Diversified Chromosome Rearrangements Detected in a Wheat‒Dasypyrum breviaristatum Substitution Line Induced by Gamma-Ray Irradiation
Abstract
1. Introduction
2. Results
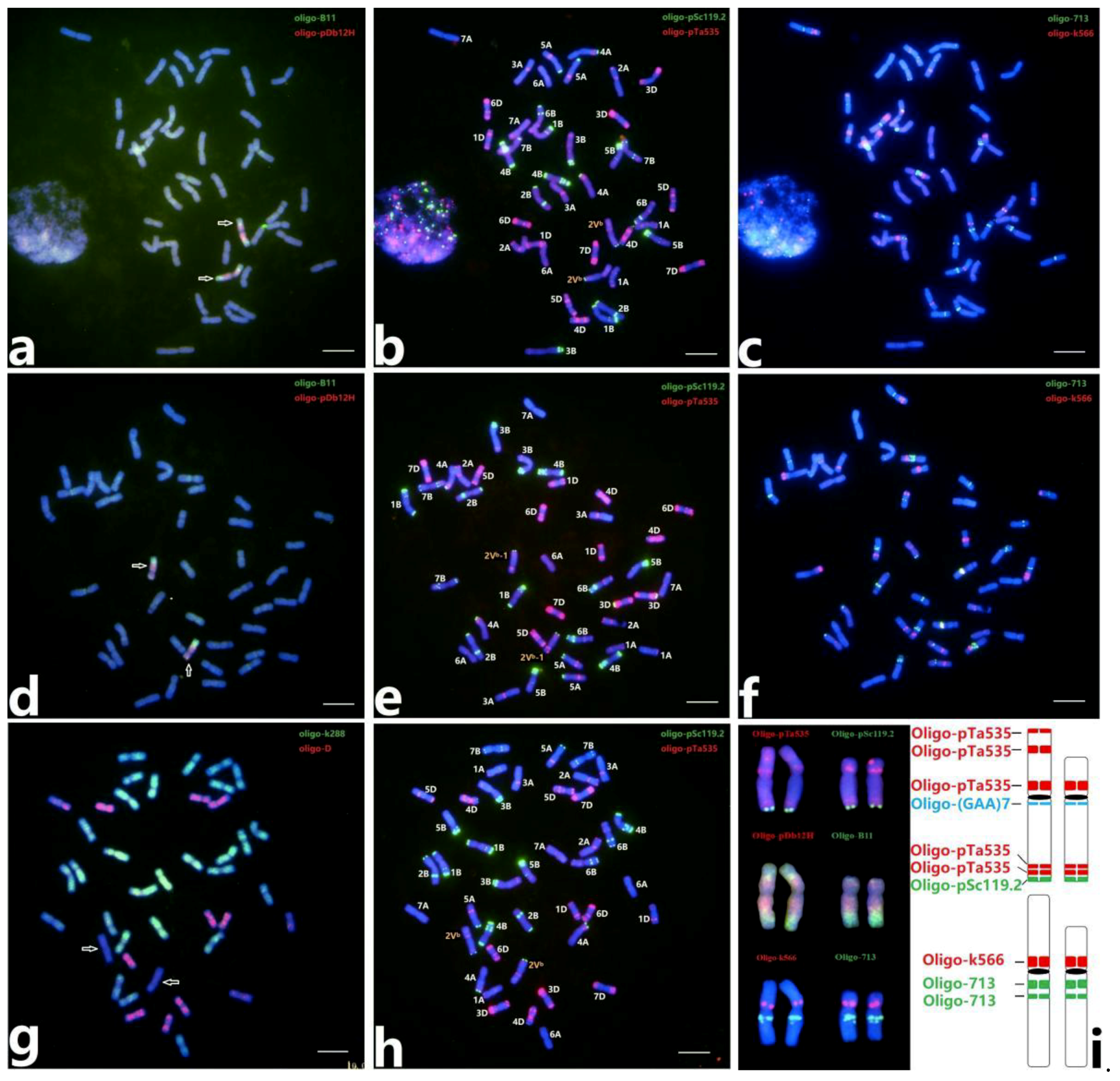
2.1. ND-FISH Karyotype of 2Vb(2D) Substitution Lines
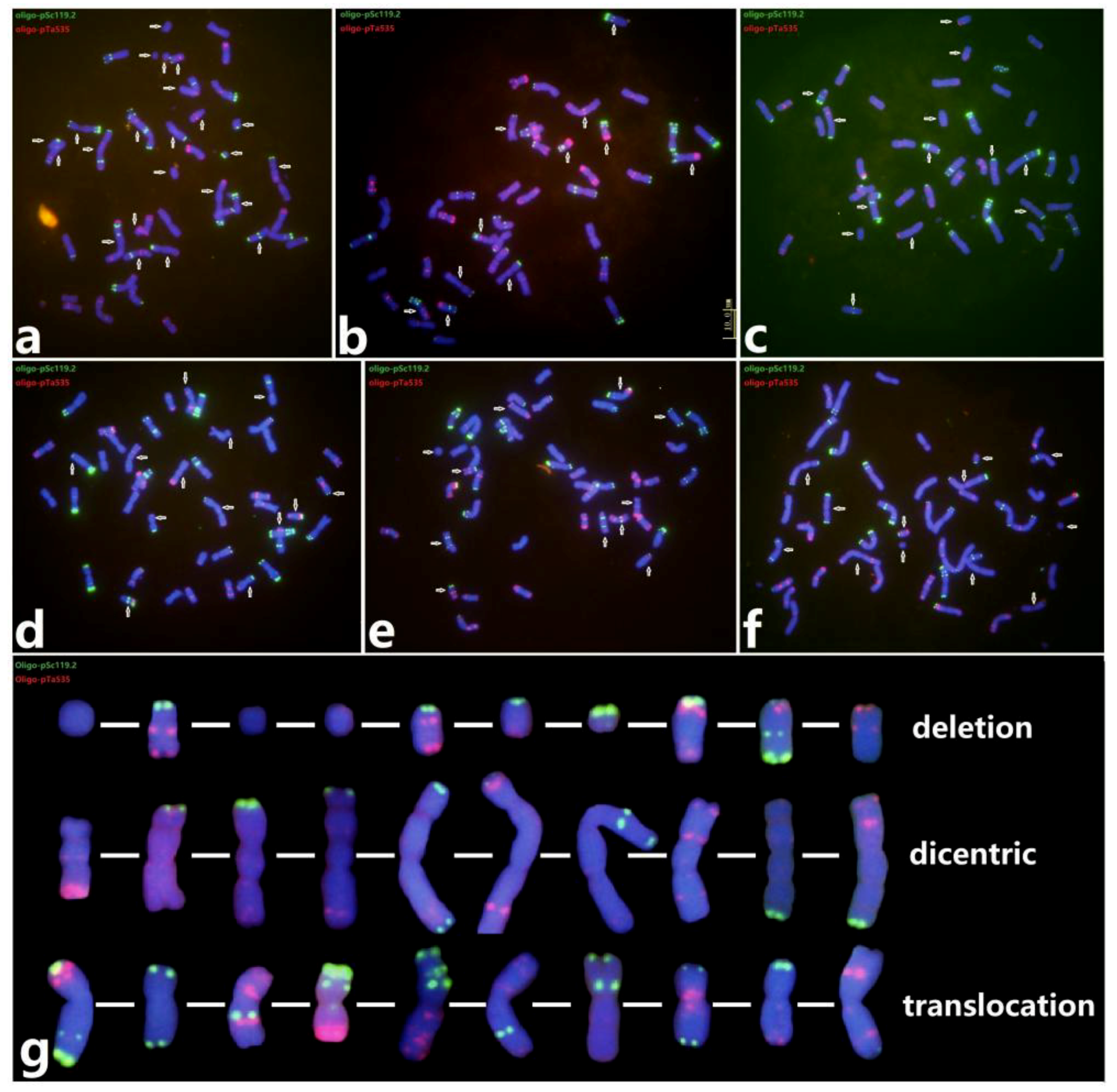
2.2. Chromosome Variations in M0 Seeds
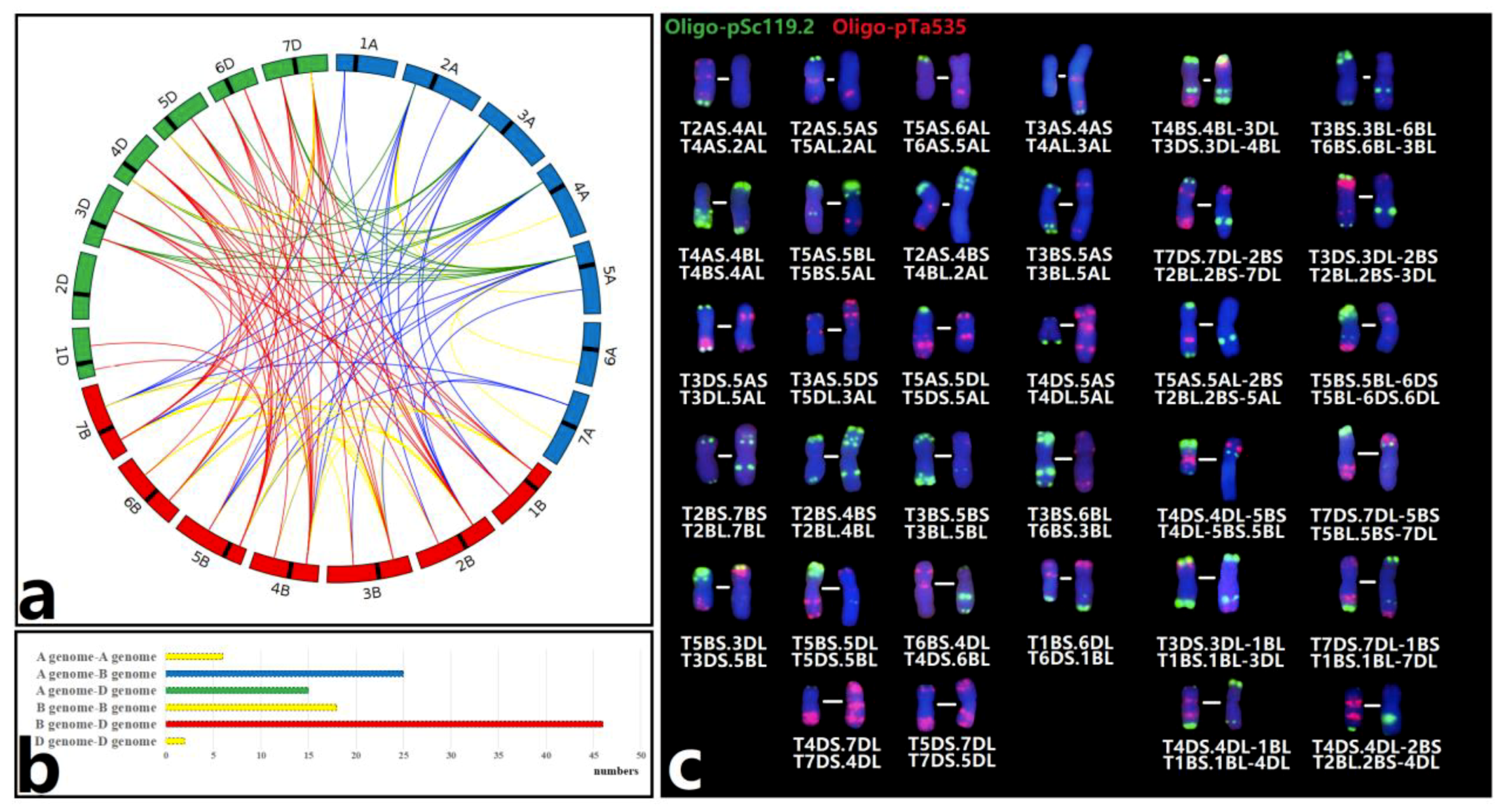
2.3. The Wheat‒Wheat Translocations in M1 and M2
2.4. Determination of Breakage Points for Non-Robertsonian Translocation
2.5. Identification of Wheat‒D. breviaristatum Translocations
2.6. Survey of Agronomic Traits
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Cytological Preparation
4.3. DNA Probes and Labeling
4.4. ND-FISH and Sequential FISH Analysis
4.5. Agronomic Traits
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sears, E.R. The transfer of leaf rust resistance from Aegilops umbellulata to wheat. Brookhaven Symp. Biol. 1956, 9, 1–22. [Google Scholar]
- Knott, D.R. Transferring alien genes to wheat. In Wheat and Wheat Improvement, Agronomy Monograph No. 13; Heyne, E.G., Ed.; American Society of Agronomy: Madison, WI, USA, 1987; pp. 462–471. [Google Scholar]
- Friebe, B.; Hatchett, J.H.; Mukai, Y.; Gill, B.S.; Sebesta, E.E. Transfer of Hessian fly resistance from rye to wheat via radiation-induced terminal and intercalary chromosomal translocations. Theor. Appl. Genet. 1991, 83, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Riera-Lizarazu, O.; Vales, M.I.; Ananiev, E.V.; Rines, H.W.; Phillips, R.L. Production and characterization of maize chromosome 9 radiation hybrids derived from an oat-maize addition line. Genetics 2000, 156, 327–339. [Google Scholar] [PubMed]
- Pu, J.; Wang, Q.; Shen, Y.F.; Zhuang, L.F.; Li, C.X.; Tan, M.F.; Bie, T.D.; Chu, C.G.; Qi, Z.J. Physical mapping of chromosome 4J of Thinopyrum bessarabicum using gamma radiation-induced aberrations. Theor. Appl. Genet. 2015, 128, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Bie, T.D.; Cao, Y.P.; Chen, P.D. Mass production of intergeneric chromosomal translocations through pollen irradiation of Triticum durum-Haynaldia villosa amphiploid. J. Integr. Plant Biol. 2007, 49, 1619–1626. [Google Scholar] [CrossRef]
- Molnár, I.; Benavente, E.; Molnár-Láng, M. Detection of intergenomic chromosome rearrangements in irradiated Triticum aestivum-Aegilops biuncialis amphiploids by multicolour genomic in situ hybridization. Genome 2009, 52, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.P.; Bie, T.D.; Wang, X.E.; Chen, P.D. Induction and transmission of wheat-Haynaldia villosa chromosomal translocations. J. Genet. Genom. 2009, 36, 313–320. [Google Scholar] [CrossRef]
- Liu, W.X.; Chen, P.D.; Liu, D.J. Development of Triticum aestivum-Leymus racemosus translocation lines by irradiating adult plants at meiosis. Acta Bot. Sin. 1999, 41, 463–467. [Google Scholar]
- Chen, P.D.; Liu, W.X.; Yuan, J.H.; Wang, X.E.; Zhou, B.; Wang, S.L.; Zhang, S.Z.; Feng, Y.G.; Yang, B.J.; Liu, G.G.; et al. Development and characterization of wheat-Leymus racemosus translocation lines with resistance to Fusarium Head Blight. Theor. Appl. Genet. 2005, 111, 941–948. [Google Scholar] [CrossRef]
- Song, L.Q.; Jiang, L.L.; Han, H.M.; Gao, A.N.; Yang, X.M.; Li, L.H.; Liu, W.H. Efficient induction of wheat-Agropyron cristatum 6P translocation lines and GISH detection. PLoS ONE 2013, 8, e69501. [Google Scholar] [CrossRef]
- Du, H.M.; Tang, Z.X.; Duan, Q.; Tang, S.Y.; Fu, S.L. Using the 6RLKu Minichromosome of rye (Secale cereale L.) to create wheat-rye 6D/6RLKu small segment translocation lines with powdery mildew resistance. Int. J. Mol. Sci. 2018, 19, 3933. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Q.; Luo, Q.L.; Li, H.W.; Li, B.; Li, Z.S.; Zheng, Q. Physical mapping of the blue-grained gene from Thinopyrum ponticum chromosome 4Ag and development of blue-grain-related molecular markers and a FISH probe based on SLAF-seq technology. Theor. Appl. Genet. 2018, 131, 2359–2370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Q.; Hou, F.; Feng, Y.G.; Zhang, W.; Zhang, M.Y.; Chen, P.D. Characterization of a Triticum aestivum-Dasypyrum villosum T2VS.2DL translocation line expressing a longer spike and more kernels traits. Theor. Appl. Genet. 2015, 128, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.W.; Chen, P.D.; Wang, X.E. Inducement of chromosome translocation with small alien segments by irradiating mature female gametes of the whole arm translocation line. Sci. China-Life Sci. 2008, 51, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.L.; Su, H.D.; Guo, X.R.; Han, F.P. Centromere structure and function analysis in wheat-rye translocation lines. Plant J. 2017, 91, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Q.; Sun, B.X.; Chen, J.; Cao, A.Z.; Xing, L.P.; Feng, Y.G.; Lan, C.X.; Chen, P.D. Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 2016, 129, 1975–1984. [Google Scholar] [CrossRef]
- Baum, B.R.; Edwards, T.; Johnson, D.A. What does the nr5S DNA multigene family tell us about the genomic relationship between Dasypyrum breviaristatum and D. villosum (Triticeae: Poaceae)? Mol. Genet. Genom. 2014, 289, 553–565. [Google Scholar] [CrossRef]
- De Pace, C.; Vaccino, P.; Giorgio, P.; Pasquini, M.; Qualset, C.O. Dasypyrum. In Wild Crop Relatives: Genomic and Breeding Resources, Cereals; Kole, C., Ed.; Springer: Berlin, Germany, 2011; Chapter 4; pp. 185–292. [Google Scholar]
- Chen, P.D.; Qi, L.L.; Zhou, B.; Zhang, S.Z.; Liu, D.J. Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 1995, 91, 1125–1128. [Google Scholar] [CrossRef]
- Zhang, R.Q.; Fan, Y.L.; Kong, L.N.; Wang, Z.J.; Wu, J.Z.; Xing, L.P.; Cao, A.Z.; Feng, Y.G. Pm62, an adul-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 2018, 131, 2613–2620. [Google Scholar] [CrossRef]
- Qi, L.L.; Pumphrey, M.O.; Friebe, B.; Zhang, P.; Qian, C.; Bowden, R.L.; Rouse, M.N.; Jin, Y.; Gill, B.S. A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor. Appl. Genet. 2011, 123, 159–167. [Google Scholar] [CrossRef]
- Jiang, H.R.; Dai, D.Q.; Sun, D.F.; Xiao, S.H. New artificial genetic resources of wheat: Several polyploids of Triticum-Dasypyrum. Sci. Agric. Sin. 1992, 25, 89. [Google Scholar]
- Yang, Z.J.; Li, G.R.; Feng, J.; Jiang, H.R.; Ren, Z.L. Molecular cytogenetic characterization and disease resistance observation of wheat-Dasypyrum breviaristatum partial amphiploid and its derivatives. Hereditas 2005, 141, 1–6. [Google Scholar]
- Li, G.R.; Zhao, J.M.; Li, D.H.; Yang, E.N.; Huang, Y.F.; Liu, C.; Yang, Z.J. A novel wheat-Dasypyrum breviaristatum substitution line with stripe rust resistance. Cytogenet. Genome Res. 2014, 143, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Li, G.R.; Gao, D.; Zhang, H.J.; Li, J.B.; Wang, H.J.; La, S.X.; Ma, J.W.; Yang, Z.J. Molecular cytogenetic characterization of Dasypyrum breviaristatum chromosomes in wheat background revealing the genomic divergence between Dasypyrum species. Mol. Cytogenet. 2016, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Yu, Z.H.; Li, B.; Lang, T.; Li, G.R.; Yang, Z.J. Characterization of new wheat-Dasypyrum breviaristatum introgression lines with superior gene(s) for spike length and stripe rust resistance. Cytogenet. Genome Res. 2018, 156, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, Á.; Golczyk, H.; Jouve, N. A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res. 2009, 17, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, Á.; Jouve, N. Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 2010, 119, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.L.; Chen, L.; Wang, Y.Y.; Li, M.; Yang, Z.J.; Qiu, L.; Yan, B.J.; Ren, Z.Z.; Tang, Z.X. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef] [PubMed]
- Delgado, A.; Carvalho, A.; Martin, A.C.; Martin, A.; Lima-Brito, J. Use of the synthetic Oligo-pTa535 and Oligo-pAs1 probes for identification of Hordeum chilense-origin chromosomes in hexaploid tritordeum. Genet. Resour. Crop Evol. 2016, 63, 945–951. [Google Scholar] [CrossRef]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.Y.; Tang, Z.X.; Qiu, L.; Yang, Z.J.; Li, G.R.; Lang, T.; Zhu, W.Q.; Zhang, J.H.; Fu, S.L. Developing new oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front. Plant Sci. 2018, 9, 1104. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.H.; Wang, H.J.; Xu, Y.F.; Li, Y.S.; Lang, T.; Yang, Z.J.; Li, G.R. Characterization of chromosomal rearrangement in new wheat-Thinopyrum intermedium addition lines carrying Thinopyrum-specific grain hardness genes. Agronomy 2019, 9, 18. [Google Scholar] [CrossRef]
- Lang, T.; Li, G.R.; Wang, H.J.; Yu, Z.H.; Chen, Q.H.; Yang, E.N.; Fu, S.L.; Tang, Z.X.; Yang, Z.J. Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 2019, 249, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosomal rearrangements in wheat: Their types and distribution. Genome 2007, 50, 907–926. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Schwarzacher, T. The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 1998, 107, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; La, S.X.; Li, B.; Yu, Z.H.; Chen, Q.H.; Li, J.B.; Yang, E.N.; Li, G.R.; Yang, Z.J. Precise identification of wheat-Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 2018, 61, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.Q.; Tang, S.Y.; Qiu, L.; Tang, Z.X.; Fu, S.L. Oligonucleotides and ND-FISH displaying different arrangements of tandem repeats and identification of Dasypyrum villosum chromosomes in wheat backgrounds. Molecules 2017, 22, 973. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Tang, Z.X.; Tang, S.Y.; Yang, Z.J.; Luo, J.; Fu, S.L. New ND-FISH-positive oligo probes for identifying Thinopyrum chromosomes in wheat backgrounds. Int. J. Mol. Sci. 2019, 20, 2031. [Google Scholar] [CrossRef]
- Komuro, S.; Endo, R.; Shikata, K.; Kato, A. Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 2013, 56, 131–137. [Google Scholar] [CrossRef]
- Liu, W.X.; Chen, P.D.; Liu, D.J. Studies of the development of Triticum aestivum-Leymus racemosus translocation lines by pollen irradiation. Acta Genet. Sin. 2000, 27, 40–49. [Google Scholar]
- Li, H.H.; Lv, M.J.; Song, L.Q.; Zhang, J.P.; Gao, A.N.; Li, L.H.; Liu, W.H. Production and identification of wheat-Agropyron cristatum 2P translocation lines. PLoS ONE 2016, 11, e0145928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, Y.; Guo, Y.L.; Li, G.R.; Yang, Z.J.; Xu, D.L.; Xuan, P. Identification of novel chromosomal aberrations induced by 60Co-γ-irradiation in wheat-Dasypyrum villosum lines. Int. J. Mol. Sci. 2015, 16, 29787–29796. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.S.; Kimber, G. Giemsa C-banding and the evolution of wheat. Proc. Natl. Acad. Sci. USA 1974, 71, 4086–4090. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, H.; Tuna, M.; Arumuganathan, K. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. J. Hered. 2003, 94, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Sears, E.R. Misdivision of univalents in common wheat. Chromosoma 1950, 4, 535–550. [Google Scholar] [CrossRef]
- Zhang, P.; Friebe, B.; Lukaszewski, A.J.; Gill, B.S. The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 2001, 110, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Künzel, G.; Gecheff, K.I.; Schubert, I. Different chromosomal distribution patterns of radiation-induced interchange breakpoints in barley: First post-treatment mitosis versus viable offspring. Genome 2001, 44, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Bedbrook, J.R.; Jones, J.; O’Dell, M.; Thompson, R.D.; Flavell, R.B. A molecular description of telometic heterochromatin in Secale species. Cell 1980, 19, 545–560. [Google Scholar] [CrossRef]
- Cuadrado, A.; Jouve, M. Distribution of highly repeated DNA sequences in species of the genus Secale. Genome 1997, 40, 309–317. [Google Scholar] [CrossRef]
- Rayburn, A.L.; Gill, B.S. Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Rep. 1986, 4, 102–109. [Google Scholar] [CrossRef]
- Gaeta, R.T.; Pires, J.C.; Iniguez-Luy, F.; Leon, E.; Osborn, T.C. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 2007, 19, 3403–3417. [Google Scholar] [CrossRef] [PubMed]
- Law, C.N.; Worland, A.J. The control of adult-plant resistance to yellow rust by the translocated chromosome 5BS-7BS of bread wheat. Plant Breed. 1997, 116, 59–63. [Google Scholar] [CrossRef]
- Su, Y.R.; Li, Y.G.; Li, S.P. Application and improvement of wheat—Rye 1BL/1RS translocation line in wheat breeding. J. Henan Agric. Sci. 2006, 35, 12. [Google Scholar]
- Li, G.P.; Chen, P.D.; Zhang, S.Z.; Zhao, H. Effects of the 6VS/6AL translocation chromosome on agronomic characteristics of wheat. J. Plant Genet. Resour. 2011, 12, 744–749. [Google Scholar]
- Qi, Z.J.; Liu, D.J.; Chen, P.D. Development and identification of T. aestivum-S. cereale-H. villosa double translocation line 1RS.1BL, 6VS.6AL via chromosome C-banding and dual color FISH. Acta Genet. Sin. 2001, 28, 267–273. [Google Scholar]
- Yang, M.Y.; Yang, Z.J.; Yang, W.Y.; Yang, E.N. Development and identification of new wheat varieties (lines) with multiple translocation chromosomes via cytogenetic method. J. Triticeae Crops 2018, 38, 127–133. [Google Scholar]
- Kato, A.; Lamb, J.C.; Birchler, J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 2004, 101, 13554–13559. [Google Scholar] [CrossRef]
- Han, F.P.; Lamb, J.C.; Birchler, J.A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 2006, 103, 3238–3243. [Google Scholar] [CrossRef]
- Tang, S.Y.; Ling, Q.; Xiao, Z.Q.; Fu, S.L.; Tang, Z.X. New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 2016, 7, 118. [Google Scholar] [CrossRef]


| Agronomic Traits | Plants without Translocations | Plants with Translocations |
|---|---|---|
| 1000-grain weight (g) | 39.96 (23.06–60.54) | 36.13 (15.68–69.33) |
| 10-kernel length (cm) | 7.84 (7.16–8.96) | 7.86 (7.00–9.42) |
| 10-kernel width (cm) | 2.82 (2.18–3.26) | 2.66 (1.85–3.25) |
| Plant height (cm) | 81.91 (60.00–103.00) | 80.03 (61.00–124.00) |
| Tiller number | 8.14 (4.00–17.00) | 9.31 (4.00–18.00) |
| Spike length (cm) | 14.29 (9.80–17.30) | 14.25 (9.00–17.00) |
| Spikelet number | 26.39 (20.00–31.00) | 26.83 (24.00–30.00) |
| Name | Sequences | Reference |
|---|---|---|
| Oligo-pSc119.2 | CCGTTTTGTGGACTATTACTCACCGCTTTGGGGTCCCATAGCTAT | [32] |
| Oligo-pTa535 | GACGAGAACTCATCTGTTACATGGGCACTTCAATGTTTTTTAAACTTATTTGAACTCCA | [32] |
| Oligo-pDb12H | TCAGAATTTTTAGGATAGCAGAAGTATTCGAAATACCCAGATTGCTACAG | [34] |
| Oligo-B11 | TCCGCTCACCTTGATGACAACATCAGGTGGAATTCCGTTCGAGGG | [40] |
| Oligo-713 | GTCGCGGTAGCGACGACGGACGCCGAGACGAGCACGTGACACCATTCCCACCCTGTCTA | [61] |
| Oligo-k566 | ATCCTACCGAGTGGAGAGCGACCCTCCCACTCGGGGGCTTAGCTGCAGTCCAGTACTCG | [61] |
| Oligo-D | TACGGGTGCCAAACGAGTGTCTGAAAGACTCCTCGAGAGGAAAATGCGAA | [33] |
| Oligo-B | GGTTCAGGAATAGCCTCAGGAATTGGCTCAATT | [33] |
| Oligo-45 | CGGCCGCTCCGCGCGTCGCCATCGGTTGGTCACCTCATCACCACT | [33] |
| Oligo-18 | GTAGCAGTAGCAGTAGTA | [33] |
| Oligo-(GAA)7 | GAAGAAGAAGAAGAAGAAGAA | [37] |
| Oligo-1D1 | CGGAGTCCGTTTTGGCTCCACAAGTAGTCAAAACGTTTGTGACGACCAGATGCT | [35] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Yu, Z.; Li, G.; Yang, Z. Diversified Chromosome Rearrangements Detected in a Wheat‒Dasypyrum breviaristatum Substitution Line Induced by Gamma-Ray Irradiation. Plants 2019, 8, 175. https://doi.org/10.3390/plants8060175
Wang H, Yu Z, Li G, Yang Z. Diversified Chromosome Rearrangements Detected in a Wheat‒Dasypyrum breviaristatum Substitution Line Induced by Gamma-Ray Irradiation. Plants. 2019; 8(6):175. https://doi.org/10.3390/plants8060175
Chicago/Turabian StyleWang, Hongjin, Zhihui Yu, Guangrong Li, and Zujun Yang. 2019. "Diversified Chromosome Rearrangements Detected in a Wheat‒Dasypyrum breviaristatum Substitution Line Induced by Gamma-Ray Irradiation" Plants 8, no. 6: 175. https://doi.org/10.3390/plants8060175
APA StyleWang, H., Yu, Z., Li, G., & Yang, Z. (2019). Diversified Chromosome Rearrangements Detected in a Wheat‒Dasypyrum breviaristatum Substitution Line Induced by Gamma-Ray Irradiation. Plants, 8(6), 175. https://doi.org/10.3390/plants8060175






