Genome-Wide Identification and Characterization of JAZ Protein Family in Two Petunia Progenitors
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of JAZ Family Proteins in P. axillaris and P. inflata
2.2. Homologous Sequence Alignment and Phylogenetic Analysis
2.3. Analysis of Conserved Motif and Gene Structure
2.4. Analysis of Cis-Acting Regulatory Elements
2.5. Calculation of Ka/Ks Ratios
2.6. Plant Materials, MeJA Treatment, RNA Isolation, and Gene Expression Analysis
2.7. Subcellular Localization Assays
2.8. Y2H Assays
3. Results
3.1. Identification of JAZ Family Proteins in P. axillaris and P. inflata
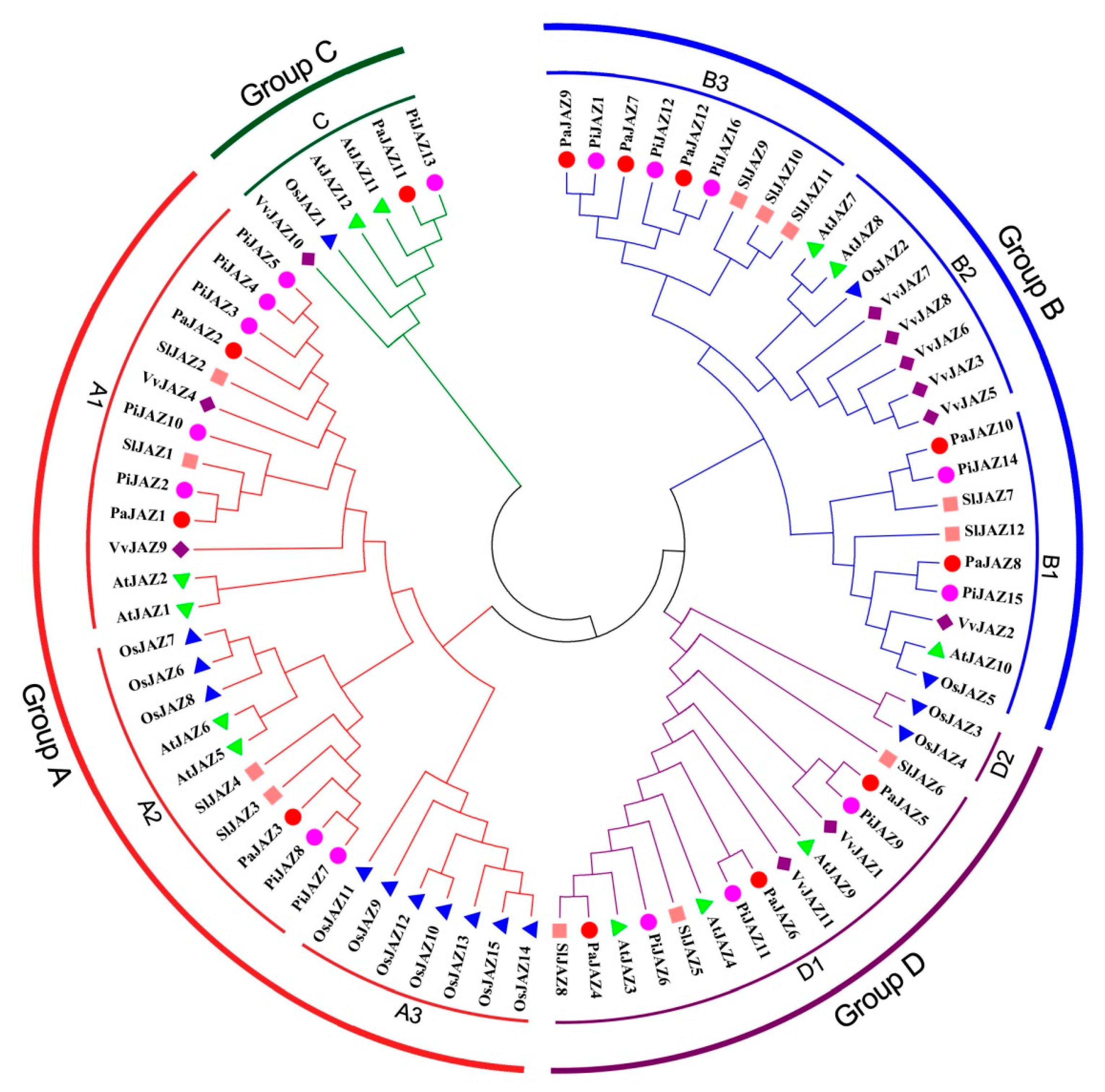
3.2. Homologous Sequences Alignment and Phylogenetic Analysis
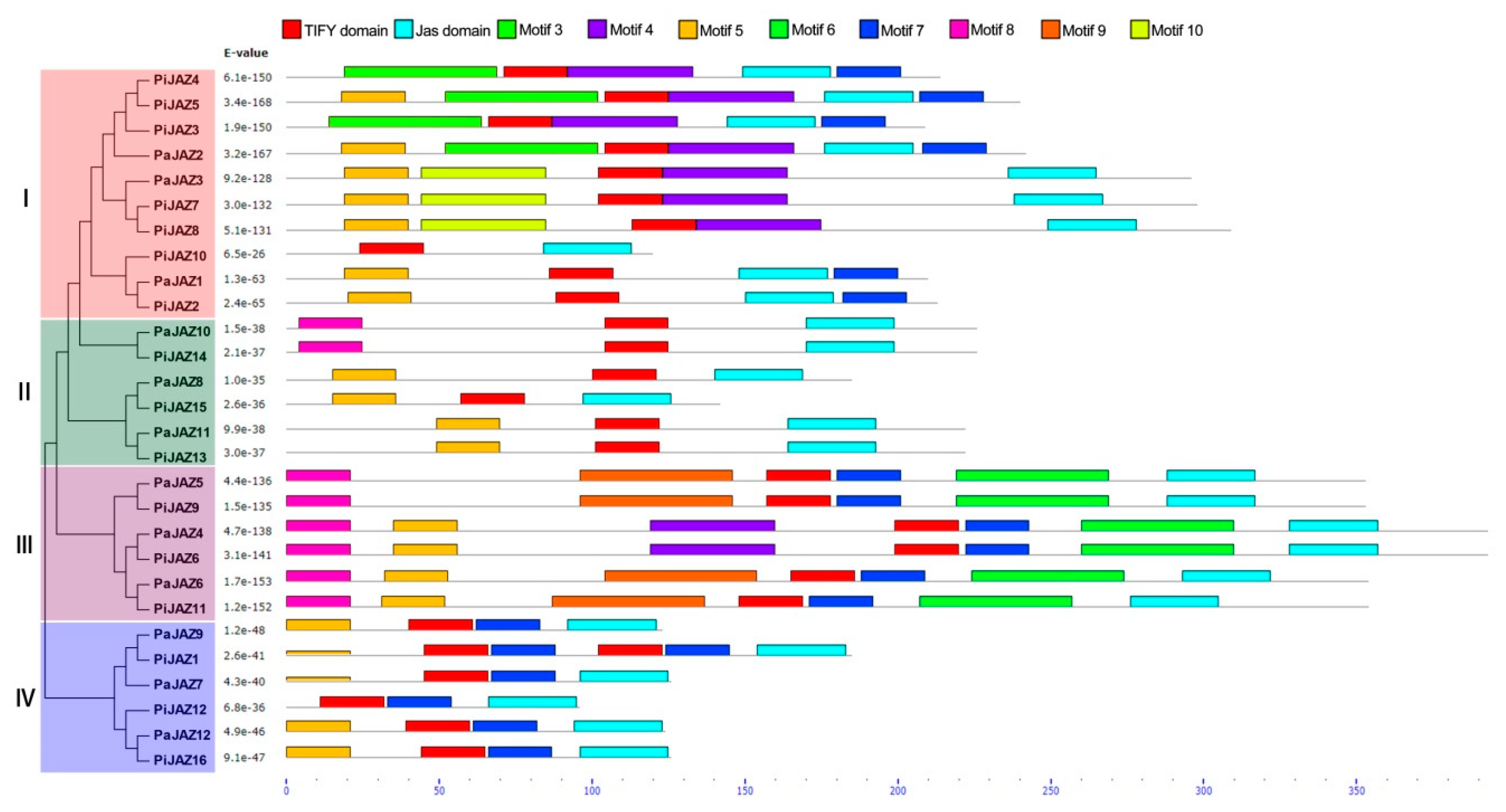
3.3. Conserved Motif and Gene Structure Analysis
3.4. Analysis of Cis-Acting Regulatory Elements
3.5. Calculation of Ka/Ks Ratios
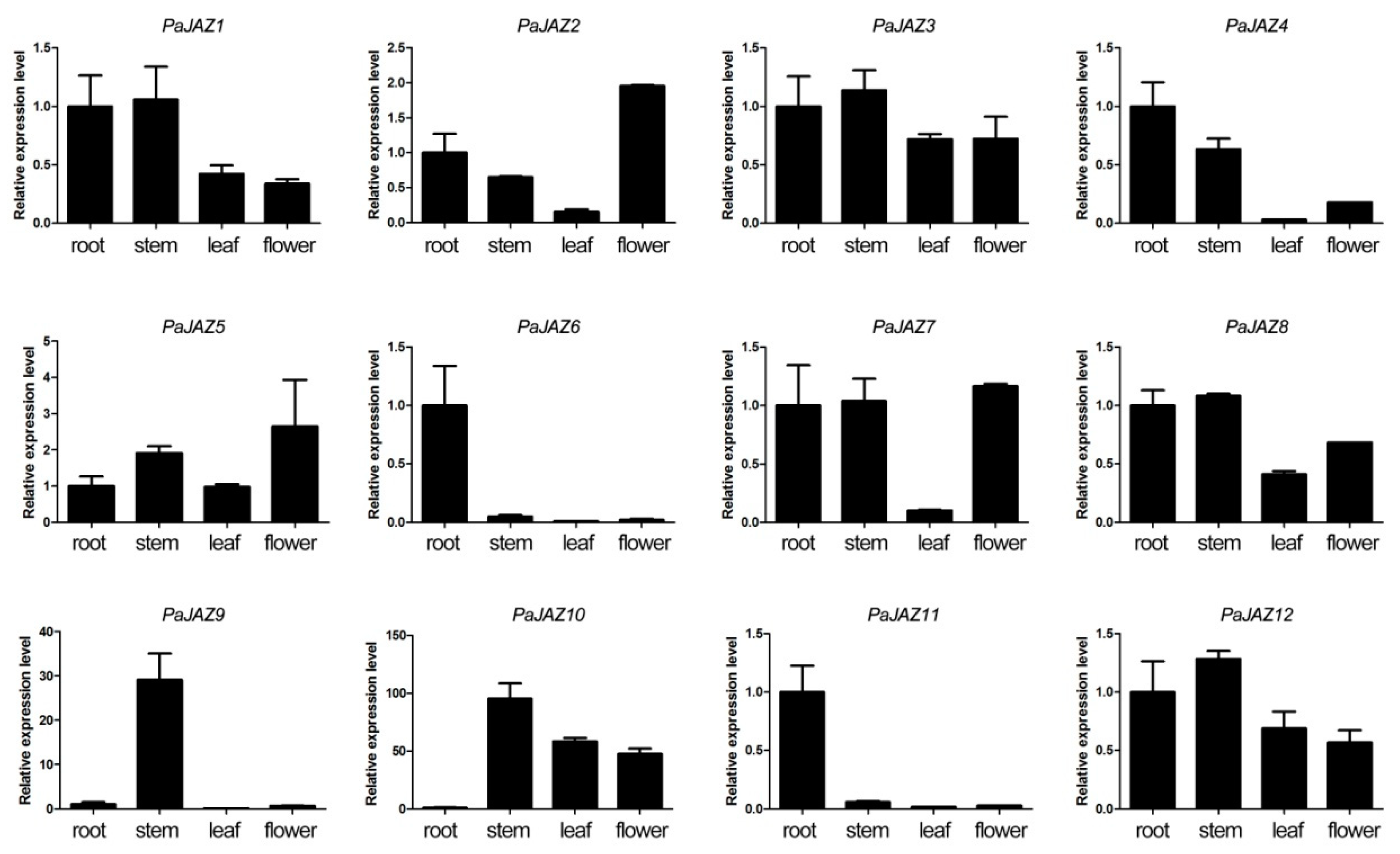
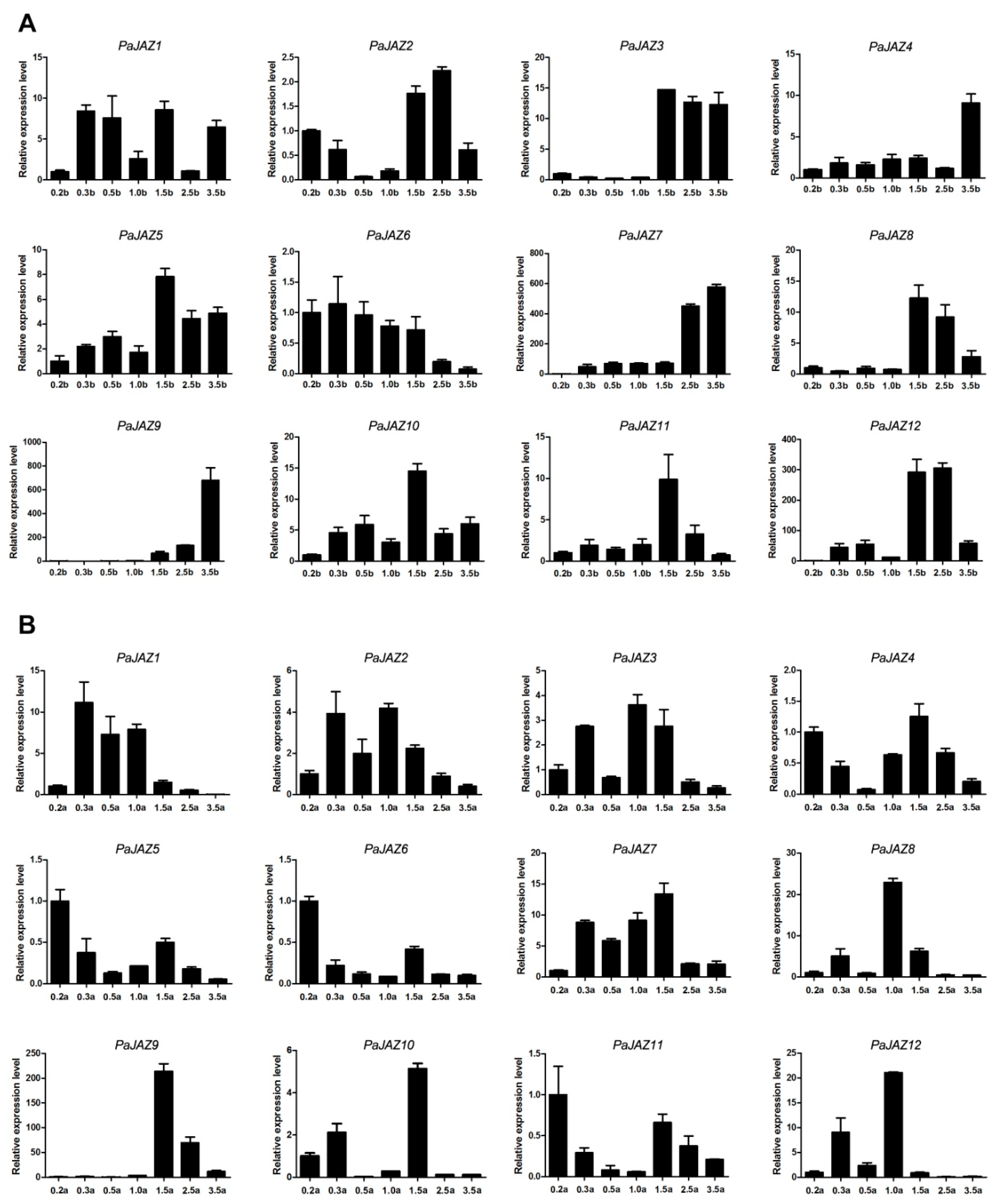
3.6. The Expression Analysis of PaJAZ Genes
3.7. Subcellular Localization of PaJAZ Proteins
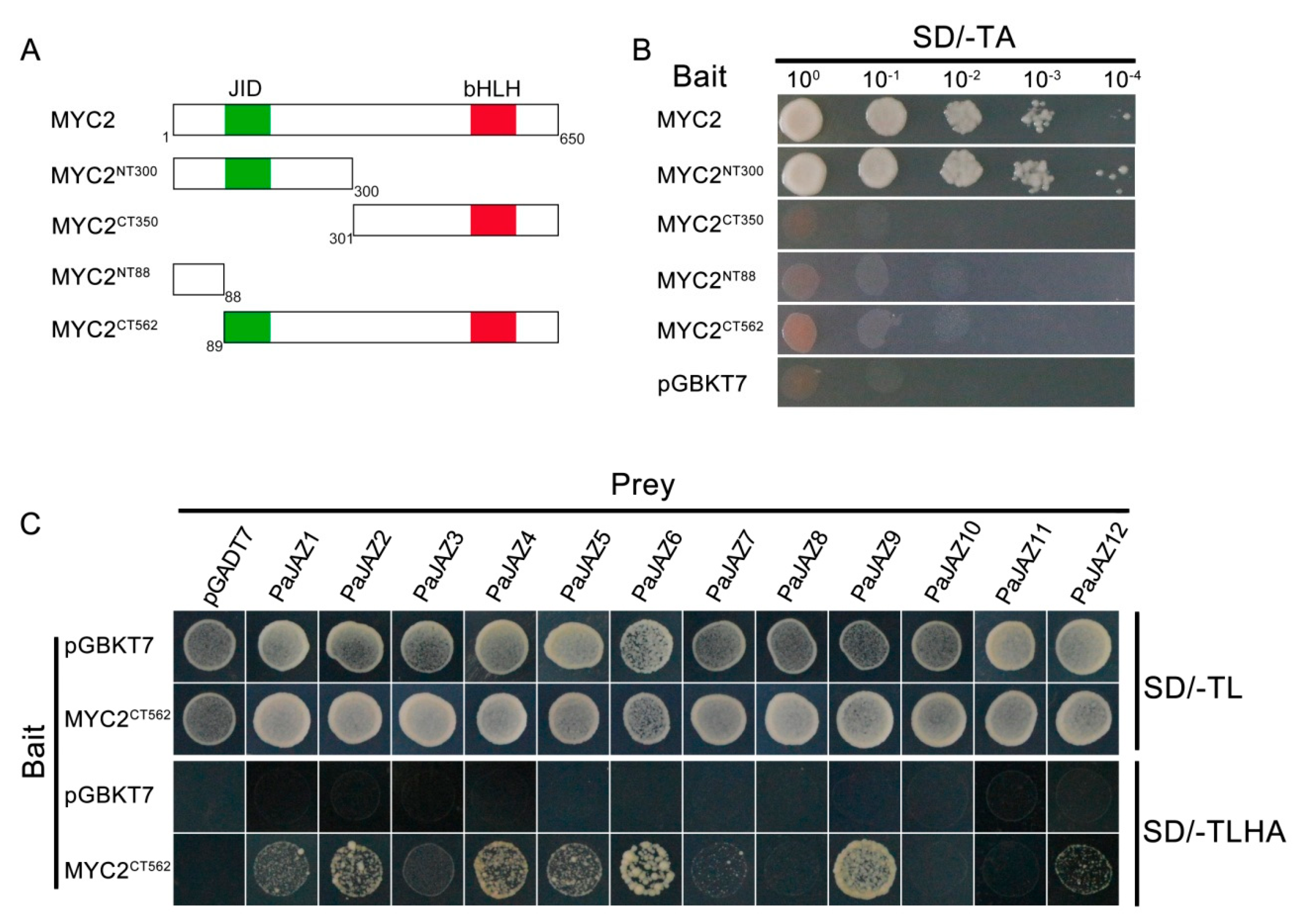
3.8. Interacion Between PaJAZ Proteins and Transcription Factor MYC2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Major, I.T.; Howe, G.A. Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 2018, 44, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreno, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Monte, I.; Franco-Zorrilla, J.M.; Garcia-Casado, G.; Zamarreno, A.M.; Garcia-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant 2019, 12, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Yoshida, Y. Evolutionary origin of JAZ proteins and jasmonate signaling. Mol. Plant 2019, 12, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yao, J.; Mei, C.; Tong, X.; Zeng, L.; Li, Q.; Xiao, L.; Sun, T.; Li, J.; Deng, X.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, E1192–E1200. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Leon-Reyes, A.; Spoel, S.H.; De Lange, E.S.; Abe, H.; Kobayashi, M.; Tsuda, S.; Millenaar, F.F.; Welschen, R.A.; Ritsema, T.; Pieterse, C.M. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 2009, 149, 1797–1809. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Niu, Y.; Browse, J.; Howe, G.A. Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 2009, 70, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernandez-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genomics 2017, 18, 152. [Google Scholar] [CrossRef] [PubMed]
- Ebel, C.; BenFeki, A.; Hanin, M.; Solano, R.; Chini, A. Characterization of wheat (Triticum aestivum) TIFY family and role of Triticum Durum TdTIFY11a in salt stress tolerance. PLoS ONE 2018, 13, e0200566. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, M.; Singer, S.D.; Fei, Z.; Wang, H.; Wang, X. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 2012, 7, e44465. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Tian, J.; Cao, L.; Chen, X.; Chen, M.; Zhang, P.; Cao, L.; Persson, S.; Zhang, D.; Yuan, Z. The OsJAZ1 degron modulates jasmonate signaling sensitivity during rice development. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, F.; Fernandez-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Guo, Y.; Yang, Q.; He, Y.; Fetouh, M.I.; Warner, R.M.; Deng, Z. Genome-wide identification of quantitative trait loci for important plant and flower traits in petunia using a high-density linkage map and an interspecific recombinant inbred population derived from Petunia integrifolia and P. axillaris. Hortic. Res. 2019, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Zenoni, S.; D’Agostino, N.; Tornielli, G.B.; Quattrocchio, F.; Chiusano, M.L.; Koes, R.; Zethof, J.; Guzzo, F.; Delledonne, M.; Frusciante, L.; et al. Revealing impaired pathways in the an11 mutant by high-throughput characterization of Petunia axillaris and Petunia inflata transcriptomes. Plant J. 2011, 68, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R.; et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef] [PubMed]
- Segatto, A.L.; Ramos-Fregonezi, A.M.; Bonatto, S.L.; Freitas, L.B. Molecular insights into the purple-flowered ancestor of garden petunias. Am. J. Bot. 2014, 101, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 2013, 25, 1641–1656. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Liu, B.; Wang, L.; Zhang, L.; Hu, J.; Chen, J.; Zheng, H.; Lu, M. Characterization of the Populus Rab family genes and the function of PtRabE1b in salt tolerance. BMC Plant Biol. 2018, 18, 124. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yue, Y.; Sun, J.; Peng, H.; Yang, Z.; Bao, M.; Hu, H. Characterization of a fertility-related SANT/MYB gene (PhRL) from petunia. Sci. Hort. 2015, 183, 152–159. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Shi, T.; Chen, M.; Li, Y.; Du, J.; Jiang, H.; Yang, X.; Hu, H.; Wang, L. Integrating transcriptomic and GC-MS metabolomic analysis to characterize color and aroma formation during tepal development in Lycoris longituba. Plants (Basel) 2019, 8, 53. [Google Scholar] [CrossRef]
- Yue, Y.; Yin, C.; Guo, R.; Peng, H.; Yang, Z.; Liu, G.; Bao, M.; Hu, H. An anther-specific gene PhGRP is regulated by PhMYC2 and causes male sterility when overexpressed in petunia anthers. Plant Cell Rep. 2017, 36, 1401–1415. [Google Scholar] [CrossRef]
- Yue, Y.; Tian, S.; Wang, Y.; Ma, H.; Liu, S.; Wang, Y.; Hu, H. Transcriptomic and GC-MS metabolomic analyses reveal the sink strength changes during petunia anther development. Int. J. Mol. Sci. 2018, 19, 955. [Google Scholar] [CrossRef]
- Byng, J.W.; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Judd, W.S.; Mabberley, D.J.; Sennikov, A.N.; Soltis, D.E.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Sagnard, F.; Deu, M.; Dembele, D.; Leblois, R.; Toure, L.; Diakite, M.; Calatayud, C.; Vaksmann, M.; Bouchet, S.; Malle, Y.; et al. Genetic diversity, structure, gene flow and evolutionary relationships within the Sorghum bicolor wild–weedy–crop complex in a western African region. Theor. Appl. Genet. 2011, 123, 1231–1246. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Nagels Durand, A.; Pauwels, L.; Goossens, A. The ubiquitin system and jasmonate signaling. Plants (Basel) 2016, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Yu, R.; Han, M.; Wu, Z. Isolation, structural analysis and expression characteristics of the maize TIFY gene family. Mol. Genet. Genomics 2015, 290, 1849–1858. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xia, X.C.; Han, L.H.; Ni, P.; Yan, J.Q.; Tao, M.; Huang, G.Q.; Li, X.B. Genome-wide identification and characterization of JAZ gene family in upland cotton (Gossypium hirsutum). Sci. Rep. 2017, 7, 2788. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Bigotes, A.; Figueroa, N.E.; Figueroa, P.M.; Figueroa, C.R. Jasmonate signalling pathway in strawberry: Genome-wide identification, molecular characterization and expression of JAZs and MYCs during fruit development and ripening. PLoS ONE 2018, e0197118. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Sun, T.; Wang, L.; Su, W.; Gao, S.; Su, Y.; Xu, L.; Que, Y. Plant jasmonate ZIM domain genes: Shedding light on structure and expression patterns of JAZ gene family in sugarcane. BMC Genomics 2017, 18, 771. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; Vanholme, B.; Pauwels, L.; Plovie, E.; Inze, D.; Gheysen, G.; Goossens, A. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009, 10, 923–928. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef]
- Lu, Z.; Ricci, W.A.; Schmitz, R.J.; Zhang, X. Identification of cis-regulatory elements by chromatin structure. Curr. Opin. Plant Biol. 2018, 42, 90–94. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Moreno, J.E.; Shyu, C.; Campos, M.L.; Patel, L.C.; Chung, H.S.; Yao, J.; He, S.Y.; Howe, G.A. Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 2013, 162, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Ruız-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef] [PubMed]
- McConn, M.; Browse, J. The critical requirement for linolenic acid is pollen develspment, not photosynthesis, in an Arabidopsis mutant. Plant Cell 1996, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Song, S.; Xiao, L.; Soo, H.M.; Cheng, Z.; Xie, D.; Peng, J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009, 5, e1000440. [Google Scholar] [CrossRef] [PubMed]
- Chua, L.; Shan, X.; Wang, J.; Peng, W.; Zhang, G.; Xie, D. Proteomics study of COI1-regulated proteins in Arabidopsis flower. J. Integr. Plant Biol. 2010, 52, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Mandaokar, A.; Thines, B.; Shin, B.; Lange, B.M.; Choi, G.; Koo, Y.J.; Yoo, Y.J.; Choi, Y.D.; Choi, G.; Browse, J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006, 46, 984–1008. [Google Scholar] [CrossRef]
- Jewell, J.B.; Browse, J. Epidermal jasmonate perception is sufficient for all aspects of jasmonate-mediated male fertility in Arabidopsis. Plant J. 2016, 85, 634–647. [Google Scholar] [CrossRef]



| Name | Protein ID in SGN 1 | Length 2 | MW 3 | pI | GRAVY 4 | TIFY 5 | Jas 6 |
|---|---|---|---|---|---|---|---|
| PaJAZ1 | Peaxi162Scf00533g00059.1 | 210 | 23.09 | 9.36 | −0.485 | 84–115 | 152–176 |
| PaJAZ2 | Peaxi162Scf00928g00130.1 | 242 | 26.98 | 9.38 | −0.596 | 101–134 | 180–204 |
| PaJAZ3 | Peaxi162Scf00038g01961.1 | 296 | 32.35 | 9.31 | −0.685 | 99–132 | 240–264 |
| PaJAZ4 | Peaxi162Scf00314g00330.1 | 393 | 41.59 | 9.36 | −0.302 | 196–229 | 332–356 |
| PaJAZ5 | Peaxi162Scf00376g00327.1 | 353 | 37.57 | 9.15 | −0.229 | 154–187 | 292–316 |
| PaJAZ6 | Peaxi162Scf00207g00008.1 | 354 | 37.45 | 9.83 | −0.364 | 164–195 | 297–321 |
| PaJAZ7 | Peaxi162Scf00317g00117.1 | 126 | 14.30 | 9.62 | −0.731 | 42–74 | 104–124 |
| PaJAZ8 | Peaxi162Scf00128g00175.1 | 185 | 20.58 | 9.46 | −0.595 | 98–128 | 144–167 |
| PaJAZ9 | Peaxi162Scf00317g00121.1 | 123 | 13.99 | 10.02 | −0.826 | 38–69 | 100–120 |
| PaJAZ10 | Peaxi162Scf00305g00018.1 | 226 | 25.74 | 5.94 | −0.767 | 101–134 | 175–198 |
| PaJAZ11 | Peaxi162Scf00011g01124.1 | 222 | 24.90 | 9.02 | −0.677 | 98–130 | 168–192 |
| PaJAZ12 | Peaxi162Scf00394g00120.1 | 124 | 14.05 | 9.67 | −0.602 | 36–68 | 102–122 |
| PiJAZ1 | Peinf101Scf01317g09018.1 | 185 | 20.81 | 9.61 | −0.744 | 42–74,100–131 | 162–182 |
| PiJAZ2 | Peinf101Scf00193g17008.1 | 213 | 23.43 | 9.38 | −0.494 | 86–117 | 154–178 |
| PiJAZ3 | Peinf101Scf03815g00017.1 | 209 | 23.56 | 9.23 | −0.512 | 63–96 | 148–172 |
| PiJAZ4 | Peinf101Ctg13655775g00003.1 | 214 | 23.99 | 9.32 | −0.602 | 68–101 | 153–177 |
| PiJAZ5 | Peinf101Scf03816g00028.1 | 240 | 26.79 | 9.24 | −0.595 | 101–134 | 180–204 |
| PiJAZ6 | Peinf101Scf01633g04032.1 | 393 | 41.65 | 9.2 | −0.284 | 196–229 | 332–356 |
| PiJAZ7 | Peinf101Scf06002g00004.1 | 298 | 32.58 | 9.33 | −0.758 | 99–132 | 242–26 |
| PiJAZ8 | Peinf101Scf03176g00010.1 | 309 | 33.77 | 9.22 | −0.745 | 110–143 | 253–277 |
| PiJAZ9 | Peinf101Scf00497g02012.1 | 353 | 37.62 | 8.84 | −0.238 | 154–187 | 292–316 |
| PiJAZ10 | Peinf101Scf00048g00001.1 | 120 | 13.39 | 9.52 | −0.592 | 22–53 | 88–103 |
| PiJAZ11 | Peinf101Scf01889g01055.1 | 354 | 37.47 | 9.69 | −0.395 | 147–178 | 280–304 |
| PiJAZ12 | Peinf101Scf00637g07016.1 | 96 | 10.85 | 10.39 | −0.822 | 8–40 | 74–94 |
| PiJAZ13 | Peinf101Scf02714g01041.1 | 222 | 24.80 | 9.21 | −0.724 | 98–130 | 168–192 |
| PiJAZ14 | Peinf101Scf00947g00007.1 | 226 | 25.85 | 6.01 | −0.735 | 101–134 | 175–198 |
| PiJAZ15 | Peinf101Scf00889g01051.1 | 142 | 16.22 | 9.44 | −0.679 | 55–85 | 101–124 |
| PiJAZ16 | Peinf101Scf00280g05010.1 | 126 | 14.44 | 9.81 | −0.66 | 41–73 | 104–124 |
| Paralogous pairs | Ka | Ks | Ka/Ks Ratio | Date (Mya) |
|---|---|---|---|---|
| PaJAZ4-PaJAZ6 | 0.0082 | 0.0626 | 0.13 | 3.44 |
| PaJAZ7-PaJAZ9 | 0.0092 | 0.0390 | 0.24 | 2.14 |
| PiJAZ2-PiJAZ10 | 0.1406 | 0.4864 | 0.29 | 26.73 |
| PiJAZ4-PiJAZ5 | 0.1792 | 0.5381 | 0.33 | 29.57 |
| PiJAZ6-PiJAZ11 | 0.3269 | 0.7209 | 0.45 | 39.61 |
| PiJAZ7-PiJAZ8 | 0.3187 | 0.6282 | 0.51 | 34.52 |
| PiJAZ12-PiJAZ16 | 0.2327 | 0.4651 | 0.50 | 25.55 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Liu, S.; Wang, Y.; Wang, K.; Yin, C.; Yue, Y.; Hu, H. Genome-Wide Identification and Characterization of JAZ Protein Family in Two Petunia Progenitors. Plants 2019, 8, 203. https://doi.org/10.3390/plants8070203
Tian S, Liu S, Wang Y, Wang K, Yin C, Yue Y, Hu H. Genome-Wide Identification and Characterization of JAZ Protein Family in Two Petunia Progenitors. Plants. 2019; 8(7):203. https://doi.org/10.3390/plants8070203
Chicago/Turabian StyleTian, Shaoze, Siyu Liu, Yu Wang, Kun Wang, Chaoqun Yin, Yuanzheng Yue, and Huirong Hu. 2019. "Genome-Wide Identification and Characterization of JAZ Protein Family in Two Petunia Progenitors" Plants 8, no. 7: 203. https://doi.org/10.3390/plants8070203
APA StyleTian, S., Liu, S., Wang, Y., Wang, K., Yin, C., Yue, Y., & Hu, H. (2019). Genome-Wide Identification and Characterization of JAZ Protein Family in Two Petunia Progenitors. Plants, 8(7), 203. https://doi.org/10.3390/plants8070203





