arvense). Biosci. Biotechnol. Biochem. 2015, 79, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Karol, K.G.; Arumuganathan, K.; Boore, J.L.; Duffy, A.M.; De Everett, K.; Hall, J.D.; Hansen, S.K.; Kuehl, J.V.; Mandoli, D.F.; Mishler, B.D.; et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: Implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol. Biol. 2010, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Rogozhin, E.A.; Tepkeeva, I.I.; Zaitsev, D.V.; Demushkin, V.P.; Smirnov, A.N. Biological activity of peptide extracts of medicinal plants against phytopathogenic fungi and oomycetes. Russ. Agric. Sci. 2011, 37, 314–317. [Google Scholar] [CrossRef]
- Odintsova, T.I.; Vassilevski, A.A.; Slavokhotova, A.A.; Musolyamov, A.K.; Finkina, E.I.; Khadeeva, N.V.; Rogozhin, E.A.; Korostyleva, T.V.; Pukhalsky, V.A.; Grishin, E.V.; et al. A novel antifungal hevein-type peptide from Triticum kiharae seeds with a unique 10-cysteine motif. FEBS J. 2009, 276, 4266–4275. [Google Scholar] [CrossRef]
- Odintsova, T.; Rogozhin, E.; Sklyar, I.; Musolyamov, A.; Kudryavtsev, A.; Pukhalsky, V.; Smirnov, A.; Grishin, E.; Egorov, T. Antifungal Activity of Storage 2S Albumins from Seeds of the Invasive Weed Dandelion Taraxacum officinale Wigg. Protein Pept. Lett. 2010, 17, 522–529. [Google Scholar] [CrossRef]
| Index/Variant | Extract | |||||||
|---|---|---|---|---|---|---|---|---|
| PE-PM | PE-Eqi | PE-Cm | PE-In | PE-Hp | PE-Ln | PE-Cs | ||
| Indirect germination | IC50, mg/mL | 0.25 | 1.0 | - | - | 2.0 | - | 2.0 |
| ICmin, mg/mL | 0.125 | 0.5 | - | - | 1.0 | - | 1.0 | |
| Direct germination | IC50, mg/mL | 0.5 | 2.0 | - | 1.0 | 0.5 | - | 0.5 |
| ICmin, mg/mL | 0.125 | 1.0 | 2.0 | 0.5 | 0.25 | 2.0 | 0.25 | |
| Morphological changes | + | - | - | - | - | - | - | |
| No Peak | Average Molecular Mass Measured, Da | N-terminal Amino Acid Sequence | Annotation |
|---|---|---|---|
| 1 | 1048.6 | 1PAVTLAFATTG11 * | Aquaporin product |
| 2 | 905.2 | 1PSGGALNY8 | Unidentified protein product |
| 3 | 1160.4 | 1PVAGEDNQFLA11 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit fragment |
| 4 | 615.2 | No sequence determined | - |
| 5 | 943.5 | 1DFYRDND7 | Possible hypothetical chloroplast RF21 protein fragment |
| 6 | 1378.2 | 1DLIRDDFIEKD11 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit fragment |
| 7 | 1471.8 | 1QYQLDGIDLDYE12 | Chitinase A fragment |
| 8 | 1010.1 | 1DFTRDDEN8 | Possible ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit fragment |
| 9 | 722.8 | No sequence determined | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rogozhin, E.A.; Vasilchenko, A.S.; Barashkova, A.S.; Smirnov, A.N.; Zavriev, S.K.; Demushkin, V.P. Peptide Extracts from Seven Medicinal Plants Discovered to Inhibit Oomycete Phytophthora infestans, a Causative Agent of Potato Late Blight Disease. Plants 2020, 9, 1294. https://doi.org/10.3390/plants9101294
Rogozhin EA, Vasilchenko AS, Barashkova AS, Smirnov AN, Zavriev SK, Demushkin VP. Peptide Extracts from Seven Medicinal Plants Discovered to Inhibit Oomycete Phytophthora infestans, a Causative Agent of Potato Late Blight Disease. Plants. 2020; 9(10):1294. https://doi.org/10.3390/plants9101294
Chicago/Turabian StyleRogozhin, Eugene A., Alexey S. Vasilchenko, Anna S. Barashkova, Alexey N. Smirnov, Sergey K. Zavriev, and Vladimir P. Demushkin. 2020. "Peptide Extracts from Seven Medicinal Plants Discovered to Inhibit Oomycete Phytophthora infestans, a Causative Agent of Potato Late Blight Disease" Plants 9, no. 10: 1294. https://doi.org/10.3390/plants9101294
APA StyleRogozhin, E. A., Vasilchenko, A. S., Barashkova, A. S., Smirnov, A. N., Zavriev, S. K., & Demushkin, V. P. (2020). Peptide Extracts from Seven Medicinal Plants Discovered to Inhibit Oomycete Phytophthora infestans, a Causative Agent of Potato Late Blight Disease. Plants, 9(10), 1294. https://doi.org/10.3390/plants9101294






