Molecular Characterization and Expression Profile of PaCOL1, a CONSTANS-like Gene in Phalaenopsis Orchid
Abstract
1. Introduction
2. Results
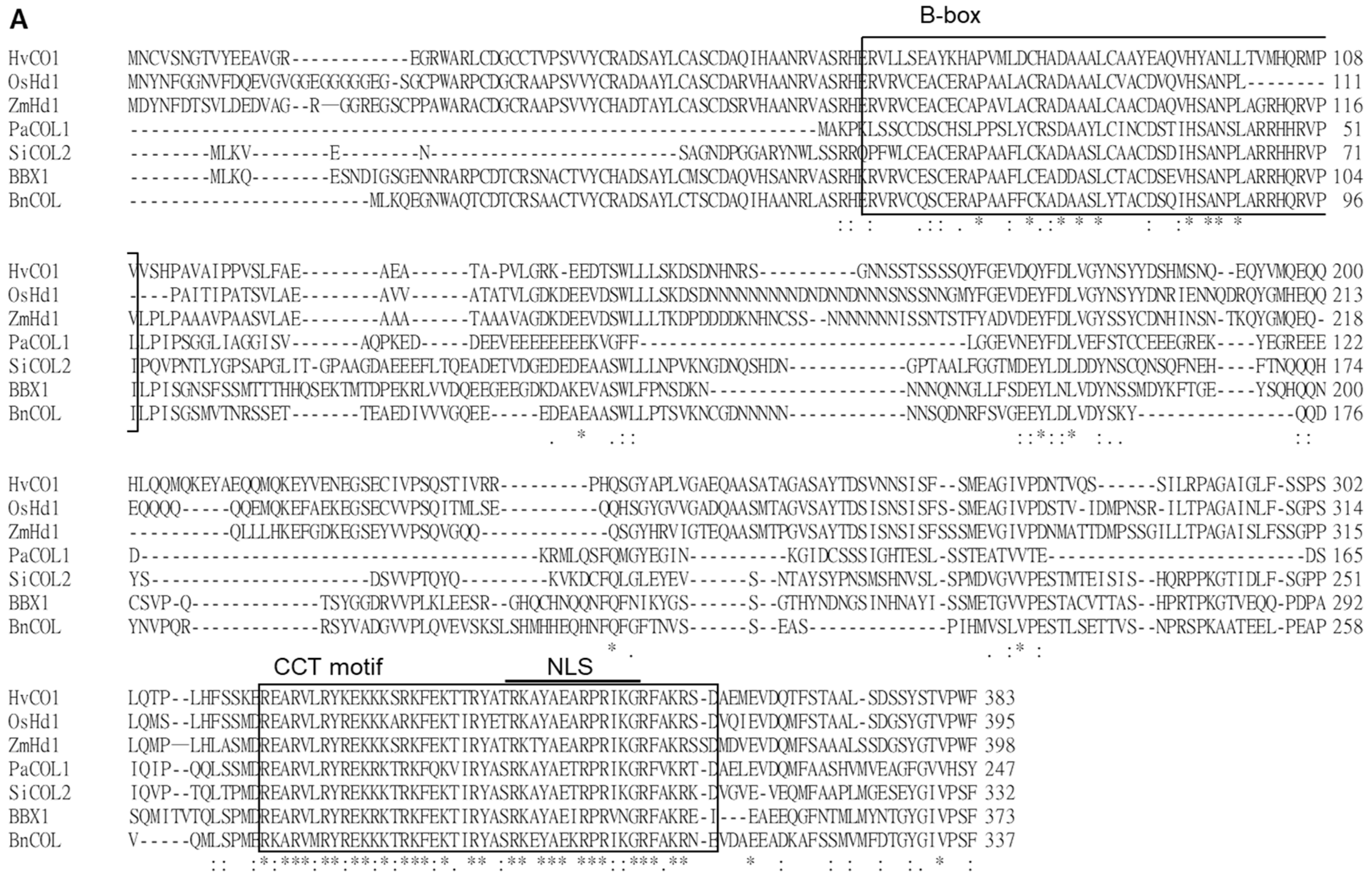
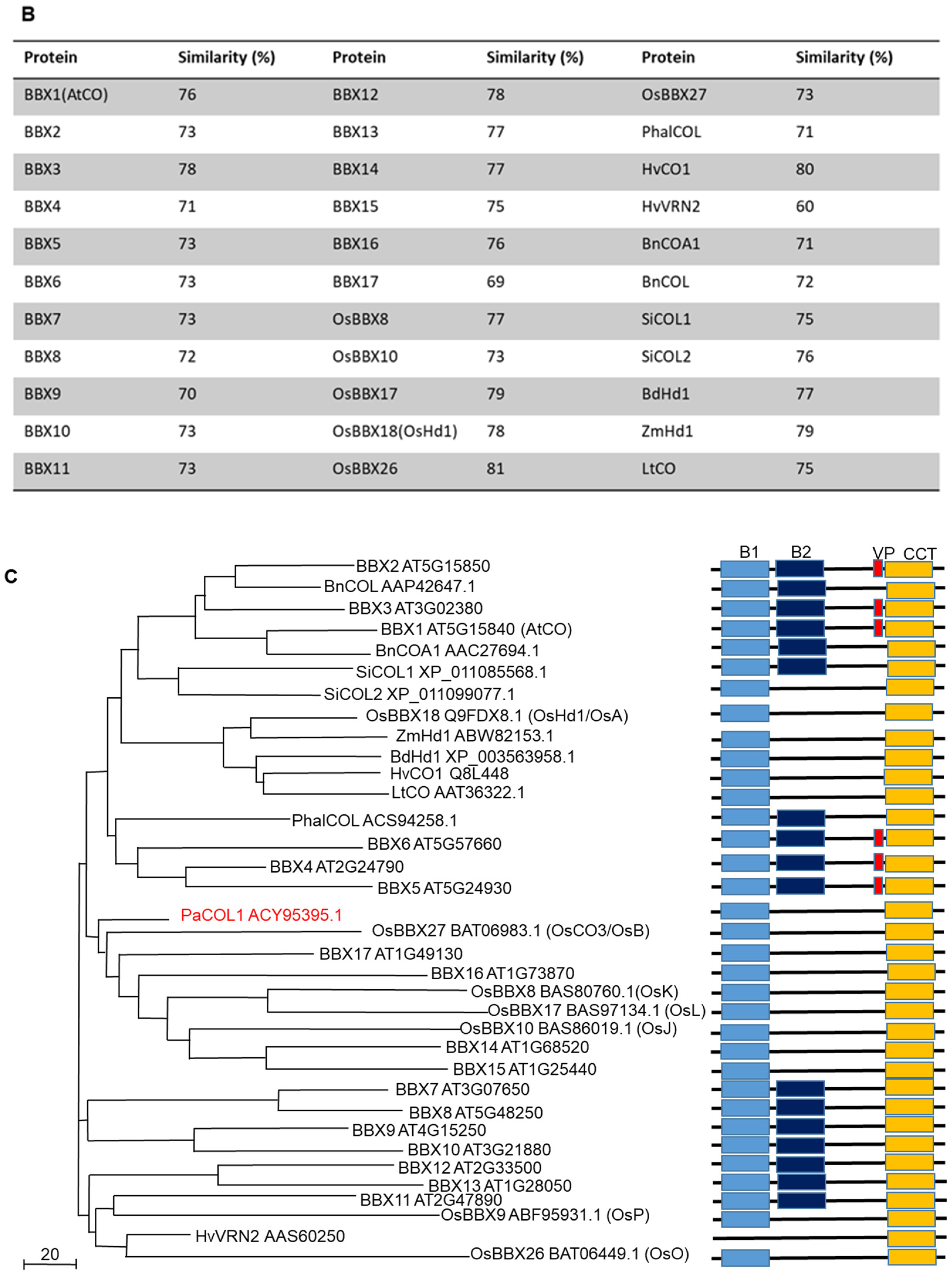
2.1. PaCOL1 Encodes a COL Protein
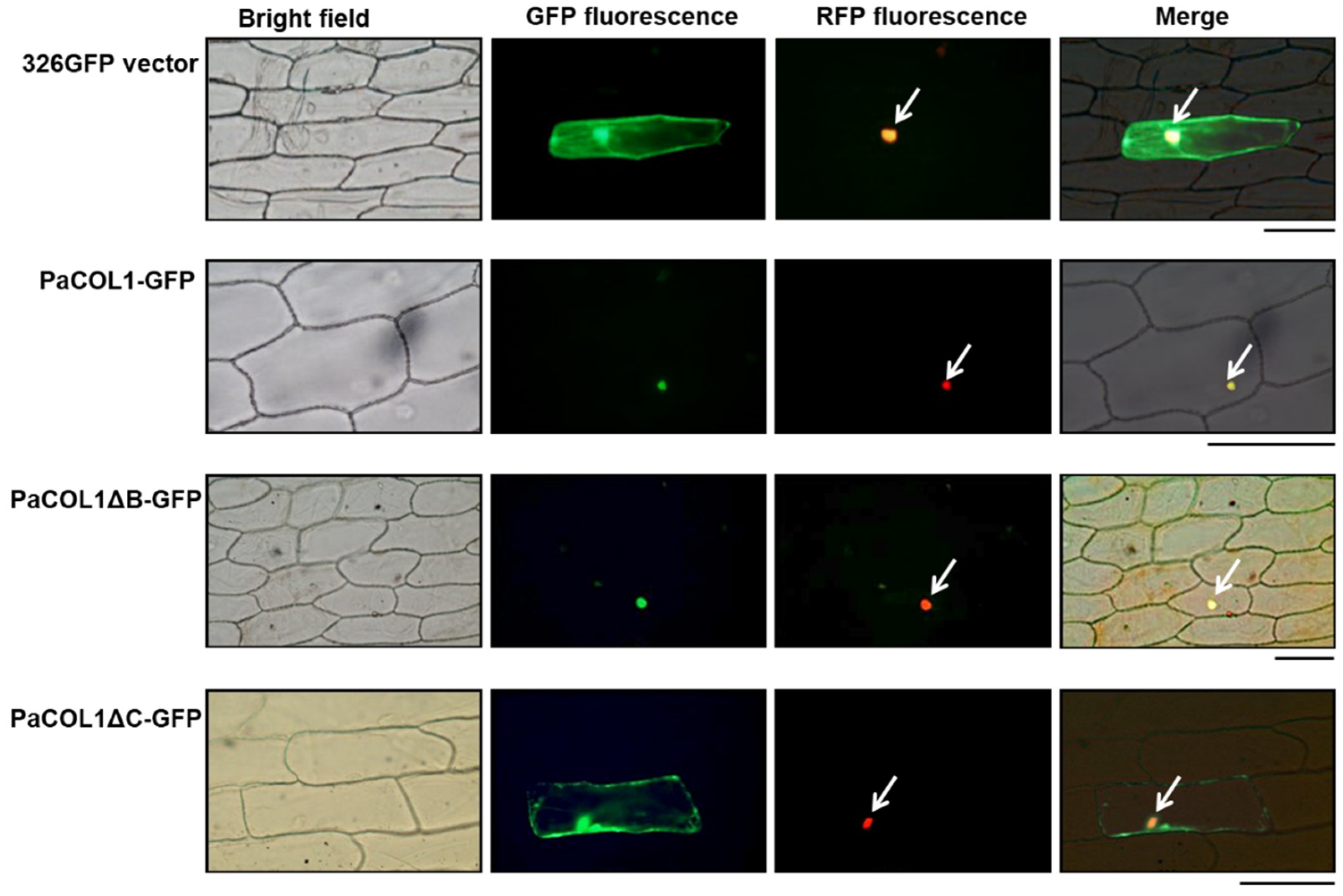
2.2. Subcellular Analysis of PaCOL1 Protein
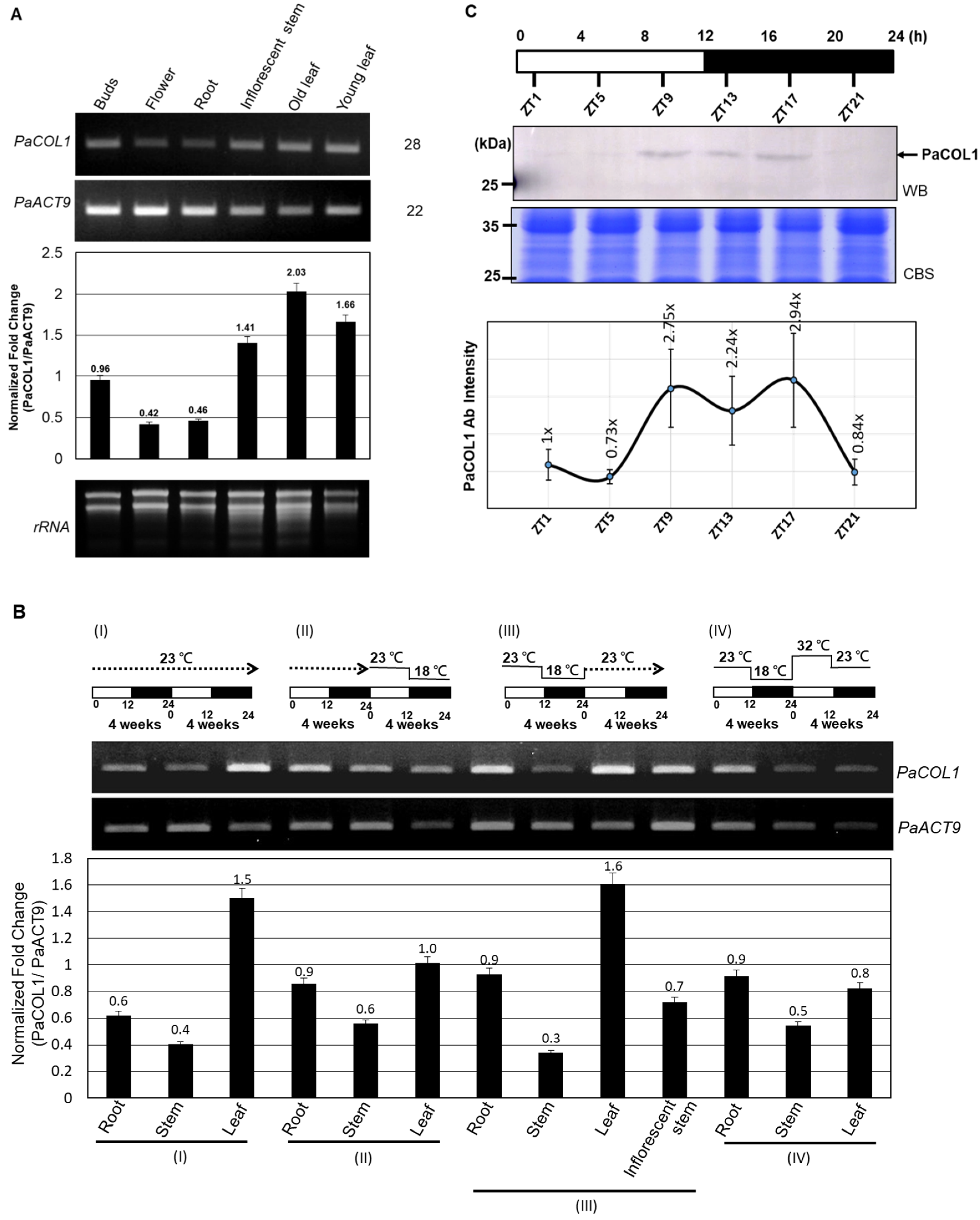
2.3. Expression Profiles of PaCOL1
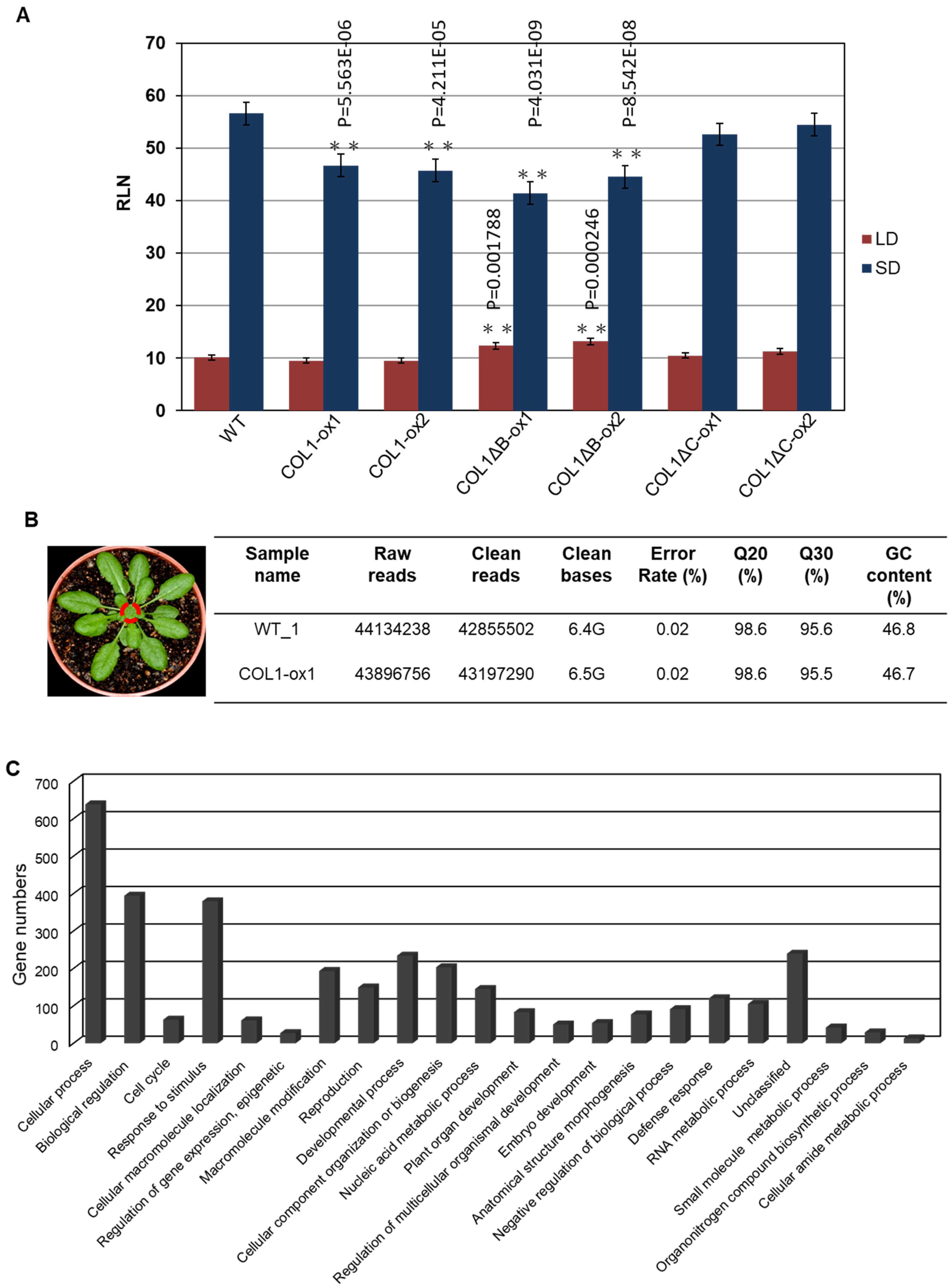
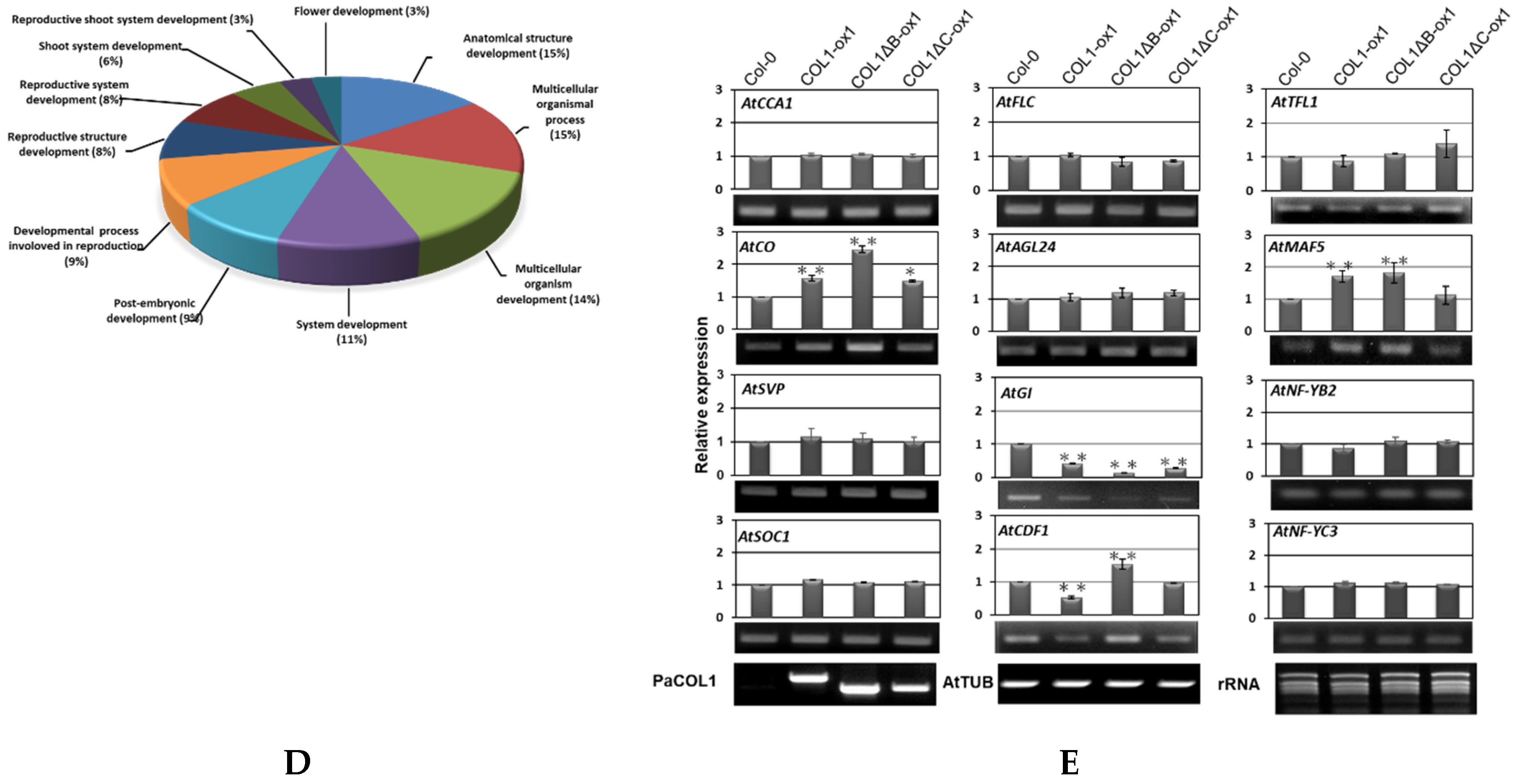
2.4. Overexpression of PaCOL1 in Arabidopsis Conferred SD Floral Inductivity
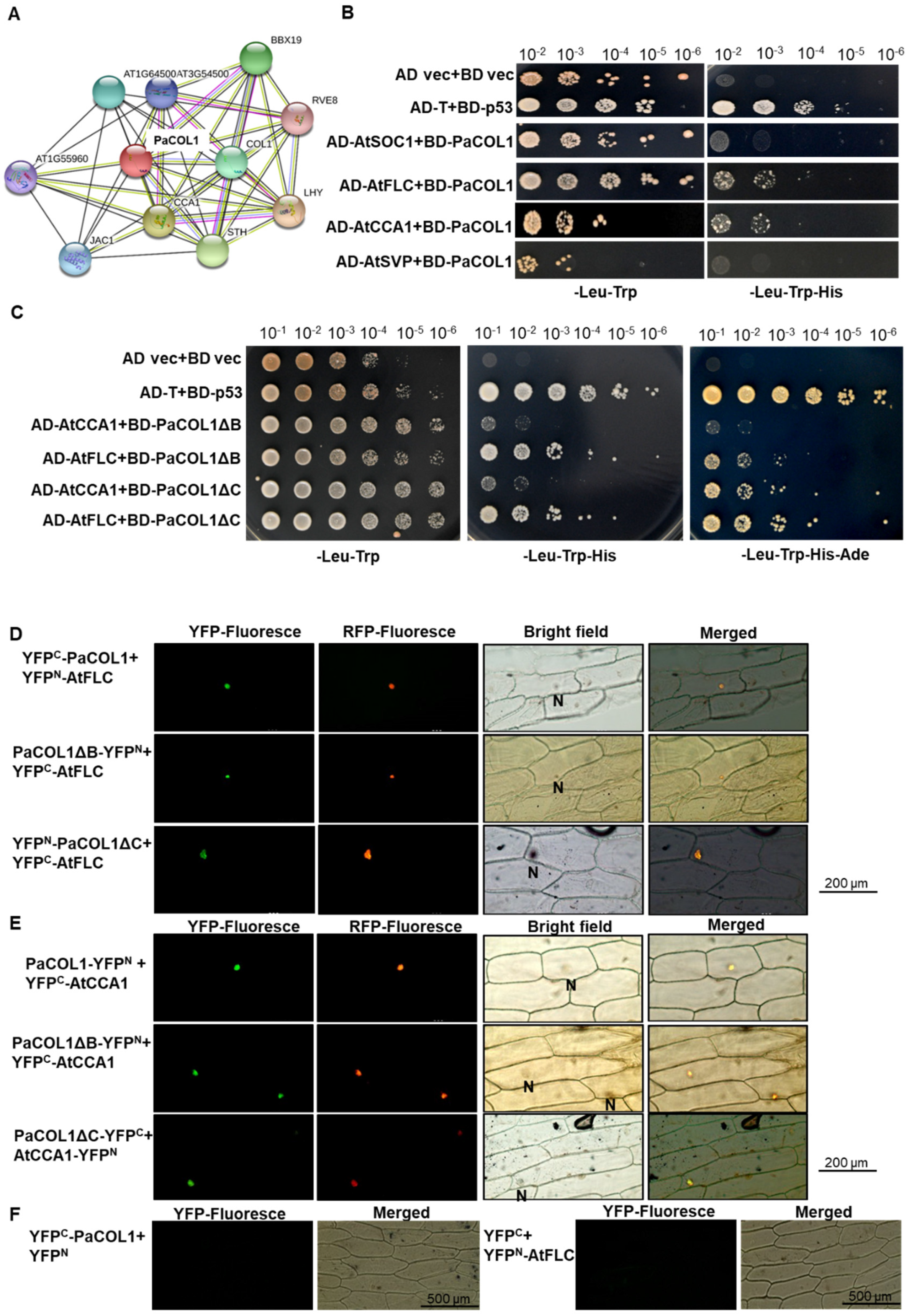
2.5. PaCOL1 Can Interact with AtCCA1 and AtFLC
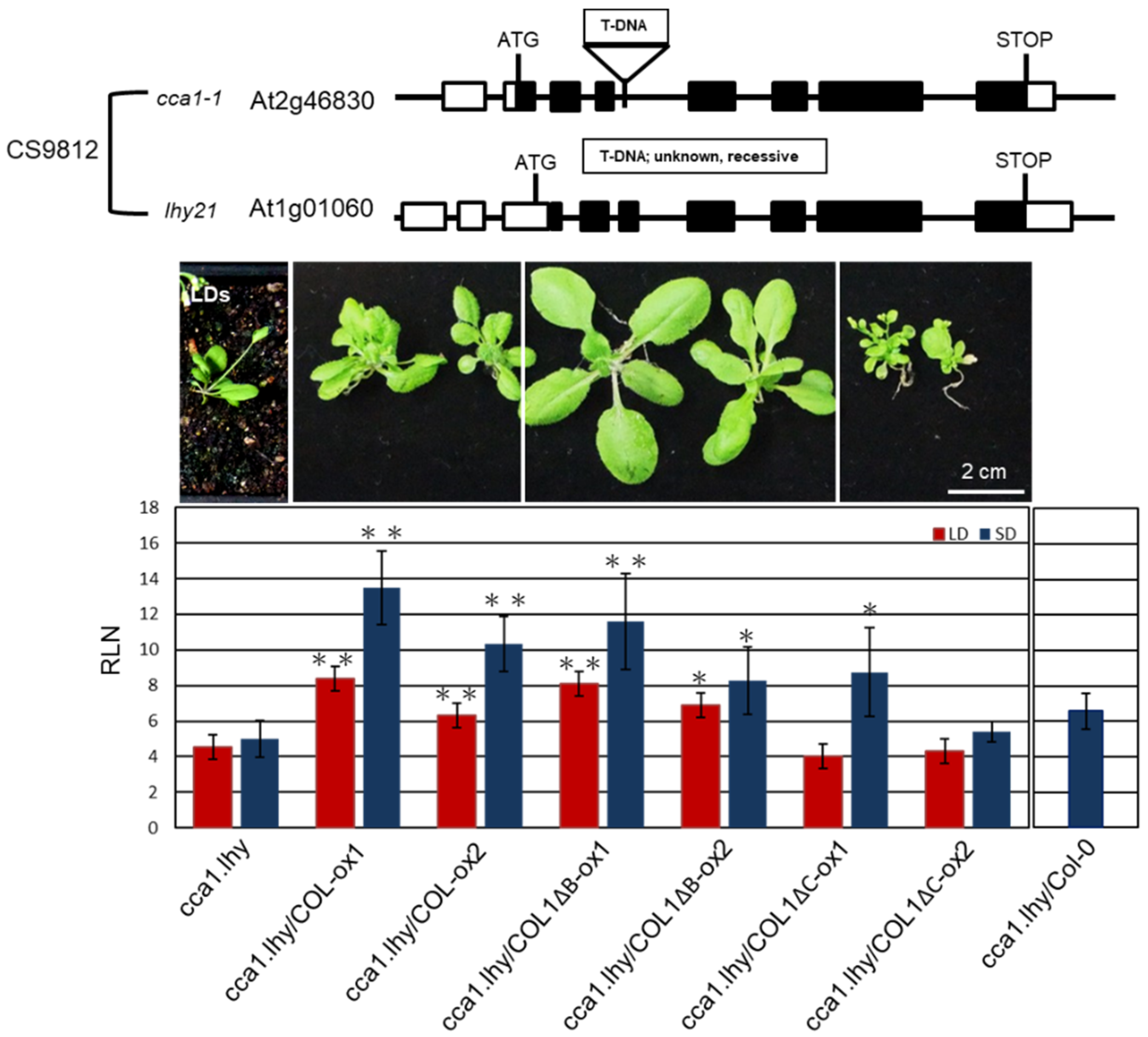
2.6. PaCOL1 Partially Complemented cca1.lhy Mutant Flowering Time under SD Conditions
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RT-PCR and qRT-PCR
4.3. Preparation of DNA Constructs and Transformation
4.4. Subcellular Localization Assays
4.5. Yeast Two-Hybrid (Y2H) Analysis
4.6. Bimolecular Fluorescence Complementation Analysis (BiFC) Assay
4.7. Transcriptome Sequencing
4.8. Electrophoresis and Immunoblotting Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Baurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Bastow, R.M.; Mylne, J.S.; Dean, C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 2004, 16, S18–S31. [Google Scholar] [CrossRef]
- Shrestha, R.; Gomez-Ariza, J.; Brambilla, V.; Fornara, F. Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 2014, 114, 1445–1458. [Google Scholar] [CrossRef]
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta stone of flowering time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef]
- Mockler, T.C.; Yu, X.; Shalitin, D.; Parikh, D.; Michael, T.P.; Liou, J.; Huang, J.; Smith, Z.; Alonso, J.M.; Ecker, J.R.; et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 12759–12764. [Google Scholar] [CrossRef]
- Purugganan, M.D.; Rounsley, S.D.; Schmidt, R.J.; Yanofsky, M.F. Molecular evolution of flower development: Diversification of the plant MADS-box regulatory gene family. Genetics 1995, 140, 345–356. [Google Scholar]
- Klug, A.; Schwabe, J.W. Protein motifs 5. Zinc fingers. FASEB J. 1995, 9, 597–604. [Google Scholar] [CrossRef]
- Klug, A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010, 79, 213–231. [Google Scholar] [CrossRef]
- Ben-Naim, O.; Eshed, R.; Parnis, A.; Teper-Bamnolker, P.; Shalit, A.; Coupland, G.; Samach, A.; Lifschitz, E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006, 46, 462–476. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Jang, S.; Marchal, V.; Panigrahi, K.C.; Wenkel, S.; Soppe, W.; Deng, X.W.; Valverde, F.; Coupland, G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008, 27, 1277–1288. [Google Scholar] [CrossRef]
- Lazaro, A.; Valverde, F.; Pineiro, M.; Jarillo, J.A. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 2012, 24, 982–999. [Google Scholar] [CrossRef]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Liu, H.; Dong, S.; Sun, D.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; Chen, Z. CONSTANS-Like 9 (OsCOL9) interacts with receptor for activated C-Kinase 1(OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS ONE 2016, 11, e0166249. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Gnesutta, N.; Kumimoto, R.W.; Swain, S.; Chiara, M.; Siriwardana, C.; Horner, D.S.; Holt, B.F., 3rd; Mantovani, R. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell 2017, 29, 1516–1532. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef]
- Ogiso, E.; Takahashi, Y.; Sasaki, T.; Yano, M.; Izawa, T. The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 2010, 152, 808–820. [Google Scholar] [CrossRef]
- Mulekar, J.J.; Bu, Q.; Chen, F.; Huq, E. Casein kinase II alpha subunits affect multiple developmental and stress-responsive pathways in Arabidopsis. Plant J. 2012, 69, 343–354. [Google Scholar] [CrossRef]
- Sun, C.; Chen, D.; Fang, J.; Wang, P.; Deng, X.; Chu, C. Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell. 2014, 5, 889–898. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef]
- Liu, L.J.; Zhang, Y.C.; Li, Q.H.; Sang, Y.; Mao, J.; Lian, H.L.; Wang, L.; Yang, H.Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef]
- Wang, C.Q.; Guthrie, C.; Sarmast, M.K.; Dehesh, K. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 2014, 26, 3589–3602. [Google Scholar] [CrossRef]
- Tripathi, P.; Carvallo, M.; Hamilton, E.E.; Preuss, S.; Kay, S.A. Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. USA 2017, 114, 172–177. [Google Scholar] [CrossRef]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.A.; Preuss, S.; et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef]
- Tan, J.; Jin, M.; Wang, J.; Wu, F.; Sheng, P.; Cheng, Z.; Wang, J.; Zheng, X.; Chen, L.; Wang, M.; et al. OsCOL10, a CONSTANS-Like gene, functions as a flowering time repressor downstream of Ghd7 in Rice. Plant Cell Physiol. 2016, 57, 798–812. [Google Scholar] [CrossRef]
- An, F.M.; Hsiao, S.R.; Chan, M.T. Sequencing-based approaches reveal low ambient temperature-responsive and tissue-specific microRNAs in phalaenopsis orchid. PLoS ONE 2011, 6, e18937. [Google Scholar] [CrossRef]
- Su, C.L.; Chao, Y.T.; Alex Chang, Y.C.; Chen, W.C.; Chen, C.Y.; Lee, A.Y.; Hwa, K.T.; Shih, M.C. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011, 52, 1501–1514. [Google Scholar] [CrossRef][Green Version]
- Jang, S.; Choi, S.C.; Li, H.Y.; An, G.; Schmelzer, E. Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS ONE 2015, 10, e0134987. [Google Scholar] [CrossRef]
- Li, D.M.; Lu, F.B.; Zhu, G.F.; Sun, Y.B.; Liu, H.L.; Liu, J.W.; Wang, Z. Molecular characterization and functional analysis of a Flowering locus T homolog gene from a Phalaenopsis orchid. Genet Mol. Res. 2014, 13, 5982–5994. [Google Scholar] [CrossRef]
- Jang, S. Functional characterization of PhapLEAFY, a FLORICAULA/LEAFY ortholog in Phalaenopsis aphrodite. Plant Cell Physiol. 2015, 56, 2234–2247. [Google Scholar]
- Chin, D.C.; Shen, C.H.; SenthilKumar, R.; Yeh, K.W. Prolonged exposure to elevated temperature induces floral transition via up-regulation of cytosolic ascorbate peroxidase 1 and subsequent reduction of the ascorbate redox ratio in Oncidium hybrid orchid. Plant Cell Physiol. 2014, 55, 2164–2176. [Google Scholar] [CrossRef] [PubMed]
- Koroleva, O.A.; Calder, G.; Pendle, A.F.; Kim, S.H.; Lewandowska, D.; Simpson, C.G.; Jones, I.M.; Brown, J.W.; Shaw, P.J. Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: Fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 2009, 21, 1592–1606. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Tseng, Y.C.; Liu, Y.C.; Chuo, C.M.; Chen, P.T.; Tseng, K.M.; Yeh, Y.C.; Ger, M.J.; Wang, H.L. Cool-night temperature induces spike emergence and affects photosynthetic efficiency and metabolizable carbohydrate and organic acid pools in Phalaenopsis aphrodite. Plant Cell Rep. 2008, 27, 1667–1675. [Google Scholar] [CrossRef]
- Ballerini, E.S.; Kramer, E.M. In the light of evolution: A reevaluation of conservation in the CO-FT Regulon and its role in photoperiodic regulation of flowering time. Front. Plant Sci. 2011, 2, 81. [Google Scholar] [CrossRef]
- Song, Y.H.; Ito, S.; Imaizumi, T. Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013, 18, 575–583. [Google Scholar] [CrossRef]
- Zeevaart, J.A. Leaf-produced floral signals. Curr. Opin. Plant Biol. 2008, 11, 541–547. [Google Scholar] [CrossRef]
- Kong, X.; Luo, X.; Qu, G.P.; Liu, P.; Jin, J.B. Arabidopsis SUMO protease ASP1 positively regulates flowering time partially through regulating FLC stability. J. Integr. Plant Biol. 2017, 59, 15–29. [Google Scholar] [CrossRef]
- Nasim, Z.; Fahim, M.; Ahn, J.H. Possible role of MADS AFFECTING FLOWERING 3 and B-BOX DOMAIN PROTEIN 19 in flowering time regulation of Arabidopsis mutants with defects in nonsense-mediated mRNA decay. Front. Plant Sci. 2017, 8, 191. [Google Scholar] [CrossRef]
- Shibuta, M.; Abe, M. FE controls the transcription of downstream flowering regulators through two distinct mechanisms in leaf phloem companion cells. Plant Cell Physiol. 2017, 58, 2017–2025. [Google Scholar] [CrossRef]
- Zeng, F.; Biligetu, B.; Coulman, B.; Schellenberg, M.P.; Fu, Y.B. RNA-Seq analysis of gene expression for floral development in crested wheatgrass (Agropyron cristatum L.). PLoS ONE. 2017, 12, e0177417. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Alabadi, D.; Yanovsky, M.J.; Mas, P.; Harmer, S.L.; Kay, S.A. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 2002, 12, 757–761. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carre, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002, 2, 629–641. [Google Scholar] [CrossRef]
- Yamamoto, K.; Suzuki, T.; Aihara, Y.; Haga, K.; Sakai, T.; Nagatani, A. The phototropic response is locally regulated within the topmost light-responsive region of the Arabidopsis thaliana seedling. Plant Cell Physiol. 2014, 55, 497–506. [Google Scholar] [CrossRef]
- Kim, S.K.; Yun, C.H.; Lee, J.H.; Jang, Y.H.; Park, H.Y.; Kim, J.K. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 2008, 228, 355–365. [Google Scholar] [CrossRef]
- Zhou, R.; Liu, P.; Li, D.; Zhang, X.; Wei, X. Photoperiod response-related gene SiCOL1 contributes to flowering in sesame. BMC Plant Biol. 2018, 18, 343. [Google Scholar] [CrossRef]
- Chao, Y.T.; Yen, S.H.; Yeh, J.H.; Chen, W.C.; Shih, M.C. Orchidstra 2.0-A Transcriptomics Resource for the Orchid Family. Plant Cell Physiol. 2017, 58, e9. [Google Scholar] [CrossRef]
- Yan, H.; Marquardt, K.; Indorf, M.; Jutt, D.; Kircher, S.; Neuhaus, G.; Rodriguez-Franco, M. Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 2011, 156, 1772–1782. [Google Scholar] [CrossRef]
- Dixon, L.E.; Karsai, I.; Kiss, T.; Adamski, N.M.; Liu, Z.; Ding, Y.; Allard, V.; Boden, S.A.; Griffiths, S. VERNALIZATION1 controls developmental responses of winter wheat under high ambient temperatures. Development 2019, 146, dev172684. [Google Scholar] [CrossRef]
- Ream, T.S.; Woods, D.P.; Schwartz, C.J.; Sanabria, C.P.; Mahoy, J.A.; Walters, E.M.; Kaeppler, H.F.; Amasino, R.M. Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 2014, 164, 694–709. [Google Scholar] [CrossRef]
- Woods, D.P.; McKeown, M.A.; Dong, Y.; Preston, J.C.; Amasino, R.M. Evolution of VRN2/Ghd7-Like Genes in Vernalization-Mediated Repression of Grass Flowering. Plant Physiol. 2016, 170, 2124–2135. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gu, L.; Gao, L.; Chen, C.; Wei, C.Q.; Qiu, Q.; Chien, C.W.; Wang, S.; Jiang, L.; Ai, L.F.; et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016, 48, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gonzalez, L.; Mouriz, A.; Narro-Diego, L.; Bustos, R.; Martinez-Zapater, J.M.; Jarillo, J.A.; Pineiro, M. Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell 2014, 26, 3922–3938. [Google Scholar] [CrossRef]
- Wagner, D.; Meyerowitz, E.M. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 2002, 12, 85–94. [Google Scholar] [CrossRef]
- Aceto, S.; Gaudio, L. The MADS and the beauty: Genes Involved in the development of orchid flowers. Curr. Genomics 2011, 12, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Enoki, S.; Fujimori, N.; Yamaguchi, C.; Hattori, T.; Suzuki, S. High constitutive overexpression of glycosyl hydrolase family 17 delays floral transition by enhancing FLC expression in transgenic Arabidopsis. Plants 2017, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Takato, H.; Fujita, K.; Suzuki, S. HSG1, a grape Bcl-2-associated athanogene, promotes floral transition by activating CONSTANS expression in transgenic Arabidopsis plant. Mol. Biol. Rep. 2012, 39, 4367–4374. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Breton, G.; Priest, H.D.; Dharmawardhana, P.; Jaiswal, P.; Fox, S.E.; Michael, T.P.; Chory, J.; Kay, S.A.; Mockler, T.C. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 2011, 6, e16907. [Google Scholar] [CrossRef]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carre, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Kenigsbuch, D.; Sun, L.; Harel, E.; Ong, M.S.; Tobin, E.M. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 1997, 9, 491–507. [Google Scholar]
- Alabadi, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012, 22, R648–R657. [Google Scholar] [CrossRef] [PubMed]
- Farre, E.M.; Harmer, S.L.; Harmon, F.G.; Yanovsky, M.J.; Kay, S.A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 2005, 15, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kita, M.; Ito, S.; Sato, E.; Yamashino, T.; Mizuno, T. The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 2005, 46, 609–619. [Google Scholar] [CrossRef]
- Toda, Y.; Kudo, T.; Kinoshita, T.; Nakamichi, N. Evolutionary insight into the clock-associated PRR5 transcriptional network of flowering plants. Sci. Rep. 2019, 9, 2983. [Google Scholar] [CrossRef]
- Daniel, X.; Sugano, S.; Tobin, E.M. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3292–3297. [Google Scholar] [CrossRef]
- Lai, A.G.; Doherty, C.J.; Mueller-Roeber, B.; Kay, S.A.; Schippers, J.H.; Dijkwel, P.P. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 17129–17134. [Google Scholar] [CrossRef]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Yoshida, R. Punctual coordination: Switching on and off for flowering during a day. Plant Signal. Behav. 2009, 4, 113–115. [Google Scholar] [CrossRef]
- Cai, J.; Liu, X.; Vanneste, K.; Proost, S.; Tsai, W.C.; Liu, K.W.; Chen, L.J.; He, Y.; Xu, Q.; Bian, C.; et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef]
- Ke, Y.T.; Lu, C.A.; Wu, S.J.; Yeh, C.H. Characterization of Rice Group 3 LEA Genes in Developmental Stages and Under Abiotic Stress. Plant Mol. Biol. Rep. 2016, 34, 1003–1015. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, D.H.; Kim, Y.W.; Hwang, I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 2001, 13, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Yeh, K.W.; Wu, S.H.; Chang, P.F.L.; Chen, Y.M.; Lin, C.Y. A recombinant rice 16.9-kDa heat-shock protein can provide thermoprotection in-vitro. Plant Cell Physio. 1995, 36, 1341–1348. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ke, Y.-T.; Lin, K.-F.; Gu, C.-H.; Yeh, C.-H. Molecular Characterization and Expression Profile of PaCOL1, a CONSTANS-like Gene in Phalaenopsis Orchid. Plants 2020, 9, 68. https://doi.org/10.3390/plants9010068
Ke Y-T, Lin K-F, Gu C-H, Yeh C-H. Molecular Characterization and Expression Profile of PaCOL1, a CONSTANS-like Gene in Phalaenopsis Orchid. Plants. 2020; 9(1):68. https://doi.org/10.3390/plants9010068
Chicago/Turabian StyleKe, Yi-Ting, Kung-Fu Lin, Chu-Han Gu, and Ching-Hui Yeh. 2020. "Molecular Characterization and Expression Profile of PaCOL1, a CONSTANS-like Gene in Phalaenopsis Orchid" Plants 9, no. 1: 68. https://doi.org/10.3390/plants9010068
APA StyleKe, Y.-T., Lin, K.-F., Gu, C.-H., & Yeh, C.-H. (2020). Molecular Characterization and Expression Profile of PaCOL1, a CONSTANS-like Gene in Phalaenopsis Orchid. Plants, 9(1), 68. https://doi.org/10.3390/plants9010068



