Characterization and Expression Analysis of the Ca2+/Cation Antiporter Gene Family in Tomatoes
Abstract
1. Introduction
2. Results and Discussion
2.1. Phylogenetic Analysis of the Tomato CaCA (SlCaCA) Gene Family
2.2. SlCaCA Protein and Gene Characterization
2.3. Protein Structure and Localization
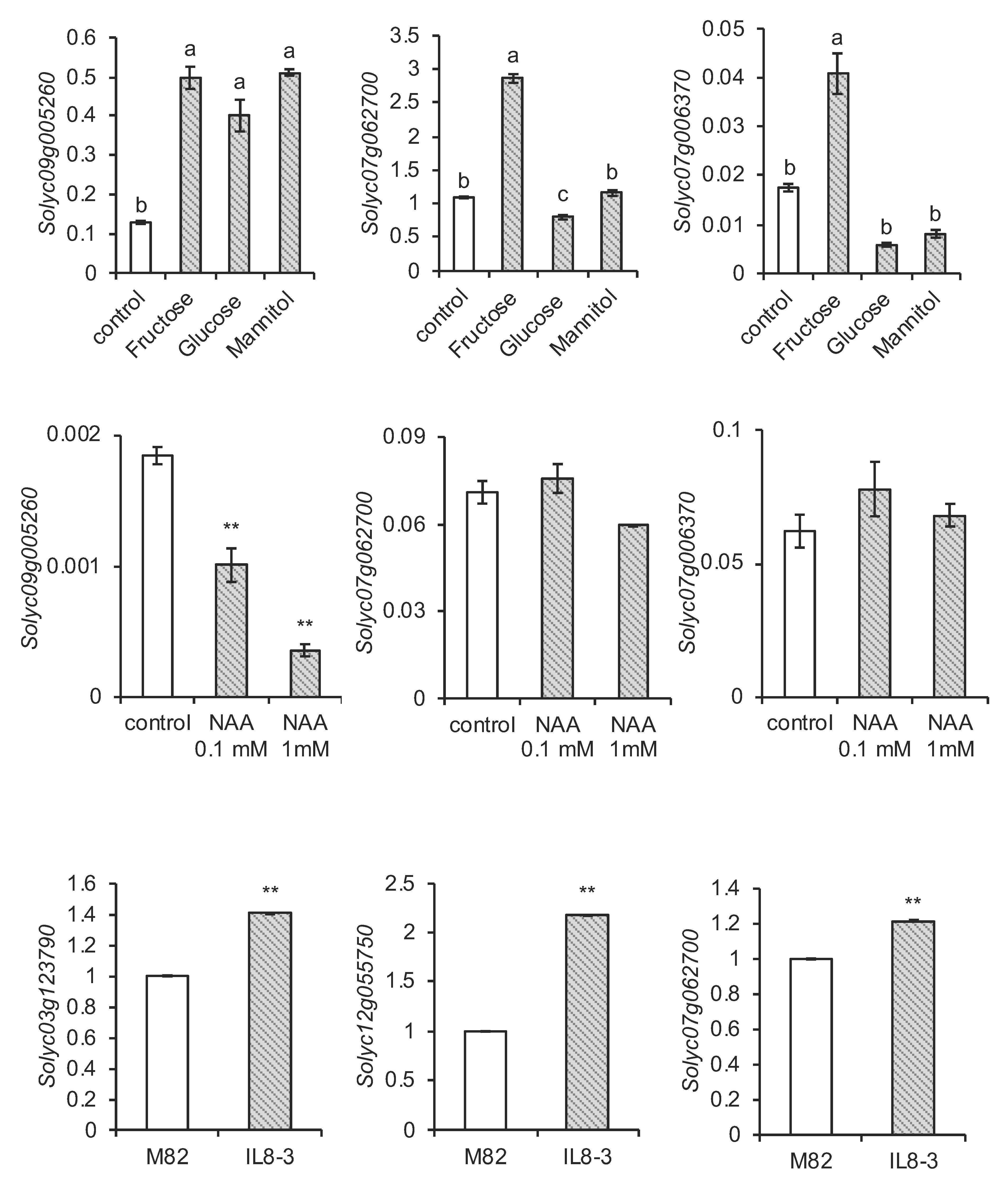
2.4. Expression Profile of SlCaCA Genes in Vegetative Organs, Flowers, and Fruit throughout Development
3. Materials and Methods
3.1. Plant Materials
3.2. RNA Extraction and cDNA Synthesis
3.3. SlCaCA Sequence Analysis
3.4. Expression Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hirschi, K.D.; Zhen, R.G.; Cunningham, K.W.; Rea, P.A.; Fink, G.R. CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Natl. Acad. Sci. USA 1996, 93, 8782–8786. [Google Scholar] [CrossRef] [PubMed]
- Hirschi, K.D.; Korenkov, V.D.; Wilganowski, N.L.; Wagner, G.J. Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 2000, 124, 125–133. [Google Scholar] [CrossRef]
- Kamiya, T.; Akahori, T.; Maeshima, M. Expression profile of the genes for rice cation/H+ exchanger family and functional analysis in yeast. Plant Cell Physiol. 2005, 46, 1735–1740. [Google Scholar] [CrossRef]
- Morris, J.; Tian, H.; Park, S.; Sreevidya, C.S.; Ward, J.M.; Hirschi, K.D. AtCCX3 is an Arabidopsis endomembrane H+-dependent K+ transporter. Plant Physiol. 2008, 148, 1474–1486. [Google Scholar] [CrossRef]
- Shaul, O.; Hilgemann, D.W.; de Almeida Engler, J.; Montagu, M.V.; Inzé, D.; Galili, G. Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J. 1999, 18, 3973–3980. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, Z.; Wei, J.; Zhao, Z.; Sun, D.; Cui, S. A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J. Biol. Chem. 2012, 287, 44062–44070. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, R.; Tripathi, A.K.; Gupta, B.K.; Pareek, A.; Singla-Pareek, S.L. Genome-wide investigation and expression analysis of Sodium/Calcium exchanger gene family in rice and Arabidopsis. Rice 2015, 8, 21. [Google Scholar] [CrossRef]
- Pittman, J.K.; Hirschi, K.D. Phylogenetic analysis and protein structure modelling identifies distinct Ca2+/Cation antiporters and conservation of gene family structure within Arabidopsis and rice species. Rice 2016, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Taneja, M.; Tyagi, S.; Sharma, S.; Upadhyay, S. Ca2+/Cation antiporters (CaCA): Identification, characterization and expression profiling in bread wheat (Triticum aestivum L.). Front. Plant Sci. 2016, 7, 1775. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.; Theerawitaya, C.; Chaum, S.; Kirdmanee, C.; Takabe, T. Expression and functional analysis of putative vacuolar Ca2+-transporters (CAXs and ACAs) in roots of salt tolerant and sensitive rice cultivars. Protoplasma 2014, 251, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.; Villieres, F.; Kroniewicz, L.; Lee, S.; Seo, Y.J.; Hirschi, K.D.; Leonhardt, N.; Kwak, J.M. Vacuolar CAX1 and CAX3 influence auxin transport in guard cells via regulation of apoplastic pH. Plant Physiol. 2012, 160, 1293–1302. [Google Scholar] [CrossRef]
- Li, P.; Zhang, G.; Gonzales, N.; Guo, Y.; Hu, H.; Park, S.; Zhao, J. Ca2+-regulated and diurnal rhythm-regulated Na+/Ca2+ exchanger AtNCL affects flowering time and auxin signalling in Arabidopsis. Plant Cell Environ. 2016, 39, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Hocking, B.; Conn, S.J.; Manohar, M.; Xu, B.; Athman, A.; Stancombe, M.A.; Webb, A.R.; Hirschi, K.D.; Gilliham, M. Heterodimerization of Arabidopsis calcium/proton exchangers contributes to regulation of guard cell dynamics and plant defense responses. J. Exp. Bot. 2017, 68, 4171–4183. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.; Hawthorne, K.; Hotze, T.; Abrams, S.; Hirschi, K.D. Nutritional impact of elevated calcium transport activity in carrots. Proc. Natl. Acad. Sci. USA 2008, 105, 431–1435. [Google Scholar] [CrossRef] [PubMed]
- Navarro-León, E.; Ruiz, J.; Graham, N.; Blasco, B. Physiological profile of CAX1a TILLING mutants of Brassica rapa exposed to different calcium doses. Plant Sci. 2018, 272, 164–172. [Google Scholar] [CrossRef]
- Park, S.; Elless, M.P.; Park, J.; Jenkins, A.; Lim, W.; Edgar, C.I.V.; Hirschi, K.D. Sensory analysis of calcium-biofortified lettuce. Plant Biotechnol. J. 2009, 7, 106–117. [Google Scholar] [CrossRef]
- Navarro-León, E.; Ruiz, J.M.; Albacete, A.; Blasco, B. Effect of CAX1a TILLING mutations and calcium concentration on some primary metabolism processes in Brassica rapa plants. J. Plant Physiol. 2019, 237, 51–60. [Google Scholar] [CrossRef]
- Qiao, K.; Wang, F.; Liang, S.; Hu, Z.; Chai, T. Heterologous expression of TuCAX1a and TuCAX1b enhances Ca2+ and Zn2+ translocation in Arabidopsis. Plant Cell Rep. 2019, 38, 597–607. [Google Scholar] [CrossRef]
- Ahmadi, H.; Corso, M.; Weber, M.; Verbruggen, N.; Clemens, S. CAX1 suppresses Cd-induced generation of reactive oxygen species in Arabidopsis halleri. Plant Cell Environ. 2018, 41, 2435–2448. [Google Scholar] [CrossRef]
- Edmond, C.; Shigaki, T.; Ewert, S.; Nelson, M.D.; Connorton, J.M.; Chalova, V.; Noordally, Z.; Pittman, J.K. Comparative analysis of CAX2-like cation transporters indicates functional and regulatory diversity. Biochem. J. 2009, 418, 145–154. [Google Scholar] [CrossRef]
- De Freitas, S.T.; Jiang, C.Z.; Mitcham, E.J. Mechanisms involved in calcium deficiency development in tomato fruit in response to gibberellins. J. Plant Growth Regul. 2012, 31, 221–234. [Google Scholar] [CrossRef]
- Ikeda, H.; Shibuya, T.; Nishiyama, M.; Nakata, Y.; Kanayama, Y. Physiological mechanisms accounting for the lower incidence of blossom-end rot in tomato introgression line IL8-3 fruit. Hort. J. 2017, 86, 327–333. [Google Scholar] [CrossRef]
- Kanayama, Y.; Kochetov, A.V. Abiotic Stress Biology in Horticultural Plants; Springer: New York, NY, USA, 2015. [Google Scholar]
- Saand, M.; Xu, Y.P.; Li, W.; Wang, J.P.; Cai, X.Z. Cyclic nucleotide gated channel gene family in tomato: Genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 2015, 6, 303. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Shirasawa, K.; Ohyama, A.; Fukuoka, H.; Aoki, K.; Rothan, C.; Sato, S.; Isobe, S.; Tabata, S. Genome-wide SNP genotyping to infer the effects on gene functions in tomato. DNA Res. 2013, 20, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Emery, L.; Whelan, S.; Hirschi, K.D.; Pittman, J.K. Protein phylogenetic analysis of Ca2+/cation antiporters and insights into their evolution in plants. Front. Plant Sci. 2012, 3, 1. [Google Scholar] [CrossRef]
- Shigaki, T.; Barkla, B.J.; Miranda-Vergara, M.C.; Zhao, J.; Pantoja, O.; Hirschi, K.D. Identification of a crucial histidine involved in metal transport activity in the Arabidopsis cation/H+ exchanger CAX1. J. Biol. Chem. 2005, 280, 30136–30142. [Google Scholar] [CrossRef]
- Shigaki, T.; Pittman, J.K.; Hirschi, K.D. Manganese specificity determinants in the Arabidopsis metal/H+ antiporter CAX2. J. Biol. Chem. 2003, 278, 6610–6617. [Google Scholar] [CrossRef]
- Conn, S.; Gilliham, M.; Athman, A.; Schreiber, A.W.; Baumann, U.; Moller, I.; Cheng, N.H.; Stancombe, M.; Hirschi, K.D.; Webb, A.; et al. Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 2011, 23, 240–257. [Google Scholar] [CrossRef]
- Mei, H.; Cheng, N.H.; Zhao, J. Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4. New Phytol. 2009, 183, 95–105. [Google Scholar] [CrossRef]
- Shang, X.; Cao, Y.; Ma, L. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, W.; Lin, Y.; Dong, Q.; Peng, X.; Jiang, H.; Zhu, S.; Cheng, B. Different evolutionary patterns among intronless genes in maize genome. Biochem. Biophys. Res. Commun. 2014, 449, 146–150. [Google Scholar] [CrossRef]
- Jain, M.; Khurana, P.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct. Integr. Genom. 2008, 8, 69–78. [Google Scholar] [CrossRef]
- Iwamoto, T.; Nakamura, T.; Pan, Y.; Uehara, A.; Imanaga, I.; Shigekawa, M. Unique topology of the internal repeats in the cardiac Na+/Ca2+ exchanger. FEBS Lett. 1999, 446, 264–268. [Google Scholar] [CrossRef]
- Nishizawa, T.; Kita, S.; Maturana, A.D.D.; Furuya, N.; Hirata, K.; Kasuya, G.; Ogasawara, S.; Dohmae, N.; Iwamoto, T.; Ishitani, R.; et al. Structural basis for the counter-transport mechanism of a H+/Ca2+ exchanger. Science 2013, 341, 168–172. [Google Scholar] [CrossRef]
- Philipson, K.D.; Nicoll, D.A. Sodium-calcium exchange: A molecular perspective. Ann. Rev. Physiol. 2000, 62, 111–133. [Google Scholar] [CrossRef]
- Waight, A.B.; Pedersen, B.; Schlessinger, A.; Bonomi, M.; Chau, B.H.; Roe-Zurz, Z.; Risenmay, A.J.; Sali, A.; Stroud, R.M. Structural basis for alternating access of a eukaryotic calcium/proton exchanger. Nature 2013, 499, 107–110. [Google Scholar] [CrossRef]
- Bickerton, P.D.; Pittman, J.K. Role of cation/proton exchangers in abiotic stress signaling and stress tolerance in plants. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives; Springer: New York, NY, USA, 2015; Volume 1, pp. 95–117. [Google Scholar]
- Kamiya, T.; Akahori, T.; Ashikari, M.; Maeshima, M. Expression of the vacuolar Ca2+/H+ exchanger, OsCAX1a, in rice: Cell and age specificity of expression, and enhancement by Ca2+. Plant Cell Physiol. 2006, 47, 96–106. [Google Scholar] [CrossRef]
- Yadav, A.K.; Shankar, A.; Jha, S.K.; Kanwar, P.; Pandey, A.; Pandey, G.K. A rice tonoplastic calcium exchanger, OsCCX2 mediates Ca2+/cation transport in yeast. Sci. Rep. 2015, 5, 17117. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, M.; Takano, T.; Liu, S. Characterization of an AtCCX5 gene from Arabidopsis thaliana that involves in high-affinity K+ uptake and Na+ transport in yeast. Biochem. Biophys. Res. Commun. 2011, 414, 96–101. [Google Scholar] [CrossRef]
- Nishio, S.; Moriguchi, R.; Ikeda, H.; Takahashi, H.; Takahashi, H.; Fujii, N.; Guilfoyle, T.J.; Kanahama, K.; Kanayama, Y. Expression analysis of the auxin efflux carrier family in tomato fruit development. Planta 2010, 232, 755–764. [Google Scholar] [CrossRef]
- Manohar, M.; Shigaki, T.; Hirschi, K.D. Plant cation/H+ exchangers (CAXs): Biological functions and genetic manipulations. Plant Biol. 2011, 13, 561–569. [Google Scholar] [CrossRef]
- Pittman, J.K. Vacuolar Ca2+ uptake. Cell Calcium 2011, 50, 139–146. [Google Scholar] [CrossRef]
- Hyodo, H.; Terao, A.; Furukawa, J.; Sakamoto, N.; Yurimoto, H.; Satoh, S.; Iwai, H. Tissue specific localization of pectin-Ca2+ cross-linkages and pectin methyl-esterification during fruit ripening in tomato (Solanum lycopersicum). PLoS ONE 2013, 8, e78949. [Google Scholar] [CrossRef]
- De Freitas, S.T.; Padda, M.; Wu, Q.; Park, S.; Mitcham, E.J. Dynamic alternations in cellular and molecular components during blossom-end rot development in tomatoes expressing sCAX1, a constitutively active Ca2+/H+ antiporter from Arabidopsis. Plant Physiol. 2011, 156, 844–855. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, Y.; Di, L.; Wang, G.; Shen, Y. The AtCCX1 transporter mediates salinity tolerance in both Arabidopsis and yeast. Plant Cell Tissue Organ Cult. 2012, 109, 91–99. [Google Scholar] [CrossRef]
- Perez, V.; Chang, E.T. Sodium-to-potassium ratio and blood pressure, hypertension, and related factors. Adv. Nutr. 2014, 5, 712–741. [Google Scholar] [CrossRef]
- Picq, C.; Asplanato, M.; Bernillon, N.; Fabre, C.; Roubeix, M.; Ricort, J.-M. Effects of water soaking and/or sodium polystyrene sulfonate addition on potassium content of foods. Int. J. Food Sci. Nutr. 2014, 65, 673–677. [Google Scholar] [CrossRef]
- Ikeda, H.; Shibuya, T.; Imanishi, S.; Aso, H.; Nishiyama, M.; Kanayama, Y. Dynamic metabolic regulation by a chromosome segment from a wild relative during fruit development in a tomato introgression line, IL8-3. Plant Cell Physiol. 2016, 57, 1257–1270. [Google Scholar] [CrossRef]
- Sagor, G.H.M.; Berberich, T.; Tanaka, S.; Nishiyama, M.; Kanayama, Y.; Kojima, S.; Muramoto, K.; Kusano, T. A novel strategy to produce sweeter tomato fruits with high sugar contents by fruit-specific expression of a single bZIP transcription factor gene. Plant Biotechnol. J. 2016, 14, 1116–1126. [Google Scholar] [CrossRef]
- Beckles, D.M.; Hong, N.; Stamova, L.; Luengwilai, K. Biochemical factors contributing to tomato fruit sugar content: A review. Fruits 2012, 67, 49–64. [Google Scholar] [CrossRef]
- Cho, Y.H.; Yoo, S.D. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genet. 2011, 7, e1001263. [Google Scholar] [CrossRef]
- Nariyama, H.; Sugiyama, Y.; Shibuya, T.; Hayashi, K.; Kanayama, Y. HPY2 gene expression analysis and the role of auxin in early fruit development in tomato. Acta Hortic. 2018, 1206, 247–252. [Google Scholar] [CrossRef]
- Ikeda, H.; Hiraga, M.; Shirasawa, K.; Nishiyama, M.; Kanahama, K.; Kanayama, Y. Analysis of a tomato introgression line, IL8-3, with increased Brix content. Sci. Hortic. 2013, 153, 103–108. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef]
- Reuscher, S.; Akiyama, M.; Yasuda, T.; Makino, H.; Aoki, K.; Shibata, D.; Shiratake, K. The sugar transporter inventory of tomato: Genome-wide identification and expression analysis. Plant Cell Physiol. 2014, 55, 1123–1141. [Google Scholar] [CrossRef]
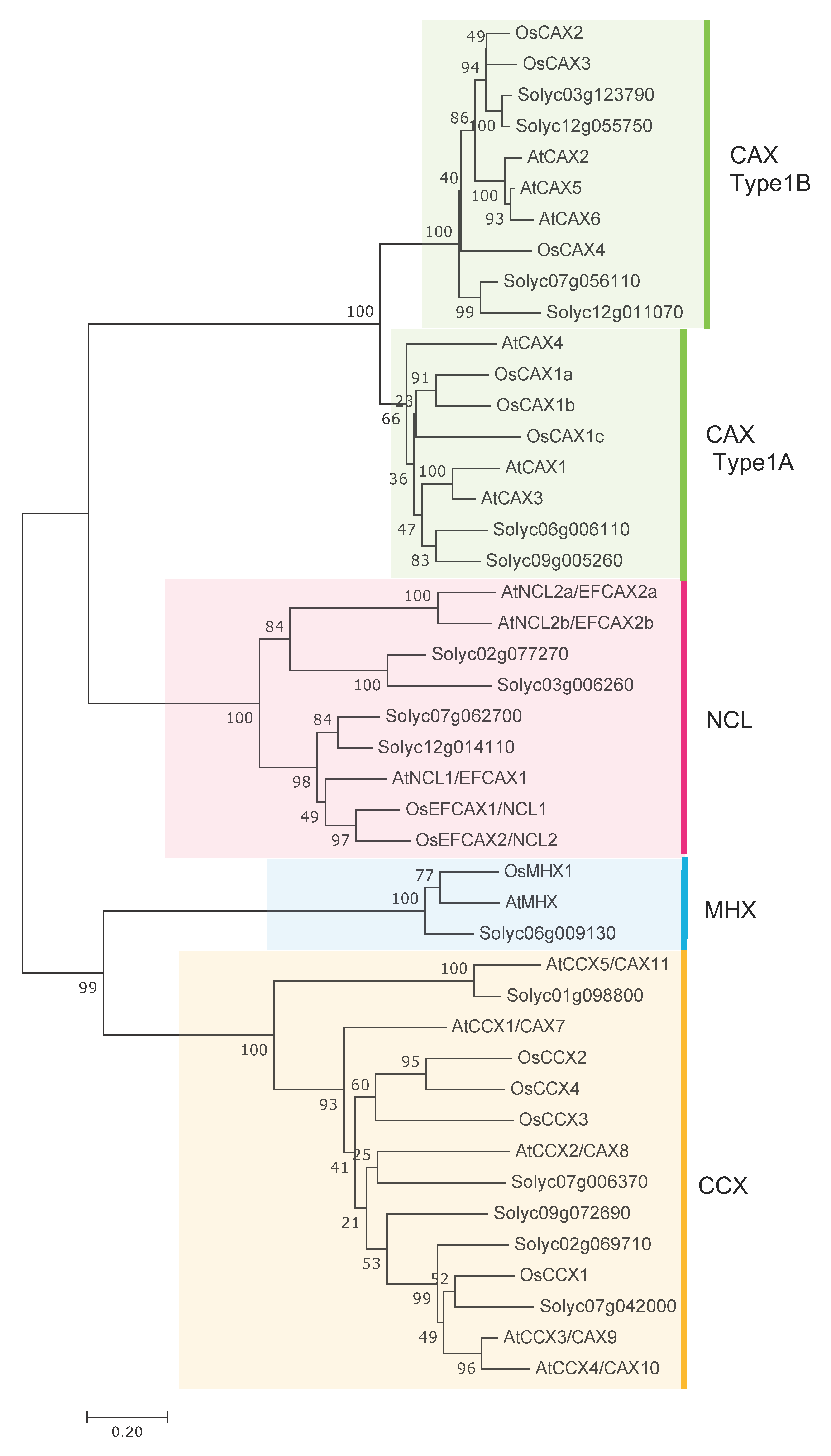
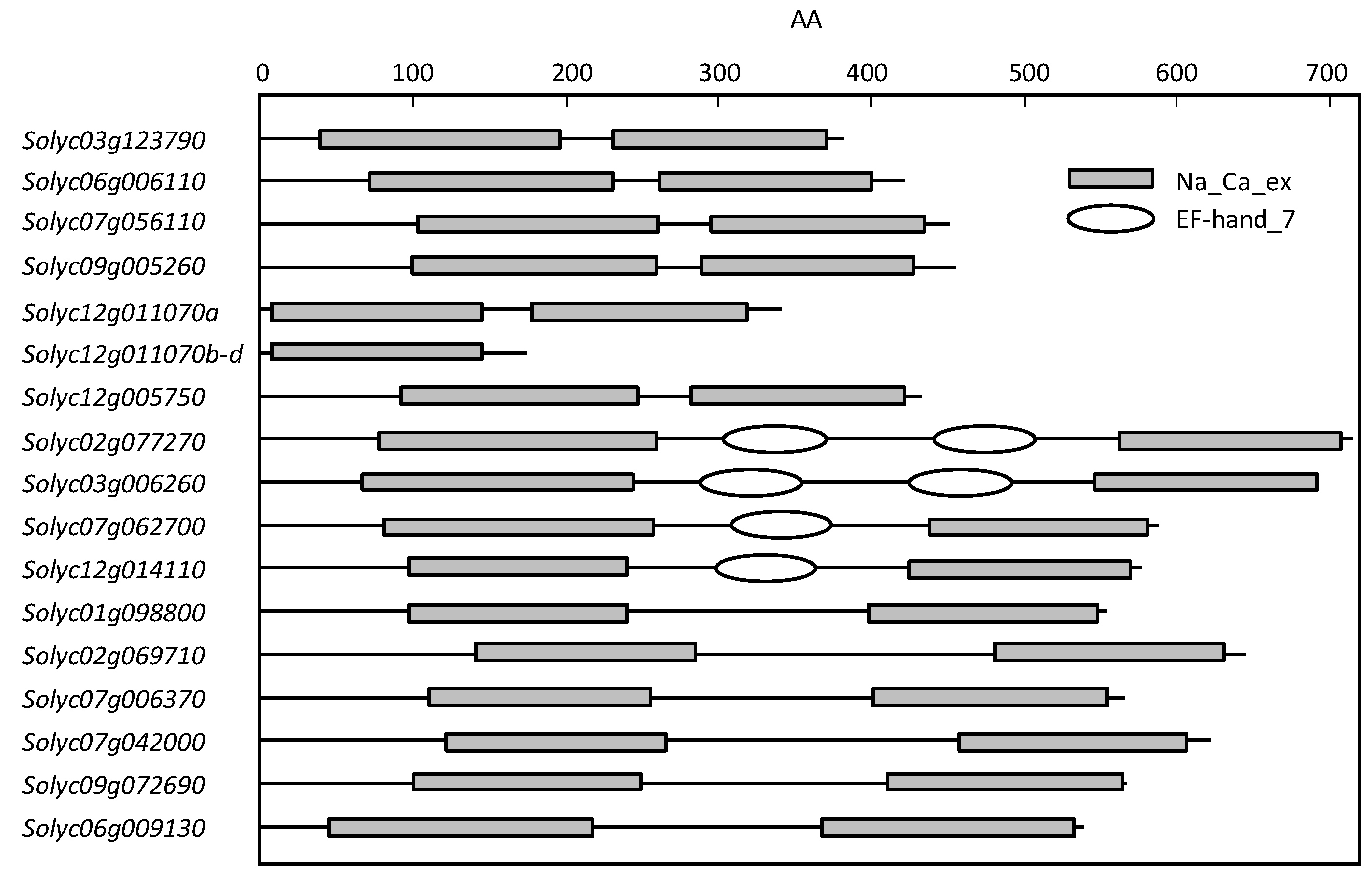
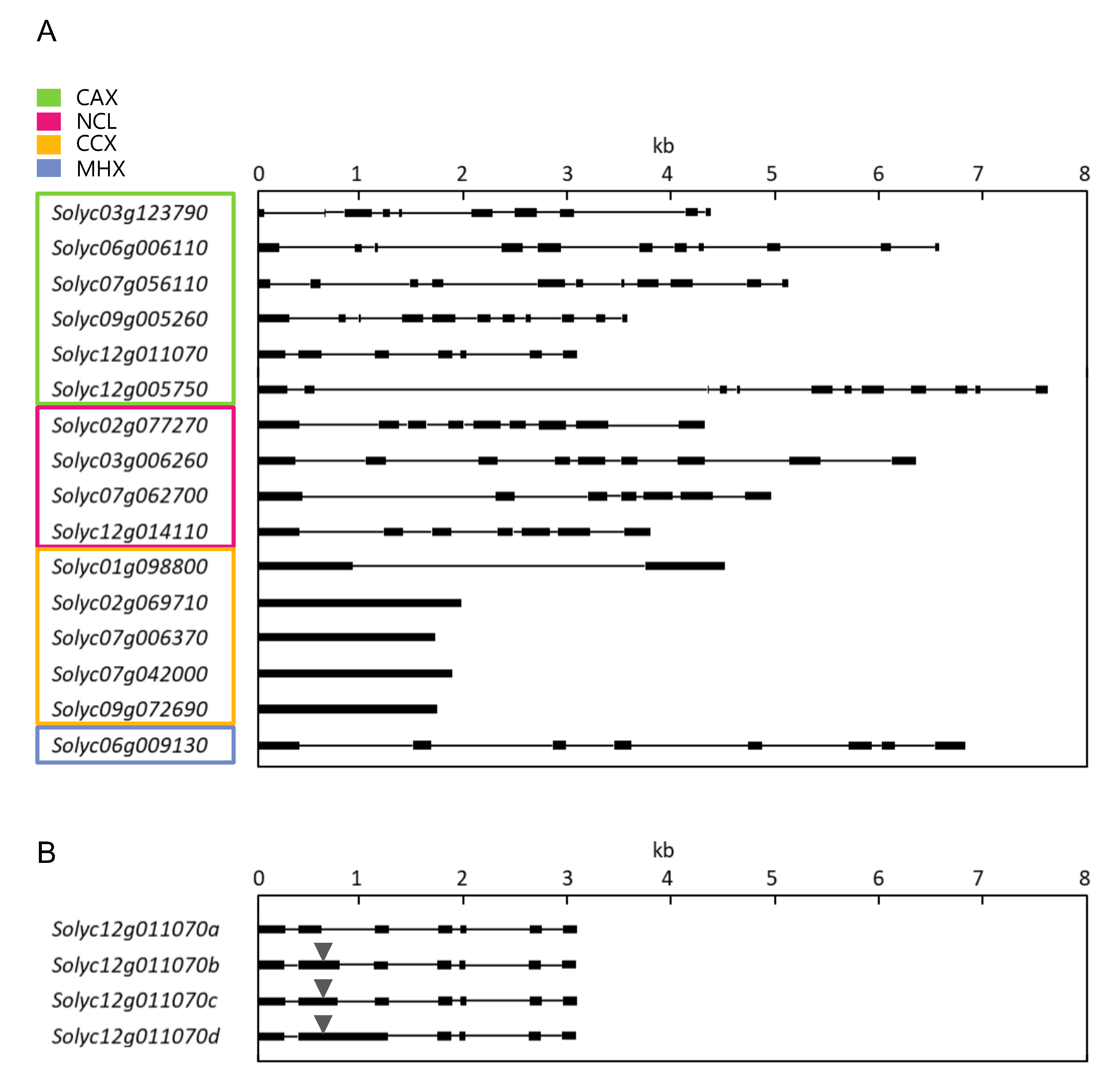
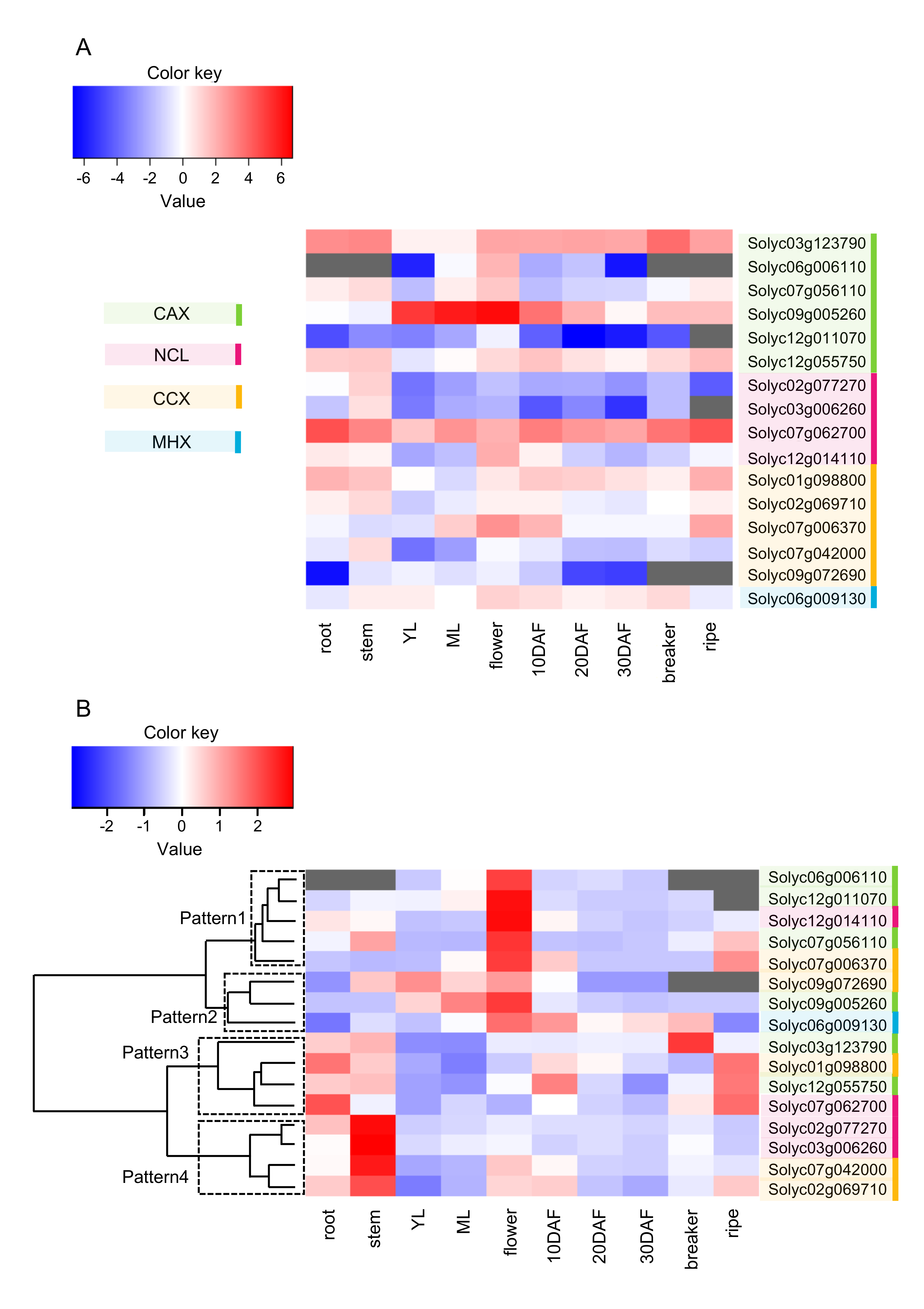
| Family | Locus a | Protein Size b | TM c | Domain d | α1-Repeat c | α2-Repeat c | Localiza-tion e |
|---|---|---|---|---|---|---|---|
| CAX | Solyc03g123790 | 383 | 11 | Na_Ca_ex | TM3-4 | TM8-9 | vacuole |
| Solyc06g006110 | 423 | 11 | Na_Ca_ex | TM3-4 | TM8-9 | vacuole | |
| Solyc07g056110 | 452 | 10 | Na_Ca_ex | TM2-3 | TM7-8 | vacuole | |
| Solyc09g005260 | 456 | 10 | Na_Ca_ex | TM2-3 | TM7-8 | vacuole | |
| Solyc12g011070a | 342 | 9 | Na_Ca_ex | TM1-2 | TM6-7 | vacuole | |
| Solyc12g011070b | 175 | 5 | Na_Ca_ex | TM1-2 | |||
| Solyc12g011070c | 175 | 5 | Na_Ca_ex | TM1-2 | |||
| Solyc12g011070d | 175 | 5 | Na_Ca_ex | TM1-2 | |||
| Solyc12g055750 | 434 | 11 | Na_Ca_ex | TM3-4 | TM8-9 | vacuole | |
| NCL | Solyc02g077270 | 716 | 10 | Na_Ca_ex, EF-hand | TM7-8 | ||
| Solyc03g006260 | 694 | 10 | Na_Ca_ex, EF-hand | TM7-8 | |||
| Solyc07g062700 | 589 | 11 | Na_Ca_ex, EF-hand | TM8-9 | |||
| Solyc12g014110 | 578 | 10 | Na_Ca_ex, EF-hand | TM7-8 | |||
| CCX | Solyc01g098800 | 555 | 11 | Na_Ca_ex | TM2-3 f | TM9-10 | plasma membrane |
| Solyc02g069710 | 646 | 11 | Na_Ca_ex | TM2-3 f | TM9-10 | vacuole | |
| Solyc07g006370 | 567 | 13 | Na_Ca_ex | TM3-4 | TM10-11 | vacuole | |
| Solyc07g042000 | 623 | 13 | Na_Ca_ex | TM3-4 | TM10-11 | vacuole | |
| Solyc09g072690 | 568 | 12 | Na_Ca_ex | TM2-3 | TM9-10 | vacuole | |
| MHX | Solyc06g009130 | 540 | 11 | Na_Ca_ex | TM2-3 | TM7-8 | vacuole |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amagaya, K.; Shibuya, T.; Nishiyama, M.; Kato, K.; Kanayama, Y. Characterization and Expression Analysis of the Ca2+/Cation Antiporter Gene Family in Tomatoes. Plants 2020, 9, 25. https://doi.org/10.3390/plants9010025
Amagaya K, Shibuya T, Nishiyama M, Kato K, Kanayama Y. Characterization and Expression Analysis of the Ca2+/Cation Antiporter Gene Family in Tomatoes. Plants. 2020; 9(1):25. https://doi.org/10.3390/plants9010025
Chicago/Turabian StyleAmagaya, Kayoko, Tomoki Shibuya, Manabu Nishiyama, Kazuhisa Kato, and Yoshinori Kanayama. 2020. "Characterization and Expression Analysis of the Ca2+/Cation Antiporter Gene Family in Tomatoes" Plants 9, no. 1: 25. https://doi.org/10.3390/plants9010025
APA StyleAmagaya, K., Shibuya, T., Nishiyama, M., Kato, K., & Kanayama, Y. (2020). Characterization and Expression Analysis of the Ca2+/Cation Antiporter Gene Family in Tomatoes. Plants, 9(1), 25. https://doi.org/10.3390/plants9010025





