Interspaced Repeat Sequences Confer the Regulatory Functions of AtXTH10, Important for Root Growth in Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Arabidopsis IRS and Functional Annotation of the IRS-Related Genes
2.2. Functional Identification of the IRSs in the CR6 and CR12 Promoters
2.3. Vector Construction and Subcellular Localization of the AtXTH10 Protein
2.4. Obtaining Transgenic AtXTH10 and Mutant Plants
2.5. Scanning Electron Microscopic Observations
2.6. Detection the Relative Expression Levels of Known Root Development-Related Genes by Reverse Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR) in the Col, AtXTH10m and AtXTH10-OE Plants
3. Results and Discussion
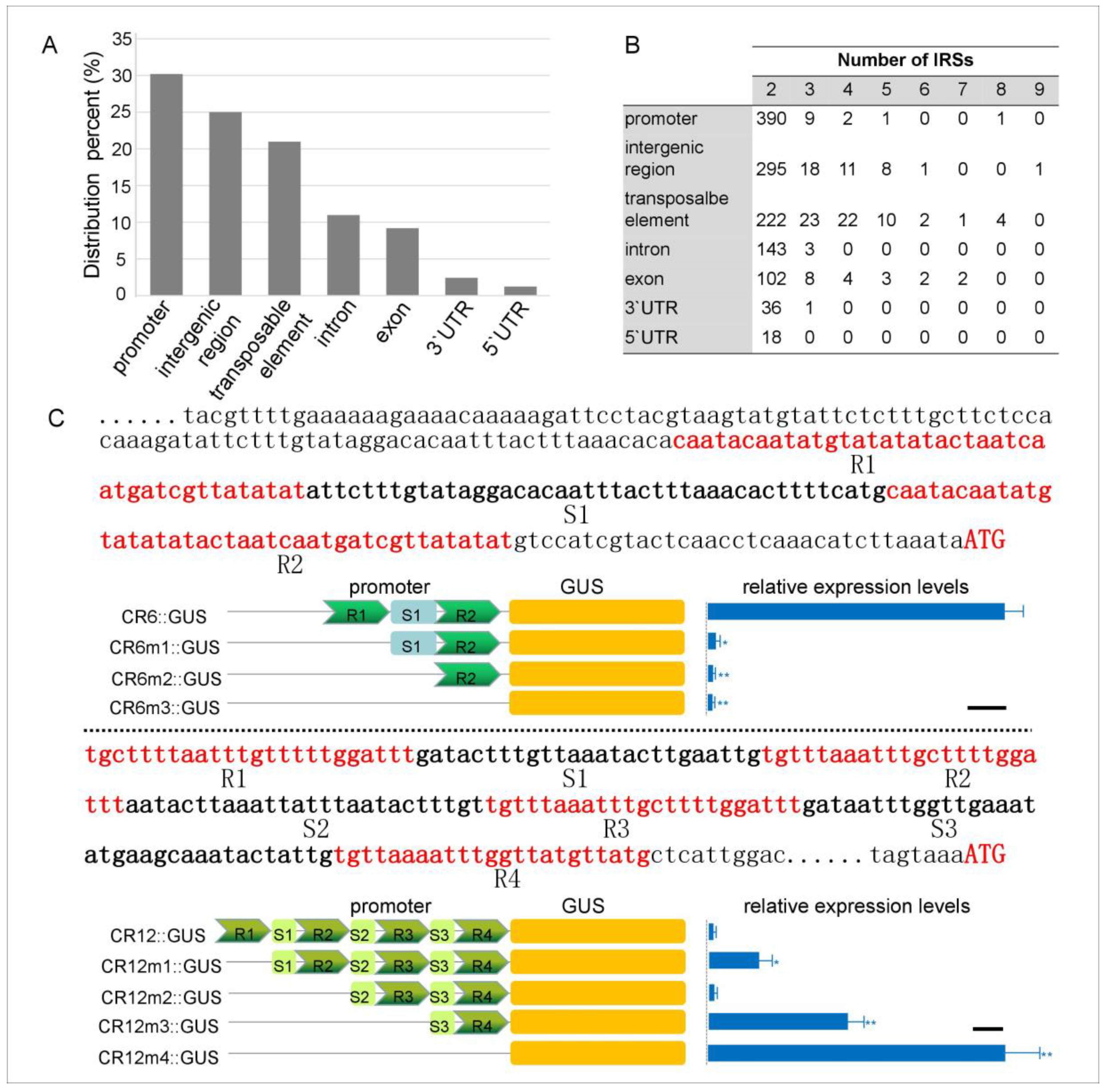
3.1. Identification of IRSs and Functional Annotation of the IRS-Related Genes
3.2. IRSs Confer a Regulatory Function of Gene Expression
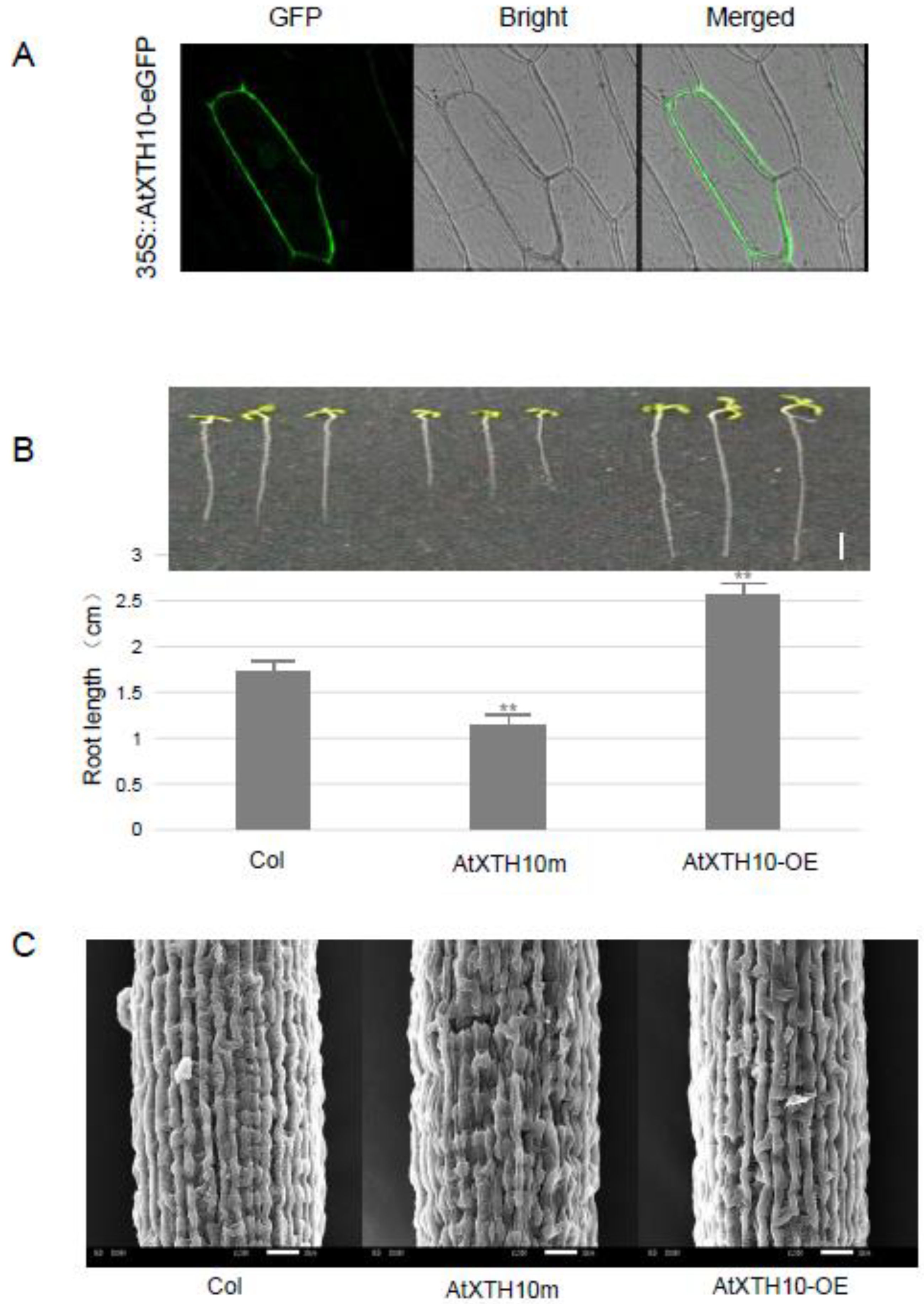
3.3. The AtXTH10 Gene Regulates the Growth and Development of Roots
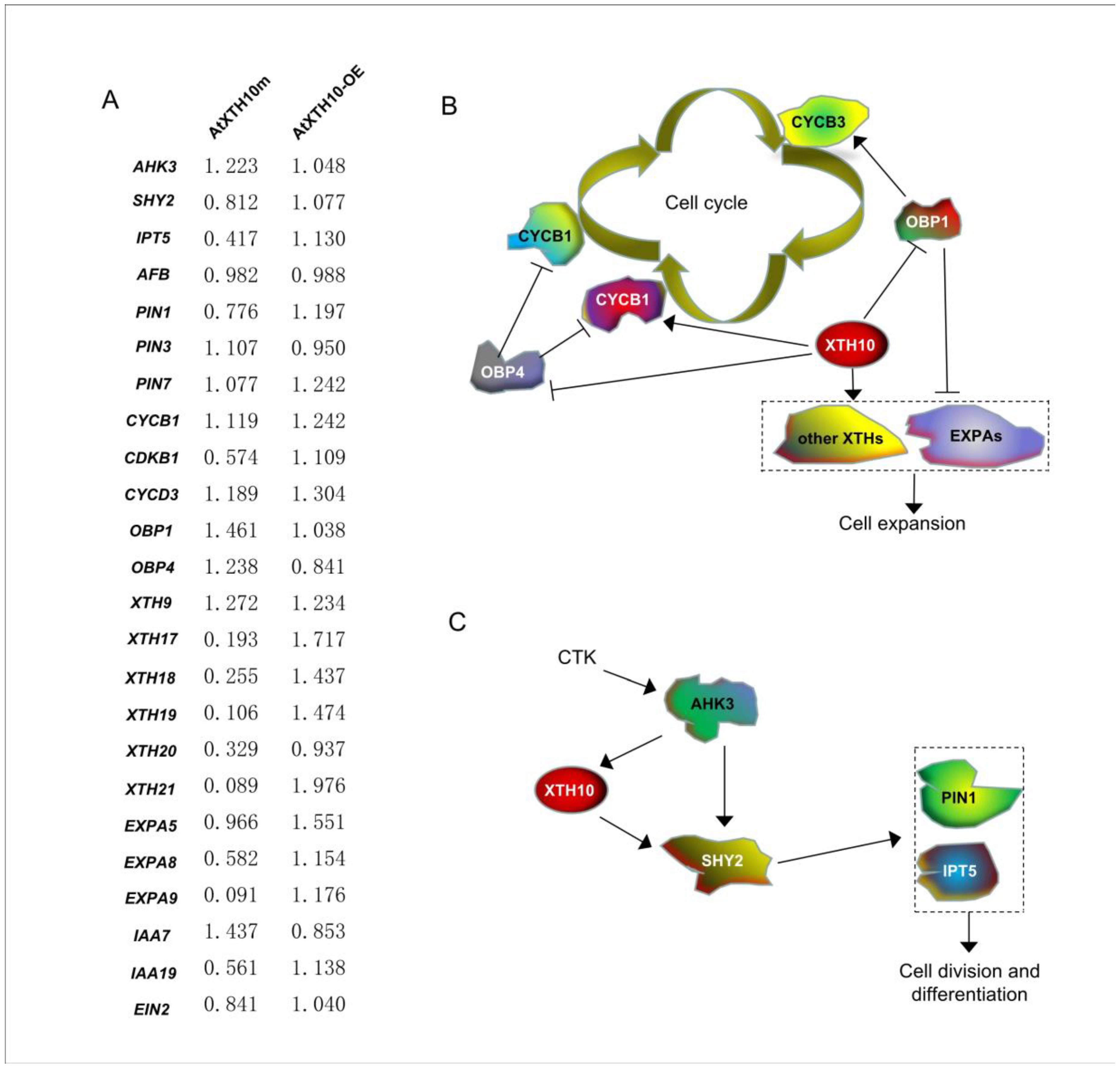
3.4. Prediction of the Root Development Signaling Pathway Involved with the AtXTH10 Gene
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Holkers, M.; de Vries, A.A.; Gonçalves, M.A. Nonspaced inverted DNA repeats are preferential targets for homology-directed gene repair in mammalian cells. Nucleic Acids Res. 2012, 40, 1984–1999. [Google Scholar] [CrossRef]
- Pearson, C.E.; Edamura, K.C.; Cleary, J.D. Repeat instability: Mechanisms of dynamic mutations. Nat. Rev. Genet. 2005, 6, 729–742. [Google Scholar] [CrossRef]
- Mizuno, K.; Lambert, S.; Baldacci, G.; Murray, J.M.; Carr, A.M. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009, 23, 2876–2886. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Venter, M. Synthetic promoters: Genetic control through cis engineering. Trends Plant Sci. 2007, 12, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Amitai, G.; Sorek, R. CRISPR-Cas adaptation: Insights into the mechanism of action. Nat. Rev. Microbiol. 2016, 14, 67–76. [Google Scholar] [CrossRef]
- Mahfouz, M.M.; Piatek, A.; Stewart, C.N., Jr. Genome engineering via TALENs and CRISPR/Cas9 systems: Challenges and perspectives. Plant Biotechnol. J. 2014, 12, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Badran, A.H.; Liu, D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017, 168, 20–36. [Google Scholar] [CrossRef]
- Takeuchi, N.; Wolf, Y.I.; Makarova, K.S.; Koonin, E.V. Nature and intensity of selection pressure on CRISPR-associated genes. J. Bacteriol. 2012, 194, 1216–1225. [Google Scholar] [CrossRef]
- Zhang, Q.; Doak, T.G.; Ye, Y. Expanding the catalog of cas genes with metagenomes. Nucleic Acids Res. 2014, 42, 2448–2459. [Google Scholar] [CrossRef]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Pandey, G.K.; Cheong, Y.H.; Kim, K.N.; Grant, J.J.; Li, L.; Hung, W.; D’Angelo, C.; Weinl, S.; Kudla, J.; Luan, S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 2004, 16, 1912–1924. [Google Scholar] [CrossRef]
- Mähs, A.; Steinhorst, L.; Han, J.P.; Shen, L.K.; Wang, Y.; Kudla, J. The calcineurin B-like Ca2+ sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis. Mol. Plant 2013, 6, 1149–1162. [Google Scholar] [CrossRef]
- Tang, R.J.; Luan, S. Regulation of calcium and magnesium homeostasis in plants: From transporters to signaling network. Curr. Opin. Plant Biol. 2017, 39, 97–105. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Kim, K.N.; Pandey, G.K.; Gupta, R.; Grant, J.J.; Luan, S. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 2003, 15, 1833–1845. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Sung, S.J.; Kim, B.G.; Pandey, G.K.; Cho, J.S.; Kim, K.N.; Luan, S. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol. Cells 2010, 29, 159–165. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, T.; Martín, I.; Labrador, E.; Dopico, B. The immunolocation of a xyloglucan endotransglucosylase/hydrolase specific to elongating tissues in Cicer arietinum suggests a role in the elongation of vascular cells. J. Exp. Bot. 2006, 57, 3979–3988. [Google Scholar] [CrossRef]
- Singh, A.P.; Tripathi, S.K.; Nath, P.; Sane, A.P. Petal abscission in rose is associated with the differential expression of two ethylene-responsive xyloglucan endotransglucosylase/hydrolase genes, RbXTH1 and RbXTH2. J. Exp. Bot. 2011, 62, 5091–5103. [Google Scholar] [CrossRef]
- Ko, J.H.; Han, K.H.; Park, S.; Yang, J. Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 2004, 135, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Zhu, X.F.; Peng, Y.X.; Zheng, C.; Li, G.X.; Liu, Y.; Shi, Y.Z.; Zheng, S.J. Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 2011, 155, 1885–1892. [Google Scholar] [CrossRef]
- Ookawara, R.; Satoh, S.; Yoshioka, T.; Ishizawa, K. Expression of alpha-expansin and xyloglucan endotransglucosylase/hydrolase genes associated with shoot elongation enhanced by anoxia, ethylene and carbon dioxide in arrowhead (Sagittaria pygmaea Miq.) tubers. Ann. Bot. 2005, 96, 693–702. [Google Scholar] [CrossRef]
- He, J.X.; Fujioka, S.; Li, T.C.; Kang, S.G.; Seto, H.; Takatsuto, S.; Yoshida, S.; Jang, J.C. Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 2003, 131, 1258–1269. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.B.; Lu, S.M.; Zhang, J.F.; Liu, S.; Lu, Y.T. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 2007, 226, 1547–1560. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Agasse, A.; Gaillard, C.; Saumonneau, A.; Delrot, S.; Atanassova, R. A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 2003, 15, 2165–2180. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Dekeyser, R.A.; Claes, B.; De Rycke, R.; Habets, M.E.; Van Montaqu, M.C.; Caplan, A.B. Transient gene expression in intact and organized rice tissues. Plant Cell 1990, 2, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Dart, P.J. Scanning electron microscopy of plant root. J. Exp. Bot. 1971, 22, 163–168. [Google Scholar] [CrossRef]
- Lincoln, C.; Britton, J.H.; Estelle, M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 1990, 2, 1071–1080. [Google Scholar] [CrossRef]
- Martinis, J.; Glauser, G.; Valimareanu, S.; Stettler, M.; Zeeman, S.C.; Yamamoto, H.; Shikanai, T.; Kessler, F. ABC1K1/PGR6 kinase: A regulatory link between photosynthetic activity and chloroplast metabolism. Plant J. 2014, 77, 269–283. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007, 8, 172. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Fry, S. The structure and functions of xyloglucan. J. Exp. Bot. 1989, 40, 1–11. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Skirycz, A.; Radziejwoski, A.; Busch, W.; Hannah, M.A.; Czeszejko, J.; Kwaśniewski, M.; Zanor, M.I.; Lohmann, J.U.; De Veylder, L.; Witt, I.; et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 2008, 56, 779–792. [Google Scholar] [CrossRef]
- Xu, P.; Chen, H.; Ying, L.; Cai, W. AtDOF5.4/OBP4, a DOF Transcription Factor Gene that Negatively Regulates Cell Cycle Progression and Cell Expansion in Arabidopsis thaliana. Sci. Rep. 2016, 6, 27705. [Google Scholar] [CrossRef] [PubMed]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Friml, J. Auxin and other signals on the move in plants. Nat. Chem. Biol. 2009, 5, 325–332. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Lv, Y.; Li, X. Interspaced Repeat Sequences Confer the Regulatory Functions of AtXTH10, Important for Root Growth in Arabidopsis. Plants 2019, 8, 130. https://doi.org/10.3390/plants8050130
Cao J, Lv Y, Li X. Interspaced Repeat Sequences Confer the Regulatory Functions of AtXTH10, Important for Root Growth in Arabidopsis. Plants. 2019; 8(5):130. https://doi.org/10.3390/plants8050130
Chicago/Turabian StyleCao, Jun, Yueqing Lv, and Xiang Li. 2019. "Interspaced Repeat Sequences Confer the Regulatory Functions of AtXTH10, Important for Root Growth in Arabidopsis" Plants 8, no. 5: 130. https://doi.org/10.3390/plants8050130



