Identification and Characterization of Phosphoproteins in Somatic Embryogenesis Acquisition during Oil Palm Tissue Culture
Abstract
1. Introduction
2. Results
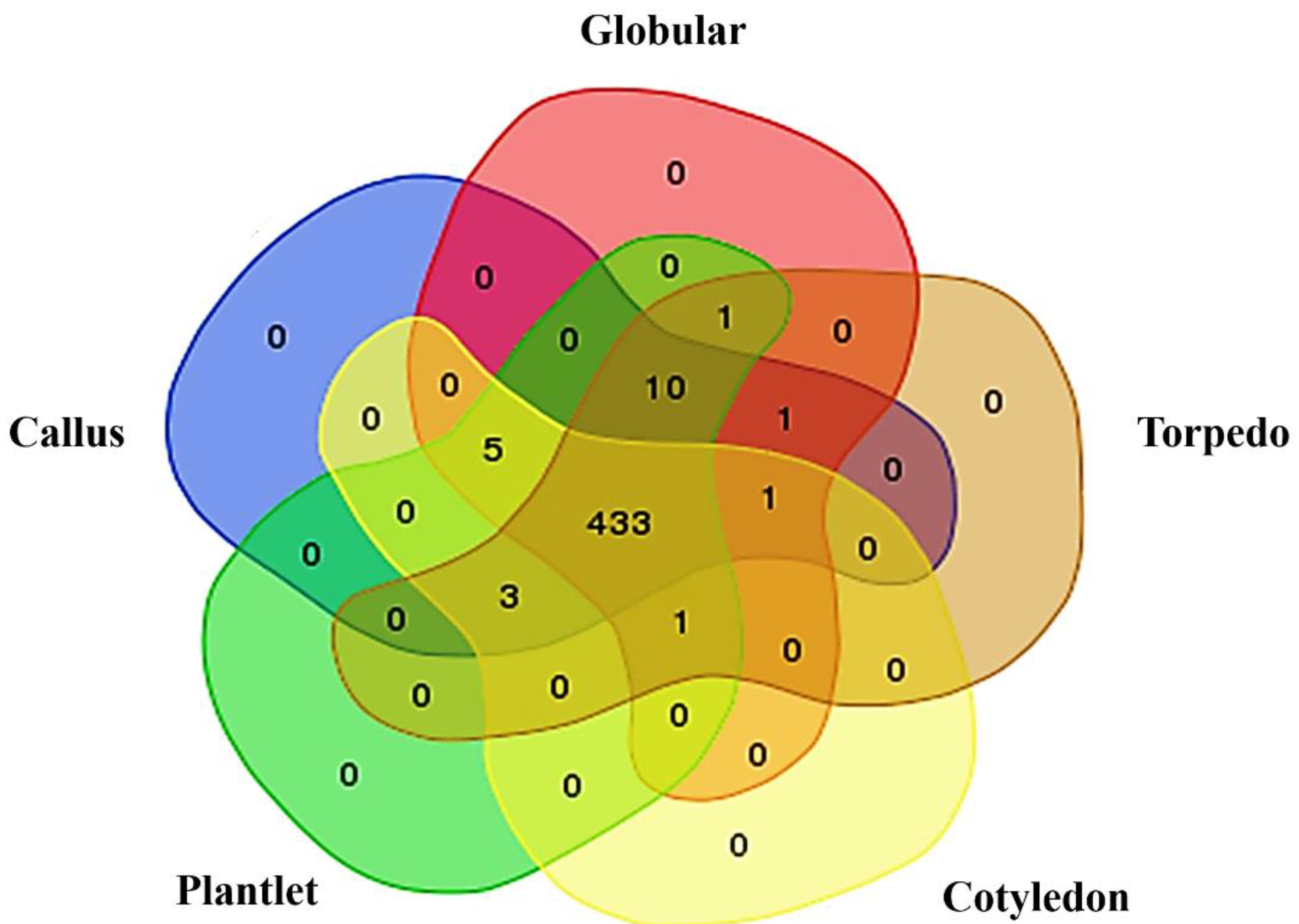
2.1. Phosphoprotein Enrichment and LC-MS/MS Identification
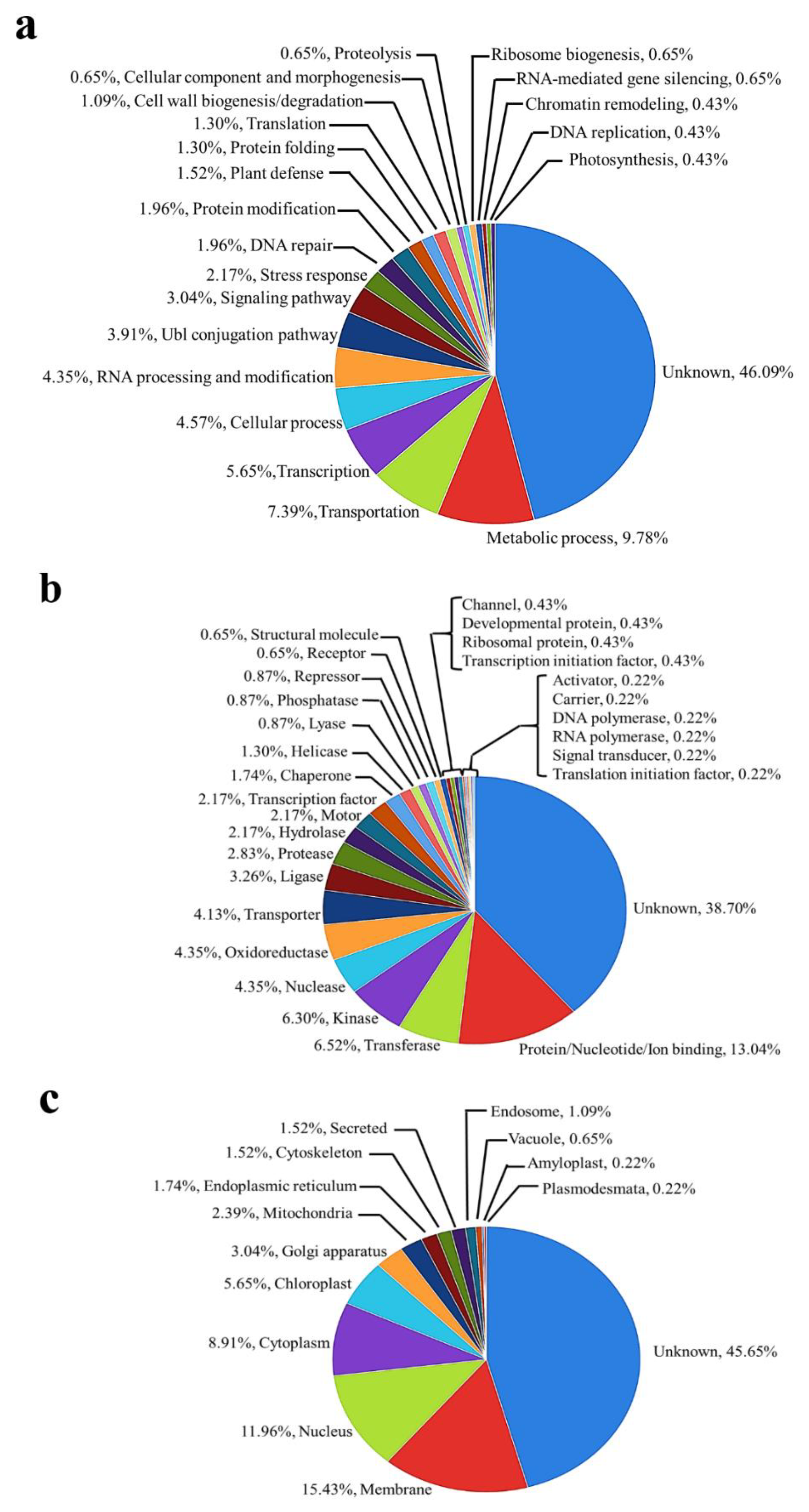
2.2. Functional Classification of Identified Phosphoproteins
2.3. Differentially Expressed Phosphoproteins during Acquisition of Oil Palm Somatic Embryo
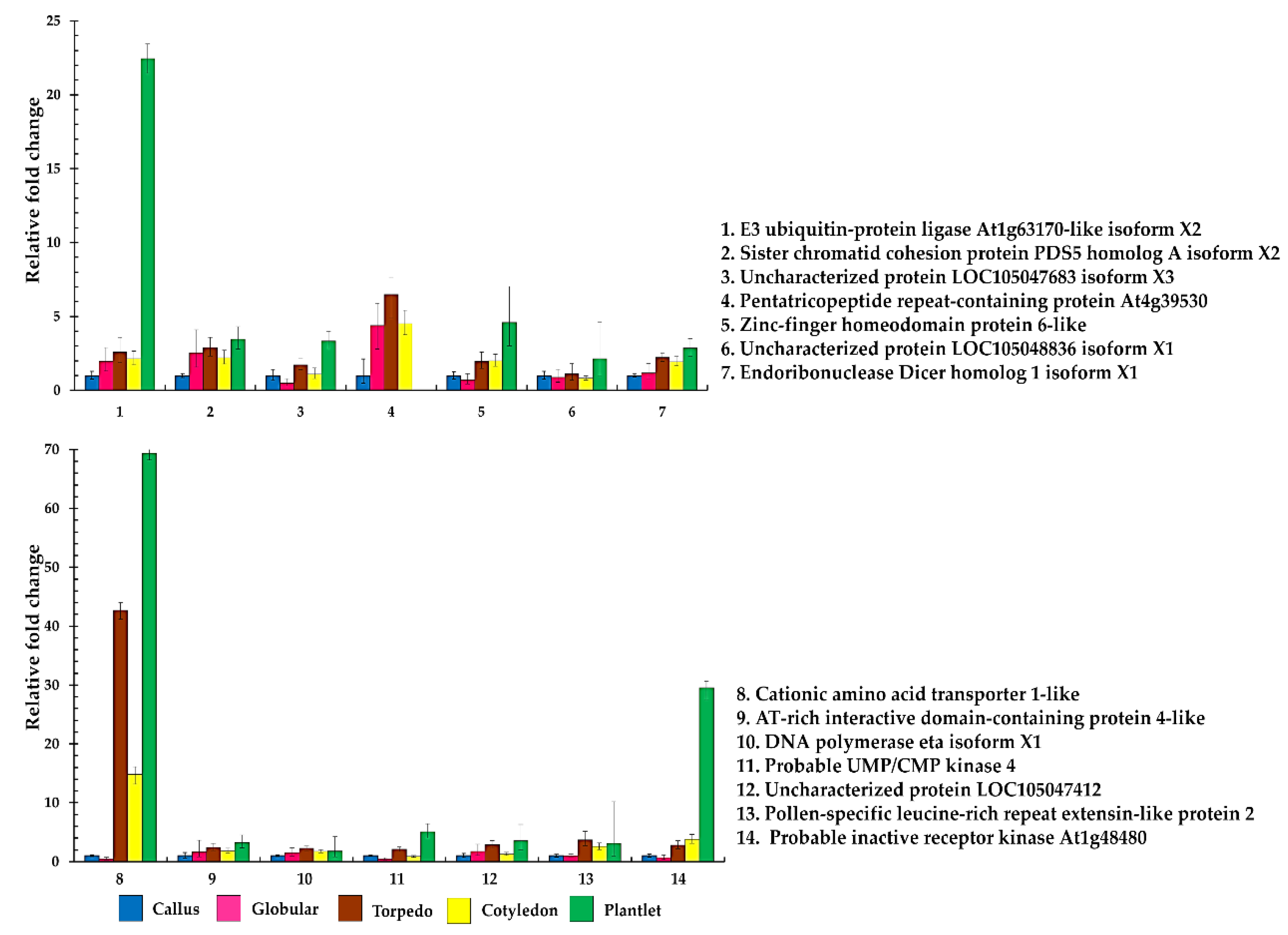
2.4. Quantitative Real-Time Reverse Transcription–PCR of Differential Phosphoproteins
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Protein Extraction and Quantification
4.3. Phosphoprotein Enrichment and Digestion
4.4. Nano-Liquid Chromatography–Mass Spectrophotometry (Nano LC-MS/MS)
4.5. Bioinformatics and Data Analysis
4.6. Quantitative Real-Time Reverse Transcription–PCR Verification
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ndon, B.A. The Oil Palm (Elaeis guineensis Jacq); Concept Publications Ltd.: Wallingford, CT, USA, 2006. [Google Scholar]
- Corley, R.H.V.; Tinker, P.B. The Oil Palm, 4th ed.; Blackwell: Oxford, UK, 2003; p. 202. [Google Scholar]
- Aratrakorn, S.; Thunhikorn, S.; Donald, P.F. Changes in bird communities following conversion of lowland forest to oil palm and rubber plantations in southern Thailand. Bird Conserv. Int. 2006, 16, 71–82. [Google Scholar] [CrossRef]
- Ummi, K.H.; Nur Shiela, M.N.; Tengku Nur Diyana, T.M.; Gunaselan, S. In vitro germination in encapsulated and non-encapsulated Elaeis guineensis Jacquin (1763) embryo. Empow. Sci. 2011, 121, 764–769. [Google Scholar]
- Thuzar, M.; Vanavichit, A.; Tragoonrung, S.; Jantasuriyarat, C. Efficient and rapid plant regeneration of oil palm zygotic embryos cv. ‘Tenera’ through somatic embryogenesis. Acta Physiol. Plant. 2011, 33, 123–128. [Google Scholar] [CrossRef]
- Thuzar, M.; Vanavichit, A.; Tragoonrung, S.; Jantasuriyarat, C. Recloning of regenerated plantlets from elite oil palm (Elaeis guineensis Jacq.) cv. Tenera. Afr. J. Biotechnol. 2012, 11, 14761–14770. [Google Scholar]
- Von Arnold, S.; Sabala, I.; Bozhkov, P.; Dyachok, J.; Filonova, L. Developmental pathways of somatic embryogenesis. Plant Cell Tissue Organ Cult. 2002, 69, 233–249. [Google Scholar] [CrossRef]
- Karami, O.; Aghavaisi, B.; Aghil Mahmoudi, P. Molecular aspects of somatic-to-embryogenic transition in plants. J. Chem. Biol. 2009, 2, 177–190. [Google Scholar] [CrossRef]
- Ledwon, A.; Gaj, M.D. LEAFY COTYLEDON1, FUSCA3 expression and auxin treatment in relation to somatic embryogenesis induction in Arabidopsis. Plant Growth Regul. 2011, 5, 157–167. [Google Scholar] [CrossRef]
- Kulinska-Lukaszek, K.; Tobojka, M.; Adamiok, A.; Kurczynska, E.U. Expression of the BBM gene during somatic embryogenesis of Arabidopsis thaliana. Biol. Plant 2012, 56, 389–394. [Google Scholar] [CrossRef]
- Takáč, T.; Pechan, T.; Samaj, J. Differential proteomics of plant development. J. Proteom. 2011, 74, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Silva Rde, C.; Carmo, L.S.; Luis, Z.G.; Silva, L.P.; Scherwinski-Pereira, J.E.; Mehta, A. Proteomic identification of differentially expressed proteins during the acquisition of somatic embryogenesis in oil palm (Elaeis guineensis Jacq.). J. Proteom. 2014, 104, 112–127. [Google Scholar] [CrossRef]
- Li, J.; Silva-Sanchez, C.; Zhang, T.; Chen, S.; Li, H. Phosphoproteomics technologies and applications in plant biology research. Front. Plant Sci. 2015, 6, 430. [Google Scholar] [CrossRef] [PubMed]
- Kline-Jonakin, K.G.; Barrett-Wilt, G.A.; Sussman, M.R. Quantitative plant phosphoproteomics. Curr. Opin. Plant Biol. 2011, 14, 507–511. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente van Bentem, S.; Hirt, H. Using phosphoproteomics to reveal signaling dynamics in plants. Trends Plant Sci. 2007, 12, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Silva-Sanchez, C.; Li, H.; Chen, S. Recent advances and challenges in plant phosphoproteomics. Proteomics 2015, 15, 1127–1141. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, S.; Chen, L.; Quatrano, R.S.; He, Y. Phospho-proteomic analysis of developmental reprogramming in the moss Physcomitrella patens. J. Proteom. 2014, 108, 284–294. [Google Scholar] [CrossRef]
- Rampitsch, C.; Bykova, N.V.; Mauthe, W.; Yakandawala, N.; Jordan, M. Phosphoproteomic profiling of wheat callus labelled in vivo. Plant Sci. 2006, 171, 488–496. [Google Scholar] [CrossRef]
- Han, C.; Yang, P.; Sakata, K.; Komatsu, S. Quantitative proteomics reveals the role of protein phosphorylation in rice embryos during early stages of germination. J. Proteome Res. 2014, 7, 1766–1782. [Google Scholar] [CrossRef]
- Meyer, L.J.; Gao, J.; Xu, D.; Thelen, J.J. Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 2012, 159, 517–528. [Google Scholar] [CrossRef]
- Loyola-Vargas, V.M.; Ochoa-Alejo, N. Somatic Embryogenesis: Fundamental Aspects and Applications, 1st ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Qiu, J.; Hou, Y.; Tong, X.; Wang, Y.; Lin, H. Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 249–265. [Google Scholar] [CrossRef]
- Mazzucotell, E.; Belloni, S.; Marone, D.; De Leonardis, A.M.; Guerra, D.; Di Fonzo, N.; Cattivelli, L.; Mastrangelo, A.M. The E3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genom. 2006, 7, 509–522. [Google Scholar] [CrossRef]
- Kelley, D.R. E3 ubiquitin ligases: Key regulators of hormone signaling in plants. Mol. Cell. Proteom. 2018, 17, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hellmann, H. Plant E3 ligases: Flexible enzymes in a sessile world. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [PubMed]
- Morel, A.; Teyssier, C.; Trontin, J.F.; Eliášová, K.; Pešek, B.; Beaufour, M.; Morabito, D.; Boizot, N.; Le Metté, C.; Belal-Bessai, L.; et al. Early molecular events involved in Pinus pinaster Ait. somatic embryo development under reduced water availability: Transcriptomic and proteomic analyses. Physiol. Plant. 2014, 152, 184–201. [Google Scholar] [CrossRef]
- Gautier, F.; Eliášová, K.; Leplé, J.C.; Vondráková, Z.; Lomenech, A.M.; Le Metté, C.; Label, P.; Costa, G.; Trontin, J.F.; Teyssier, C.; et al. Repetitive somatic embryogenesis induced cytological and proteomic changes in embryogenic lines of Pseudotsuga menziesii [Mirb.]. BMC Plant Biol. 2018, 18, 164. [Google Scholar] [CrossRef] [PubMed]
- Pradillo, M.; Knoll, A.; Oliver, C.; Varas, J.; Corredor, E.; Puchta, H.; Santos, J.L. Involvement of the Cohesin cofactor PDS5 (SPO76) during meiosis and DNA repair in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1034. [Google Scholar] [CrossRef]
- Hammani, K.; Gobert, A.; Hleibieh, K.; Choulier, L.; Small, I.; Giegé, P. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 2011, 23, 730–740. [Google Scholar] [CrossRef]
- Kurihara, Y.; Takashi, Y.; Watanabe, Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 2006, 12, 206–212. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Hong, L.; Fitzpatrick, K.E.; Amasino, R.M. DICER-LIKE 1 and DICER-LIKE 3 redundantly act to promote flowering via repression of FLOWERING LOCUS C in Arabidopsis thaliana. Genetics 2007, 176, 1359–1362. [Google Scholar] [CrossRef]
- Xu, Y.; Zong, W.; Hou, X.; Yao, J.; Liu, H.; Li, X.; Zhao, Y.; Xiong, L. OsARID3, an AT-rich interaction domain-containing protein, is required for shoot meristem development in rice. Plant J. 2015, 83, 806–817. [Google Scholar] [CrossRef]
- Jesús Santiago, M.; Alejandre-Durán, E.; Muñoz-Serrano, A.; Ruiz-Rubio, M. Two translation synthesis DNA polymerase genes, AtPOLH and AtREV1, are involved in development and UV light resistance in Arabidopsis. J. Plant Physiol. 2008, 165, 1582–1591. [Google Scholar] [CrossRef]
- Curtis, M.J.; Hays, J.B. Cooperative responses of DNA-damage-activated protein kinases ATR and ATM and DNA translesion polymerases to replication-blocking DNA damage in a stem-cell niche. DNA Repair 2011, 10, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Lacroute, F.; Thornburg, R. Cloning, expression in Escherichia coli, and characterization of Arabidopsis thaliana UMP/CMP kinase. Plant Physiol. 1998, 117, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Stratford, S.; Barne, W.; Hohorst, D.L.; Sagert, J.G.; Cotter, R. A leucine-rich repeat region is conserved in pollen extensin-like (Pex) proteins in monocots and dicots. Plant Mol. Biol. 2001, 46, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Bayer, M.; Nawy, T.; Giglione, C.; Galli, M.; Meinnel, T.; Lukowitz, W. Paternal control of embryonic patterning in Arabidopsis thaliana. Science 2009, 323, 1485–1488. [Google Scholar] [CrossRef] [PubMed]
- Pattarapimol, T.; Thuzar, M.; Vanavichit, A.; Tragoonrung, S.; Roytrakul, S.; Jantasuriyarat, C. Identification of genes involved in somatic embryogenesis development in oil palm (Elaeis guineensis Jacq.) using cDNA AFLP. J. Oil Palm Res. 2015, 27, 1–11. [Google Scholar]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef]
- Kaweewong, K.; Garnjanagoonchorn, W.; Jirapakkul, W.; Roytrakul, S. Solubilization and identification of hen eggshell membrane proteins during different times of chicken embryo development using the proteomic approach. Protein J. 2013, 32, 297–308. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Cantin, G.T.; Yi, W.; Lu, B.; Park, S.K.; Xu, T.; Lee, J.D.; Yates, J.R. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 2008, 7, 1346–1351. [Google Scholar] [CrossRef]
- Nakagami, H.; Sugiyama, N.; Ishihama, Y.; Shirasu, K. Shotguns in the Front Line: Phosphoproteomics in Plants. Plant Cell Physiol. 2012, 53, 118–124. [Google Scholar] [CrossRef]

| NCBI Accession Number | Protein Name | Peptide Sequence and Predicted Phosphorylation Site (*) | MOWSE Scores | Log2 Abundance | ||||
|---|---|---|---|---|---|---|---|---|
| Callus | Globular | Torpedo | Cotyledon | Plantlet | ||||
| gi|743754692 | E3 Ubiquitin-Protein Ligase | GAET *NS *ENICNGGTVAS *GT *DR | 12.71 | 0 | 16.51 | 15.48 | 16.50 | 17.08 |
| gi|743807360 | sister chromatid cohesion protein PDS5 | KNS *MKKS *S *S *S *K | 8.51 | 0 | 16.01 | 14.62 | 0 | 17.66 |
| gi|743797598 | uncharacterized protein | LVLRS *LR | 1.36 | 15.42 | 15.05 | 12.95 | 0 | 16.23 |
| gi|743821075 | pentatricopeptide repeat-containing protein | MPERNMISWSS*MISMY *TQHNR | 10.70 | 15.09 | 15.66 | 14.32 | 0 | 17.13 |
| gi|743851198 | zinc-finger homeodomain protein 6-like | GSNS *T *GGGAGGGPMT *ES *S *S *EER | 16.71 | 15.33 | 15.42 | 14.49 | 0 | 18.64 |
| gi|743802384 | uncharacterized protein | RAWMPTGCLK | 8.43 | 15.72 | 16.48 | 15.18 | 0 | 18.13 |
| gi|743828992 | endoribonuclease Dicer | GVS *Y *CK | 4.14 | 15.97 | 16.55 | 15.26 | 0 | 19.01 |
| gi|743856309 | cationic amino acid transporter 1-like | RGAT *GVAS *AEK | 9.96 | 14.42 | 17.73 | 15.59 | 0 | 18.29 |
| gi|743774060 | AT-rich interactive domain-containing protein 4-like | T *DGLEY *ICPHCSLS *NYK | 6.62 | 15.72 | 16.83 | 15.98 | 0 | 17.38 |
| gi|743882271 | DNA polymerase beta X1 | IQEDAMKLFVSGLR | 12.10 | 16.93 | 18.02 | 16.36 | 0 | 18.83 |
| gi|743855052 | UMP/CMP kinase 4 | MVDAS *KDVDGSFPGEK | 5.19 | 16.20 | 17.54 | 16.80 | 0 | 19.48 |
| gi|743796037 | uncharacterized protein | DAPFPVTTS *FYFHAKSLDPDR | 13.44 | 18.02 | 17.81 | 18.19 | 0 | 17.66 |
| gi|743880173 | pollen-specific leucine-rich repeat extensin-like protein | GS *S *AMKSS *PLAS *S *AAS *S *GGSTNY *T *R | 7.82 | 15.40 | 15.54 | 13.69 | 15.75 | 0 |
| gi|743771831 | inactive receptor kinase | VAGY *RAPEVTDARKVS *QK | 9.61 | 15.84 | 16.27 | 14.72 | 0 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aroonluk, S.; Roytrakul, S.; Jantasuriyarat, C. Identification and Characterization of Phosphoproteins in Somatic Embryogenesis Acquisition during Oil Palm Tissue Culture. Plants 2020, 9, 36. https://doi.org/10.3390/plants9010036
Aroonluk S, Roytrakul S, Jantasuriyarat C. Identification and Characterization of Phosphoproteins in Somatic Embryogenesis Acquisition during Oil Palm Tissue Culture. Plants. 2020; 9(1):36. https://doi.org/10.3390/plants9010036
Chicago/Turabian StyleAroonluk, Suvichark, Sittiruk Roytrakul, and Chatchawan Jantasuriyarat. 2020. "Identification and Characterization of Phosphoproteins in Somatic Embryogenesis Acquisition during Oil Palm Tissue Culture" Plants 9, no. 1: 36. https://doi.org/10.3390/plants9010036
APA StyleAroonluk, S., Roytrakul, S., & Jantasuriyarat, C. (2020). Identification and Characterization of Phosphoproteins in Somatic Embryogenesis Acquisition during Oil Palm Tissue Culture. Plants, 9(1), 36. https://doi.org/10.3390/plants9010036





