Transcriptome Analysis of Young Ovaries Reveals Candidate Genes Involved in Gamete Formation in Lantana camara
Abstract
:1. Introduction
2. Results
2.1. Sample Collection, Illumina Sequencing, and De Novo Assembly
2.2. Functional Annotation of the Unique Transcripts
2.3. Gene Expression in the Ovaries of GGO and LWL Lantana
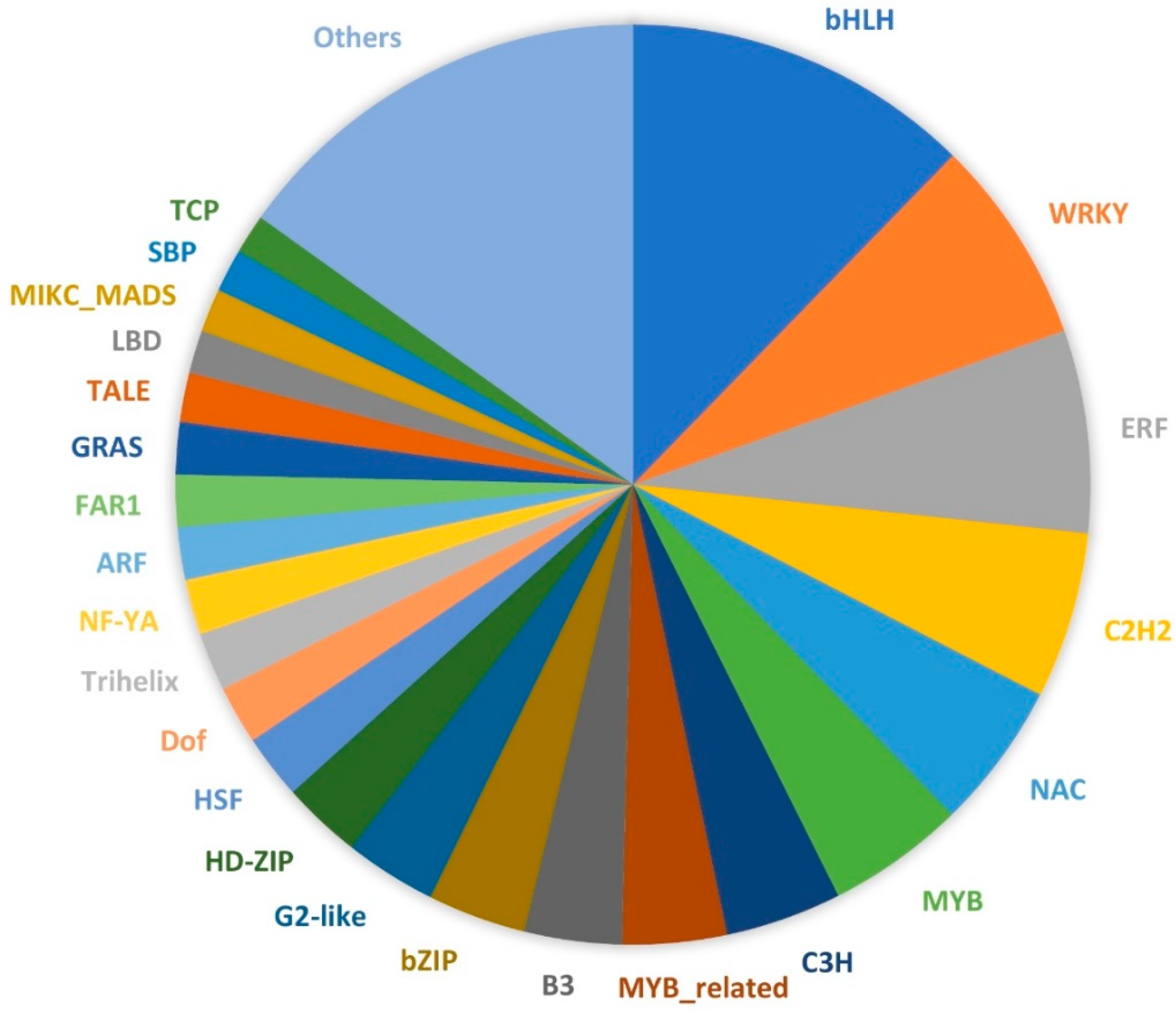
2.4. Lantana Gene Families
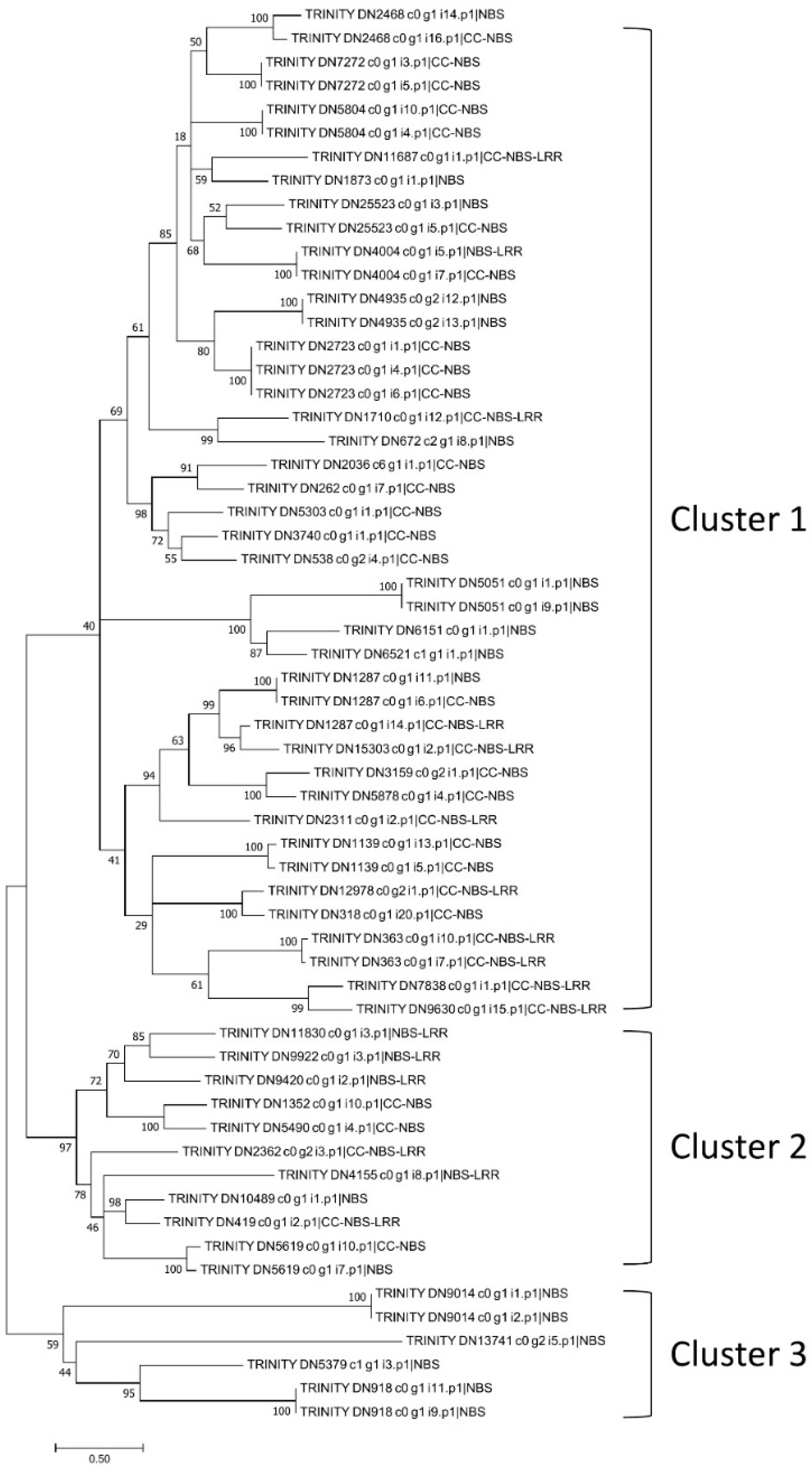
2.5. Identification and Classification of NBS Genes
2.6. Discovery of Simple Sequence Repeats, Single Nucleotide Polymorphisms, and Insertion or Deletions
3. Discussion
4. Materials and Methods
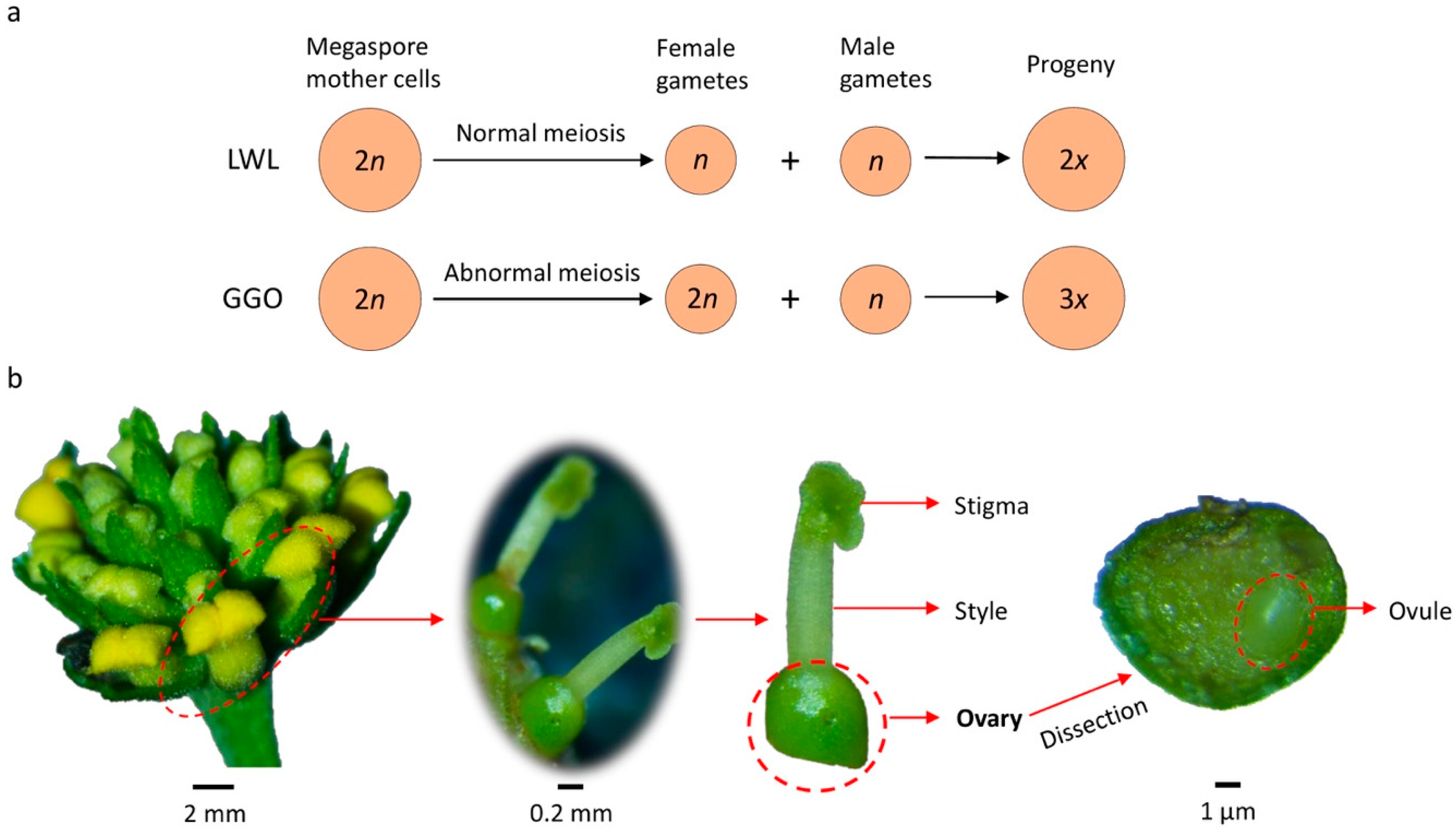
4.1. Plant Materials, RNA Extraction, and Sequencing
4.2. Sequence Trimming, De Novo Assembly, and Clustering
4.3. Function Annotation
4.4. Mapping and Gene Expression
4.5. Lantana Gene Families Involved in Unreduced Gamete Production, Drought Tolerance, Salt Tolerance, and Allelopathy
4.6. Identification of NBS Genes and Phylogenetic Analysis
4.7. Identification and Annotation of SSR, SNP, and Indel Variants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wirth, F.F.; Davis, K.J.; Wilson, S.B. Florida nursery sales and economic impacts of 14 potentially invasive landscape plant species. J. Environ. Hortic. 2004, 22, 12–16. [Google Scholar]
- Sharma, G.R.; Raghubanshi, A.S.; Singh, J.S. Lantana invasion: An overview. Weed Biol. Manag. 2005, 5, 157–165. [Google Scholar] [CrossRef]
- Gunasekara, C.J.; Ranwala, S.M.W. Growth responses of lantana (Lantana camara L.) varieties to varying water availability and light conditions. J. Natl. Sci. Found. Sri Lanka 2018, 46, 69–79. [Google Scholar] [CrossRef]
- Munir, A.A. A taxonomic review of Lantana camara L. and L. montevidensis (Spreng.) Briq. (Verbenaceae) in Australia. J. Adelaide Bot. Gard. 1996, 17, 1–27. [Google Scholar]
- Sanders, R.W. The genera of Verbenaceae in the southeastern United States. Harv. Pap. Bot. 2001, 5, 303–358. [Google Scholar]
- Howard, R.A. A checklist of cultivar names used in the genus Lantana. Arnoldia 1969, 29, 73–109. [Google Scholar]
- Ray, A.; Quader, S. Genetic diversity and population structure of Lantana camara in India indicates multiple introductions and gene flow. Plant Biol. 2014, 16, 651–658. [Google Scholar] [CrossRef]
- Czarnecki, D.M., II; Deng, Z.N. Occurrence of unreduced female gametes leads to sexual polyploidization in Lantana. J. Am. Soc. Hortic. Sci. 2009, 134, 560–566. [Google Scholar] [CrossRef]
- Czarnecki, D.M., II; Hershberger, A.J.; Robacker, C.D.; Clark, D.G.; Deng, Z. Ploidy levels and pollen stainability of Lantana camara cultivars and breeding lines. HortScience 2014, 49, 1271–1276. [Google Scholar] [CrossRef]
- Czarnecki, D.M., II; Wilson, S.B.; Knox, G.W.; Freyre, R.; Deng, Z.N. UF-T3 and UF-T4: Two sterile Lantana camara cultivars. HortScience 2012, 47, 132–137. [Google Scholar] [CrossRef]
- Deng, Z.; Wilson, S.B.; Ying, X.; Czarnecki, D.M., II. Infertile Lantana camara cultivars UF-1011-2 and UF-1013A-2A. HortScience 2017, 52, 652–657. [Google Scholar] [CrossRef]
- Bretagnolle, F.; Thompson, J.D. Gametes with the somatic chromosome-number—Mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 1995, 129, 1–22. [Google Scholar] [CrossRef]
- Brownfield, L.; Kohler, C. Unreduced gamete formation in plants: Mechanisms and prospects. J. Exp. Bot. 2011, 62, 1659–1668. [Google Scholar] [CrossRef]
- Ramanna, M.S.; Jacobsen, E. Relevance of sexual polyploidization for crop improvement—A review. Euphytica 2003, 133, 3–18. [Google Scholar] [CrossRef]
- Consiglio, F.; Carputo, D.; Monti, L.; Conicella, C. Exploitation of genes affecting meiotic non-reduction and nuclear restitution: Arabidopsis as a model? Sex. Plant Reprod. 2004, 17, 97–105. [Google Scholar] [CrossRef]
- De Storme, N.; Geelen, D. Sexual polyploidization in plants cytological mechanisms and molecular regulation. New Phytol. 2013, 198, 670–684. [Google Scholar] [CrossRef]
- Ravi, M.; Marimuthu, M.P.A.; Siddiqi, I. Gamete formation without meiosis in Arabidopsis. Nature 2008, 451, 1121–1124. [Google Scholar] [CrossRef]
- d’Erfurth, I.; Cromer, L.; Jolivet, S.; Girard, C.; Horlow, C.; Sun, Y.J.; To, J.P.C.; Berchowitz, L.E.; Copenhaver, G.P.; Mercier, R. The CYCLIN-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with osd1 for the prophase to first meiotic division transition. PLoS Genet. 2010, 6, e1000989. [Google Scholar] [CrossRef]
- d’Erfurth, I.; Jolivet, S.; Froger, N.; Catrice, O.; Novatchkova, M.; Mercier, R. Turning meiosis into mitosis. PLoS Biol. 2009, 7, e1000124. [Google Scholar] [CrossRef]
- d’Erfurth, I.; Jolivet, S.; Froger, N.; Catrice, O.; Novatchkova, M.; Simon, M.; Jenczewski, E.; Mercier, R. Mutations in AtPS1 (Arabidopsis thaliana parallel spindle 1) lead to the production of diploid pollen grains. PLoS Genet. 2008, 4, e1000274. [Google Scholar] [CrossRef]
- Yang, C.Y.; Spielman, M.; Coles, J.P.; Li, Y.; Ghelani, S.; Bourdon, V.; Brown, R.C.; Lemmon, B.E.; Scott, R.J.; Dickinson, H.G. Tetraspore encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J. 2003, 34, 229–240. [Google Scholar] [CrossRef]
- Sharma, O.P.; Makkar, H.P.S.; Dawra, R.K. A review of the noxious plant Lantana camara. Toxicon 1988, 26, 975–987. [Google Scholar] [CrossRef]
- Ramaswami, G.; Sukumar, R. Long-term environmental correlates of invasion by Lantana camara (Verbenaceae) in a seasonally dry tropical forest. PLoS ONE 2013, 8, e76995. [Google Scholar] [CrossRef]
- Wu, L.; Guo, X.; Hunter, K.; Zagory, E.; Waters, R.; Brown, J. Studies of salt tolerance of landscape plant species and California native grasses for recycled water irrigation. Slosson Rep. 2001, 1–14. Available online: https://pdfs.semanticscholar.org/8c08/e9051bd0cc67c6c8c6a9fff12fa346db5f4a.pdf (accessed on 10 April 2019).
- Gentle, C.; Duggin, J. Allelopathy as a competitive strategy in persistent thickets of Lantana camara L. In three Australian forest communities. Plant Ecol. 1997, 132, 85–95. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef]
- Czarnecki, D.M. Genetic Sterilization and Reproductive Biology of Lantana camara. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2011. [Google Scholar]
- Vriezen, W.H.; Feron, R.; Maretto, F.; Keijman, J.; Mariani, C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 2008, 177, 60–76. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, M.; Qiu, Z.; Wang, K.; Du, Y.; Gu, L.; Cui, X. Spatiotemporal transcriptome provides insights into early fruit development of tomato (Solanum lycopersicum). Sci. Rep. 2016, 6, 23173. [Google Scholar] [CrossRef]
- Pattison, R.J.; Csukasi, F.; Zheng, Y.; Fei, Z.; van der Knaap, E.; Catalá, C. Comprehensive tissue-specific transcriptome analysis reveals distinct regulatory programs during early tomato fruit development. Plant Physiol. 2015, 168, 1684–1701. [Google Scholar] [CrossRef]
- Uchiumi, T.; Okamoto, T. Rice fruit development is associated with an increased IAA content in pollinated ovaries. Planta 2010, 232, 579–592. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, P.; Xia, H.; Zhao, C.; Hou, L.; Li, C.; Gao, C.; Zhao, S.; Wang, X. Comparative transcriptome analysis of basal and zygote-located tip regions of peanut ovaries provides insight into the mechanism of light regulation in peanut embryo and pod development. BMC Genom. 2016, 17, 606. [Google Scholar]
- Mousavi, S.; Alisoltani, A.; Shiran, B.; Fallahi, H.; Ebrahimie, E.; Imani, A.; Houshmand, S. De novo transcriptome assembly and comparative analysis of differentially expressed genes in Prunus dulcis Mill. in response to freezing stress. PLoS ONE 2014, 9, e104541. [Google Scholar] [CrossRef]
- Xu, L.; Yang, P.; Yuan, S.; Feng, Y.; Xu, H.; Cao, Y.; Ming, J. Transcriptome analysis identifies key candidate genes mediating purple ovary coloration in Asiatic hybrid lilies. Int. J. Mol. Sci. 2016, 17, 1881. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. Crabs claw and spatula genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 2002, 163, 17–41. [Google Scholar] [CrossRef]
- Gremski, K.; Ditta, G.; Yanofsky, M.F. The hecate genes regulate female reproductive tract development in Arabidopsis thaliana. Development 2007, 134, 3593–3601. [Google Scholar] [CrossRef]
- Lee, C.Y.; Conrad, M.N.; Dresser, M.E. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 2012, 8, e1002730. [Google Scholar] [CrossRef]
- Bass, H.W.; Marshall, W.F.; Sedat, J.W.; Agard, D.A.; Cande, W.Z. Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 1997, 137, 5–18. [Google Scholar] [CrossRef]
- Naranjo, T.; Valenzuela, N.T.; Perera, E. Chiasma frequency is region specific and chromosome conformation dependent in a rye chromosome added to wheat. Cytogenet. Genome Res. 2010, 129, 133–142. [Google Scholar] [CrossRef]
- Wang, Y.S.; Zhou, L.J.; Li, D.Z.; Dai, L.Y.; Lawton-Rauh, A.; Srimani, P.K.; Duan, Y.P.; Luo, F. Genome-wide comparative analysis reveals similar types of NBS genes in hybrid Citrus sinensis genome and original Citrus clementine genome and provides new insights into non-TIR NBS genes. PLoS ONE 2015, 10, e0121893. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef]
- Hammer, R.L. The lantana mess—A critical look at the genus in Florida. Palmetto 2004, 23, 21–23. [Google Scholar]
- Cao, Z.; Deng, Z. De novo assembly, annotation, and characterization of root transcriptomes of three caladium cultivars with a focus on necrotrophic pathogen resistance/defense-related genes. Int. J. Mol. Sci. 2017, 18, 712. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 7 March 2019).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Conesa, A.; Stefan, G.; Juan Miguel, G.-G.; Terol, J.; Manuel, T.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Feng, J.; Meyer, C.A.; Wang, Q.; Liu, J.S.; Shirley Liu, X.; Zhang, Y. Gfold: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012, 28, 2782–2788. [Google Scholar] [CrossRef]
- Kreiner, J.M.; Kron, P.; Husband, B.C. Evolutionary dynamics of unreduced gametes. Trends Genet. 2017, 33, 583–593. [Google Scholar] [CrossRef]
- Loginova, D.; Silkova, O. Mechanisms of unreduced gamete formation in flowering plants. Russ. J. Genet. 2017, 53, 741–756. [Google Scholar] [CrossRef]
- Fischer, S.; Brunk, B.P.; Chen, F.; Gao, X.; Harb, O.S.; Iodice, J.B.; Shanmugam, D.; Roos, D.S.; Stoeckert, C.J., Jr. Using orthoMCL to assign proteins to orthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. 2011, 35, 6.12.1–6.12.19. [Google Scholar]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinf. 2008, 9, 253. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]


| Trinity | CD-HIT-EST | TransDecoder (Full-Length) | |
|---|---|---|---|
| No. of sequences | 112,505 | 90,641 | 29,383 |
| Total length (bp) | 126,460,570 | 94,042,530 | 57,219,462 |
| Average length (bp) | 1124 | 1037.53 | 1947.40 |
| N50 (bp) | 1787 | 1692 | 2206 |
| Minimum length (bp) | 201 | 201 | 327 |
| Maximum length (bp) | 12,218 | 12,218 | 12,083 |
| Biological Process | Gene Ontology (Including Child Terms) | No. of Transcripts |
|---|---|---|
| Cell cycle | GO:0007049 | 57 |
| Regulation of cell cycle | GO:0051726 | 25 |
| Positive regulation of cell cycle | GO:0045787 | 16 |
| Negative regulation of cell cycle | GO:0045786 | 5 |
| Meiotic cell cycle | GO:0051321 | 22 |
| Cytokinesis | GO:0000910 | 53 |
| Chromosome organization | GO:0051276 | 34 |
| Chromosome segregation | GO:0007059 | 18 |
| Chromosome organization involved in meiotic cell cycle | GO:0070192 | 16 |
| Gamete generation | GO:0007276 | 8 |
| Total (unique) | 214 |
| NBS Gene Class | Number of Genes |
|---|---|
| CC-NBS-LRR | 12 |
| CC-NBS | 29 |
| NBS-LRR | 7 |
| NBS | 43 |
| Total | 91 |
| SSRs | Repeat Motif Length | Total | ||||
|---|---|---|---|---|---|---|
| Di- | Tri- | Tetra- | Penta- | Hexa- | ||
| SSRs identified in transcript assembly | 7886 | 2118 | 101 | 30 | 55 | 10,190 |
| SSRs designed with primers | 7325 | 2007 | 97 | 30 | 54 | 9513 |
| SNP Type | SNP Number | INDEL Number | GGO Genotype | LWL Genotype |
|---|---|---|---|---|
| Homozygous | 11,865 | 1014 | 0/0 | 1/1 |
| 13,115 | 1070 | 1/1 | 0/0 | |
| Heterozygous | 54,628 | 3052 | 0/0 | 0/1 |
| 74,210 | 4194 | 0/1 | 0/0 | |
| 6494 | 377 | 0/1 | 1/1 | |
| 4917 | 277 | 1/1 | 0/1 | |
| Total | 165,229 | 9984 |
| Variant Effects | Count | Percentage (%) | Variant Type |
|---|---|---|---|
| Synonymous variant | 51,878 | 33.67 | SNP |
| Missense variant | 45,326 | 29.42 | SNP |
| 3′ UTR variant | 31,145 | 20.22 | SNP |
| 5′ UTR variant | 21,390 | 13.88 | SNP |
| 5′ UTR premature start codon gain variant | 3568 | 2.32 | SNP |
| Stop gained | 529 | 0.34 | SNP |
| Stop lost | 119 | 0.08 | SNP |
| Stop retained variant | 62 | 0.04 | SNP |
| Start lost | 42 | 0.03 | SNP |
| Initiator codon variant | 9 | 0.01 | SNP |
| 3′ UTR variant | 3497 | 39.34 | Indel |
| 5′ UTR variant | 2720 | 30.60 | Indel |
| Frameshift variant | 1029 | 11.58 | Indel |
| Conservative inframe deletion | 516 | 5.80 | Indel |
| Conservative inframe insertion | 471 | 5.30 | Indel |
| Disruptive inframe deletion | 309 | 3.48 | Indel |
| Disruptive inframe insertion | 245 | 2.76 | Indel |
| Stop gained | 49 | 0.55 | Indel |
| Stop lost | 30 | 0.34 | Indel |
| Start lost | 23 | 0.26 | Indel |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Z.; Bhattarai, K.; Parajuli, S.; Cao, Z.; Deng, Z. Transcriptome Analysis of Young Ovaries Reveals Candidate Genes Involved in Gamete Formation in Lantana camara. Plants 2019, 8, 263. https://doi.org/10.3390/plants8080263
Peng Z, Bhattarai K, Parajuli S, Cao Z, Deng Z. Transcriptome Analysis of Young Ovaries Reveals Candidate Genes Involved in Gamete Formation in Lantana camara. Plants. 2019; 8(8):263. https://doi.org/10.3390/plants8080263
Chicago/Turabian StylePeng, Ze, Krishna Bhattarai, Saroj Parajuli, Zhe Cao, and Zhanao Deng. 2019. "Transcriptome Analysis of Young Ovaries Reveals Candidate Genes Involved in Gamete Formation in Lantana camara" Plants 8, no. 8: 263. https://doi.org/10.3390/plants8080263
APA StylePeng, Z., Bhattarai, K., Parajuli, S., Cao, Z., & Deng, Z. (2019). Transcriptome Analysis of Young Ovaries Reveals Candidate Genes Involved in Gamete Formation in Lantana camara. Plants, 8(8), 263. https://doi.org/10.3390/plants8080263






