High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily
Abstract
1. Introduction
2. Results and Discussion
2.1. Morphological and Grain Quality Traits
2.2. SNP Analysis and Genetic Characterization
3. Conclusions
4. Materials and Methods
4.1. Plant Material and Experimental Conditions
4.2. Morphological Traits Characterization
4.3. Grain and Wholemeal Flour Quality-related Traits
4.4. Statistical Analysis for Morphological and Quality-Related Traits
4.5. DNA Extraction and SNP Genotyping
4.6. 90K Chip Array SNP-data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef]
- Bozzini, A.; Corazza, L.; D’Egidio, M.G.; Di Fonzo, N.; La Fiandra, D.; Pogna, N.E.; Poma, I. Durum wheat. In Italian Contribution to Plant Genetics and Breeding; Scarascia Mugnozza, G.T., Pagnotta, M.A., Eds.; Tipolitografia Quatrini: Viterbo, Italy, 1998; pp. 181–194. [Google Scholar]
- Lorenzetti, R. The Wheat Science: The Green Revolution of Nazareno Strampelli; MiBAC: Roma, Italy, 2000; pp. 1–230. [Google Scholar]
- Royo, C.; Maccaferri, M.A.; Alvaro, F.; Moragues, M.; Sanguineti, M.C.; Tuberosa, R.; Maaloufc, F.; García del Moral, L.F.; Demontis, A.; Rhouma, S.; et al. Understanding the relationships between genetic and phenotypic structures of a collection of elite durum wheat accessions. Field Crop. Res. 2010, 119, 91–105. [Google Scholar] [CrossRef]
- Prohens, J.; Nuez, F. In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution. In Proceedings of the International Congress, Bologna, Italy, 27–31 May 2003; Scarascia Mugnozza, G.T., Tuberosa, R., Phillips, R.L., Gale, M., Eds.; Avenue Media: Bologna, Italy, 2005; pp. 53–75. [Google Scholar]
- De Cillis, U. I frumenti siciliani; Tipografia Zuccarello & Izzi: Catania, Italy, 1942; ISBN 88-7751-229-6. [Google Scholar]
- Colomba, M.S.; Gregorini, A. Genetic diversity analysis of the durum wheat Graziella Ra, Triticum turgidum L. subsp. durum (Desf.) Husn. (Poales, Poaceae). Biodivers. J. 2011, 2, 73–84. [Google Scholar]
- Newton, A.C.; Begg, G.; Swanston, J.S. Deployment of diversity for enhanced crop function. Ann. Appl. Biol. 2009, 154, 309–322. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Di Loreto, A.; Bosi, S.; Montero, L.; Bregola, V.; Marotti, I.; Sferrazza, R.E.; Dinelli, G.; Herrero, M.; Cifuentes, A. Determination of phenolic compounds in ancient and modern durum wheat genotypes. Electrophoresis 2018, 39, 2001–2010. [Google Scholar] [CrossRef]
- Lo Bianco, M.; Siracusa, L.; Dattilo, S.; Venora, G.; Ruberto, G. Phenolic fingerprint of Sicilian modern varieties and old durum wheat landraces: A tool to assess with diversity. Cereal Chem. 2017, 94, 1045–1051. [Google Scholar]
- International Board for Plant Genetic Resources, (IBPGR). Descriptors for Wheat (Revised), IPBGR, 1985 Rome, Italy. Available online: https://www.bioversityinternational.org (accessed on 15 January 2019).
- International Union for the Protection of New Varieties of Plants (UPVO) Durum Wheat. 2012. Available online: https://www.upov.int (accessed on 15 December 2018).
- Brookes, A.J. The essence of SNPs. Gene 1999, 234, 177–186. [Google Scholar] [CrossRef]
- Deschamps, S.; Campbell, M.A. Utilization of next-generation sequencing platforms in plant genomics and genetic variant discovery. Mol. Breed. 2010, 25, 553–570. [Google Scholar] [CrossRef]
- Liao, P.Y.; Lee, K.H. From SNPs to functional polymorphism: The insight into biotechnology applications. Biochem. Eng. J. 2010, 49, 149–158. [Google Scholar] [CrossRef]
- Zhao, H.; Pfeiffer, R.; Gail, M.H. Haplotype analysis in population genetics and association studies. Pharmacogenomics 2003, 4, 171–178. [Google Scholar] [CrossRef]
- Bardel, C.; Danjean, V.; Hugot, J.P.; Darlu, P.; Genin, E. On the use of haplotype phylogeny to detect disease susceptibility loci. BMC Genet. 2005, 6, 24. [Google Scholar] [CrossRef][Green Version]
- Trebbi, D.; Maccaferri, M.; de Heer, P.; Sørensen, A.; Giuliani, S.; Salvi, S.; Sanguineti, M.C.; Massi, A.; van der Vossen, E.A.G.; Tuberosa, R. High-throughput SNP discovery and genotyping in durum wheat (Triticum durum Desf.). Theor. Appl. Genet. 2011, 123, 555–569. [Google Scholar] [CrossRef]
- Oliveira, H.R.; Hagenblad, J.; Leino, M.W.; Leigh, F.J.; Lister, D.L.; Penã-Chocarro, L.; Jones, M.K. Wheat in the Mediterranean revisited – tetraploid wheat landraces assessed with elite bread wheat Single Nucleotide Polymorphism markers. BMC Genet. 2014, 15, 54. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotech. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Gao, L.F.; Jia, J.Z.; Kong, X.Y. A SNP-Based Molecular Barcode for characterization of common wheat. PLoS ONE 2016, 11, e0150947. [Google Scholar] [CrossRef]
- Burridge, A.J.; Winfield, M.O.; Allen, A.M.; Wilkinson, P.; Barker, G.L.A.; Coghill, J.; Waterfall, C.; Edwards, K.J. High-Density SNP Genotyping Array for Hexaploid Wheat and Its Relatives. Methods Mol. Biol. 2017, 1679, 293–306. [Google Scholar]
- Lucas, S.J.; Salantur, A.; Yazar, S.; Budak, H. High-throughput SNP genotyping of modern and wild emmer wheat for yield and root morphology using a combined association and linkage analysis. Funct. Int. Genom. 2017, 17, 667–685. [Google Scholar] [CrossRef]
- Edwards, K.J.; Reid, A.L.; Coghill, J.A.; Berry, S.T.; Barker, G.L.A. Multiplex single nucleotide polymorphism (SNP)-based genotyping in allohexaploid wheat using padlock probes. Plant Biotech. J. 2009, 7, 375–390. [Google Scholar] [CrossRef]
- Ganal, M.W.; Altmann, T.; Röder, M.S. SNP identification in crop plants. Curr. Opin. Plant Biol. 2009, 12, 211–217. [Google Scholar] [CrossRef]
- Bozzini, A. Genetics and breeding of durum wheats. Genet. Agraria 1970, 24, 145–193. [Google Scholar]
- Motzo, R.; Giunta, F. The effect of breeding on the phenology of Italian durum wheats: From landraces to modern cultivars. Eur. J. Agron. 2007, 26, 462–470. [Google Scholar] [CrossRef]
- Cauvain, S.P.; Young, L.S. The ICC Handbook of Cereals, Flour, Dough and Product Testing: Methods and Applications; International Association for Cereal Science and Technology (ICC): Vienna, Austria, 2009. [Google Scholar]
- Dexter, J.E.; Matsuo, R.R.; Martin, D.G. The relationship of durum wheat test weight to milling performance and spaghetti quality. Cereal Foods World 1987, 32, 772–777. [Google Scholar]
- Giunta, F.; Motzo, R.; Pruneddu, G. Trends since 1900 in the yield potential of Italian-bred durum wheat cultivars. Eur. J. Agron. 2007, 27, 12–24. [Google Scholar] [CrossRef]
- Dexter, J.E.; Matsuo, R.R. Effect of starchy kernels, immaturity and shrunken kernels on durum wheat quality. Cereal Chem. 1981, 58, 395–400. [Google Scholar]
- Gallo, G.; Lo Bianco, M.; Bognanni, R.; Saimbene, G.; Orlando, A.; Grillo, O.; Saccone, R.; Venora, G. Durum wheat bread: Old sicilian varieties and improved ones. J. Agric. Sci. and Techn. 2010, 4, 10–17. [Google Scholar]
- Peǹa, R.J.; Zarco-Hernandez, J.; Amaya-Celis, A.; Mujeeb-Kazi, A. Relationships between chromosome 1B-encoded glutenin subunit compositions and bread-making quality characteristics of some durum wheat (Triticum turgidum) cultivars. J. Cereal Sci. 1994, 19, 243–249. [Google Scholar] [CrossRef]
- Sissons, M. Role of Durum Wheat Composition on the Quality of Pasta and Bread. Food 2008, 2, 75–90. [Google Scholar]
- Autran, C.; Galterio, G. Associations between electrophoretic composition of proteins, quality characteristics and agronomic attributes of durum wheats II. Protein—Quality associations. J. Cereal Sci. 1989, 9, 195–215. [Google Scholar] [CrossRef]
- Ruiz, M.; Vázquez, J.; Carrillo, J. Genetic bases of grain quality. In Durum Wheat Breeding: Current Approaches and Future Strategies; Royo, C., Nachit, M.M., Di Fonzo, N., Araus, J., Pfeiffer, W., Slafer, G., Eds.; Food Product Press: New York, NY, USA, 2005; pp. 349–378. [Google Scholar]
- Boggini, G.; Doust, M.A.; Annichiarico, P.; Pecetti, L. Yielding ability, yield stability, and quality of exotic durum wheat germplasm in Sicily. Plant Breed. 1997, 116, 541–545. [Google Scholar] [CrossRef]
- Moragues, M.; Zarco-Hernandez, J.; Moralejo, M.A.; Royo, C. Genetic diversity of glutenin protein subunits composition in durum wheat landraces [Triticum turgidum ssp. turgidum convar. durum (Desf.) MacKey] from the Mediterranean basin. Genet. Res. Crop Evol. 2006, 53, 993–1002. [Google Scholar]
- Nazco, R.; Villegas, D.; Ammar, K.; Peña, R.J.; Moragues, M.; Royo, C. Can Mediterranean durum wheat landraces contribute to improved grain quality attributes in modern cultivars? Euphytica 2012, 185, 1–17. [Google Scholar] [CrossRef]
- De Vita, P.; Li Destri Nicosia, O.; Nigro, F.; Platani, C.; Riefolo, C.; Di Fonzo, N.; Cattivelli, L. Breeding progress in morpho-physiological, agronomical and qualitative traits of durum wheat cultivars released in Italy during the 20th century. Eur. J. Agron. 2007, 26, 39–53. [Google Scholar] [CrossRef]
- Nazco, R.; Peña, R.J.; Ammar, K.; Villegas, D.; Crossa, J.; Moragues, M.; Royo, C. Variability in glutenin subunit composition of Mediterranean durum wheat germplasm and its relationship with gluten strength. J. Agric. Sci. 2014, 152, 379–393. [Google Scholar] [CrossRef]
- Janni, M.; Cadonici, S.; Bonas, U.; Grasso, A.; Dahab, A.A.; Visioli, G.; Pignone, D.; Ceriotti, A.; Marmiroli, N. Gene-ecology of durum wheat HMW glutenin reflects their diffusion from the center of origin. Sci. Rep. 2018, 8, 16929. [Google Scholar] [CrossRef]
- De Santis, M.A.; Giuliani, M.M.; Giuzio, L.; De Vita, P.; Lovegrove, A.; Shewry, P.R.; Flagella, Z. Differences in gluten protein composition between old and modern durum wheat genotypes in relation to 20th century breeding in Italy. Eur. J. Agron. 2017, 87, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Spina, A.; Guarnaccia, P.; Blangiforti, S.; Giannone, V.; Amato, C.; Venora, G.; Caruso, P.; Anastasi, U. Popolazioni locali siciliane di frumento tenero: Caratteristiche bioagronomiche e qualitative. In Atti dell’11° Convegno Aistec: I cereali per un sistema agroalimentare di qualità; Roma, 22–24 November 2017; Associazione Italiana di Scienza e Tecnologia dei Cereali - AISTEC: Roma, Italy, 2018; pp. 264–267. [Google Scholar]
- Anastasi, U.; Spina, A.; Amato, C.; Blangiforti, S.; Venora, G.; Giannone, V.; Caruso, P.; Guarnaccia, P. Caratteristiche bioagronomiche e qualitative di popolazioni locali siciliane di frumento duro. In Atti dell’11° Convegno Aistec: I cereali per un sistema agroalimentare di qualità; Roma, 22–24 November 2017; Associazione Italiana di Scienza e Tecnologia dei Cereali - AISTEC: Roma, Italy, 2018; pp. 68–72. [Google Scholar]
- Guarnaccia, P.; Blangiforti, S.; Spina, A.; Caruso, P.; Amato, C.; Mattiolo, E.; Anastasi, U. Old Sicilian wheat landraces as a tool to optimize organic and low-input farming systems. In Proceedings of the ICC/AISTEC Conference “Grains for feeding the world”, EXPO 2015, Milan, Italy, 1–3 July 2015; Italian Association for Cereal Science and Technology: Rome, Italy, 2015; pp. 116–119. [Google Scholar]
- Palumbo, M.; Spina, A.; Boggini, G. Bread-making quality of Italian durum wheat (Triticum durum Desf.) cultivars. Ital. J. Food Sci. 2002, 14, 123–133. [Google Scholar]
- Spina, A.; Sciacca, F.; Palumbo, M.; Boggini, G.; Romano, E. Attitudine panificatoria di varietà di frumento duro. Tec. Molit. 2001, 11, 1096–1106. [Google Scholar]
- Spina, A.; Boggini, G.; Palumbo, M.; Romano, E. Attitudine panificatoria di nuove varietà di frumento duro coltivate in Sicilia. Tec. Molit. 2001, 8, 788–792. [Google Scholar]
- Ficco, D.B.M.; Mastrangelo, A.M.; Trono, D.; Borrelli, G.M.; De Vita, P.; Fares, C.; Beleggia, R.; Platani, C.; Papa, R. The colours of durum wheat: A review. Crop Pasture Sci. 2014, 65, 1–15. [Google Scholar] [CrossRef]
- Digesù, A.M.; Platani, C.; Cattivelli, L.; Mangini, G.; Blanco, A. Genetic variability in yellow pigment components in cultivated and wild tetraploid wheats. J. Cereal Sci. 2009, 50, 210–218. [Google Scholar] [CrossRef]
- Huang, X.; Borner, A.; Roder, M.; Ganal, M. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2002, 105, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Sanguineti, M.C.; Donini, P.; Tuberosa, R. Microsatellite analysis reveals a progressive widening of the genetic basis in the elite durum wheat germplasm. Theor. Appl. Genet. 2003, 107, 783–797. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannik, J.K. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef] [PubMed]
- Riefolo, C.; Ficco, D.; Cattivelli, L.; De Vita, P. Genetic diversity of gluten proteins in T. turgidum L. Cereal Res. Commun. 2011, 39, 405–414. [Google Scholar] [CrossRef]
- O’Neill, R.; Snowdon, R.; Kohler, W. Population genetics: Aspects of biodiversity. Prog. Bot. 2003, 64, 115–137. [Google Scholar]
- ISO (2010). ISO 12099:2010. Animal Feeding Stuffs, Cereals and Milled Cereal Products. Guidelines for the Application of Near Infrared Spectrometry. Geneva, Switzerland: ISO. Available online: https://www.iso.org/standard/51197.html (accessed on 12 March 2019).
- ICC. International Assoc. Cereal Chem. No. 107. Vienna. Austria. 1995. Available online: https://www.icc.or.at/publications/icc-standards/standards-overview/158-gluten-index-method-for-assessing-gluten-strength-in-durum-wheat-triticum-durum (accessed on 19 March 2019).
- AACC International. Method 38-12.02 Wet Gluten, Dry Gluten, Water-Binding Capacity, and Gluten Index. In Approved Methods of Analysis, 11th ed.; AACC International: St. Paul, MN, USA, 1999. [Google Scholar] [CrossRef]
- Sgrulletta, D.; De Stefanis, E.; Conciatori, A. Analisi veloce del colore giallo sulla granella. Il Sistema qualità nella filiera frumento duro. Tec. Molit. 1999, 8, 861–866. [Google Scholar]
- Robertson, A.R. The CIE 1976 Color Difference Formulae. Color Res. Appl. 1977. [Google Scholar] [CrossRef]
- Gomez, K.A.; Gomez, A.A. Statistical Procedures for Agricultural Research, 2nd ed.; J. Wiley & Sons: New York, NY, USA, 1984; pp. 467–471. [Google Scholar]
- Husson, F.; Josse, J.; Lê, S. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Soft. 2008, 25, 1–18. [Google Scholar]
- Sunseri, F.; Lupini, A.; Mauceri, A.; De Lorenzis, G.; Araniti, F.; Brancadoro, L.; Dattola, A.; Gullo, G.; Zappia, R.; Mercati, F. Single nucleotide polymorphism profiles reveal an admixture genetic structure of grapevine germplasm from Calabria, Italy, uncovering its key role for the diversification of cultivars in the Mediterranean Basin. Aust. J. Grape Wine Res. 2018, 24, 345–359. [Google Scholar] [CrossRef]
- Purcell, S.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; Sham, P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Clayton, D. snpStats: SnpMatrix and XSnpMatrix classes and methods. R package version 1.32.0. 2018. [Google Scholar]
- Goudet, G. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol. Ecol. Res. 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Mercati, F.; Riccardi, P.; Leebens-Mack, J.; Abenavoli, M.R.; Falavigna, A.; Sunseri, F. Single nucleotide polymorphism isolated from a novel EST dataset in garden asparagus (Asparagus officinalis L.). Plant Sci. 2013, 203–204, 115–123. [Google Scholar] [CrossRef]
- Yang, M.H.; Fu, Y.B. AveDissR: An R function for assessing genetic distinctness and genetic redundancy. Appl. Plant Sci. 2017, 5, 1700018. [Google Scholar] [CrossRef] [PubMed]
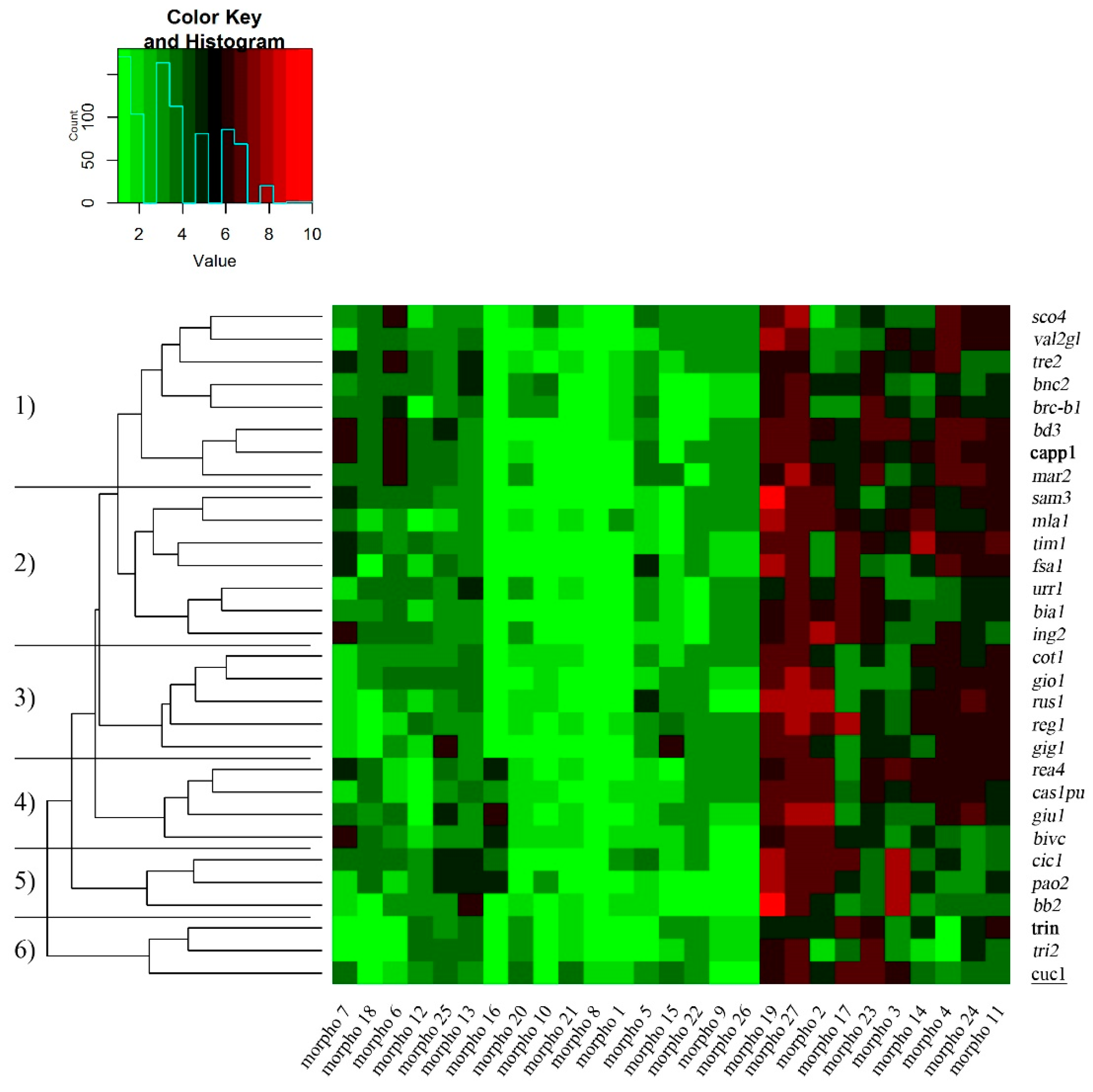
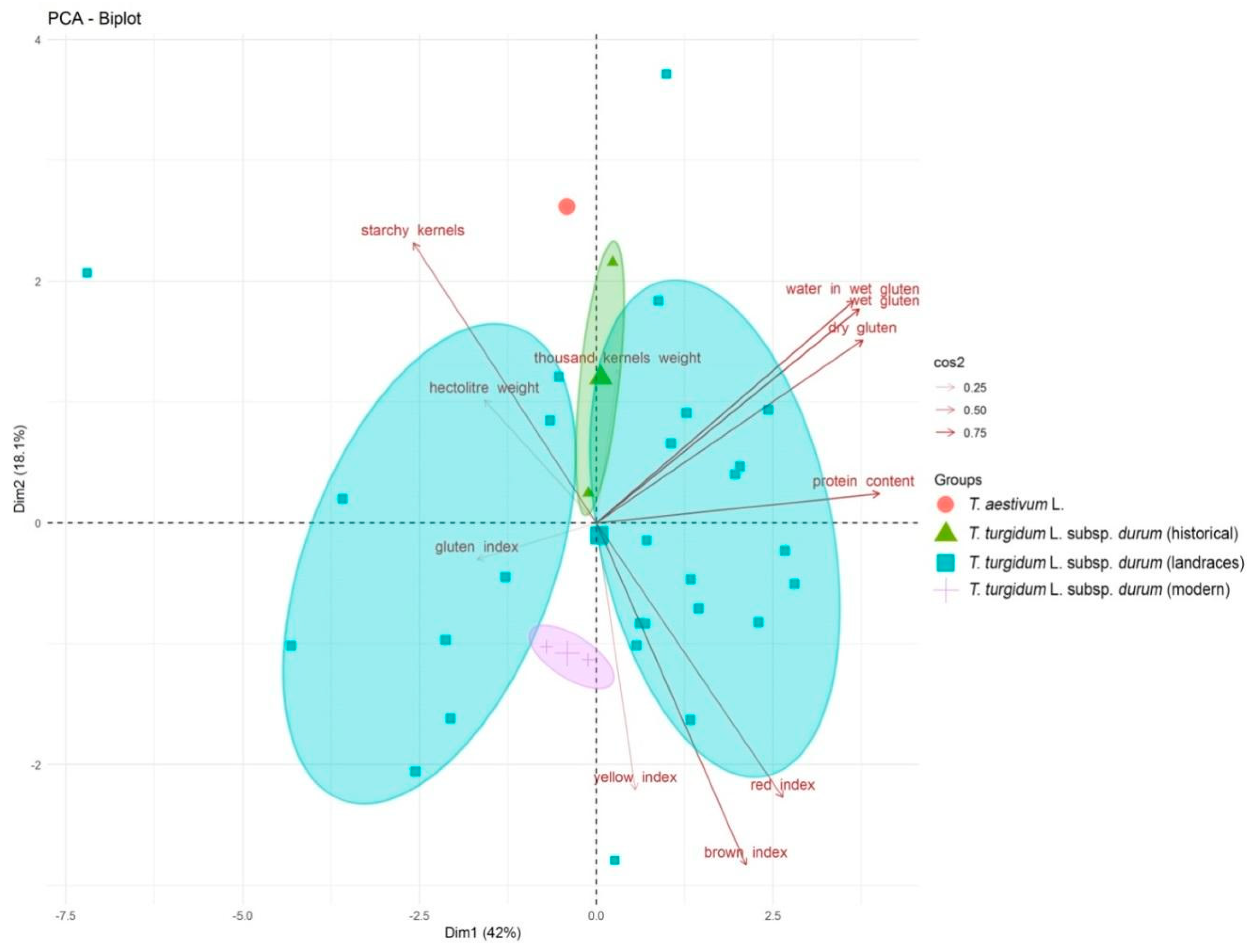
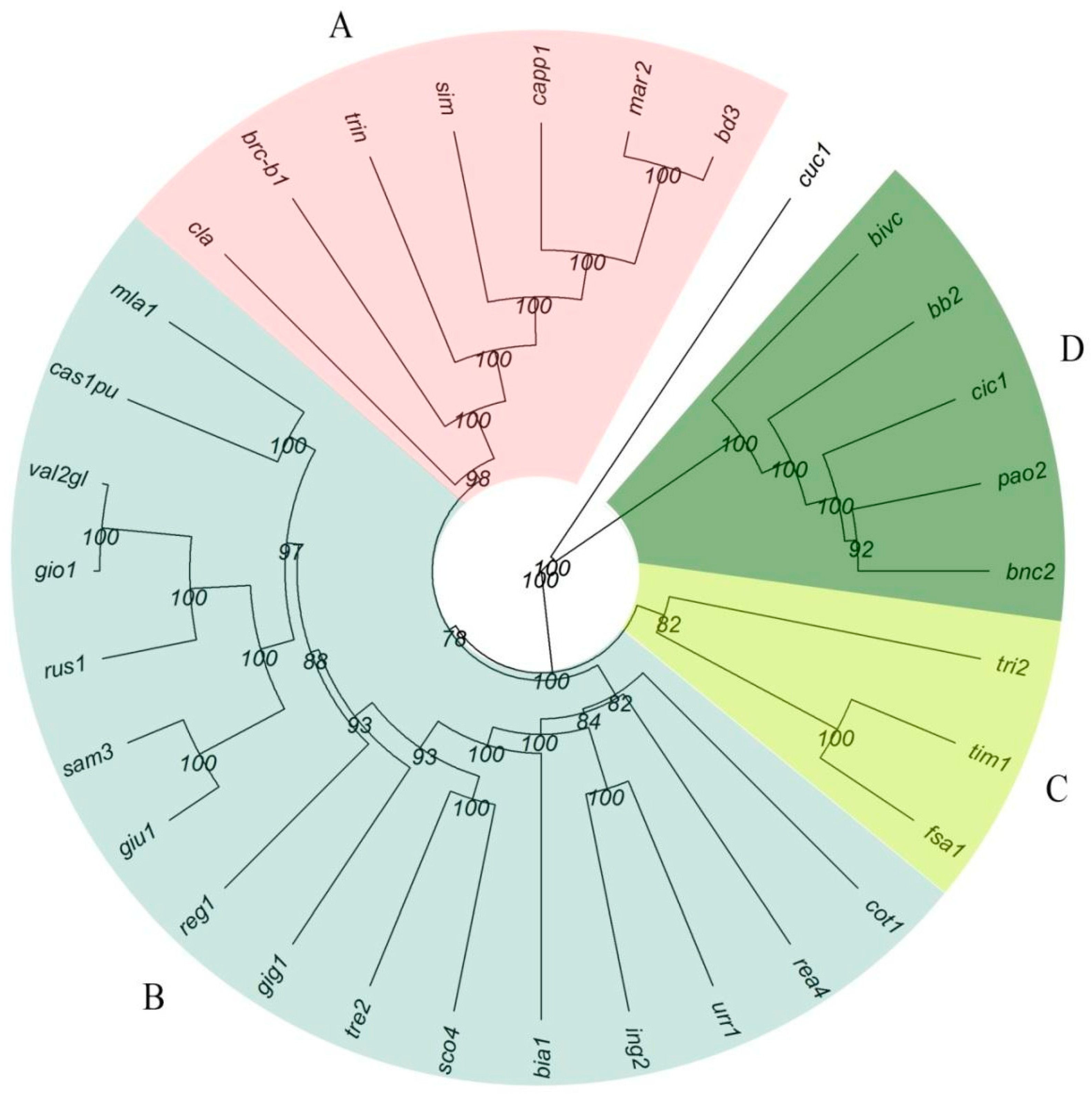
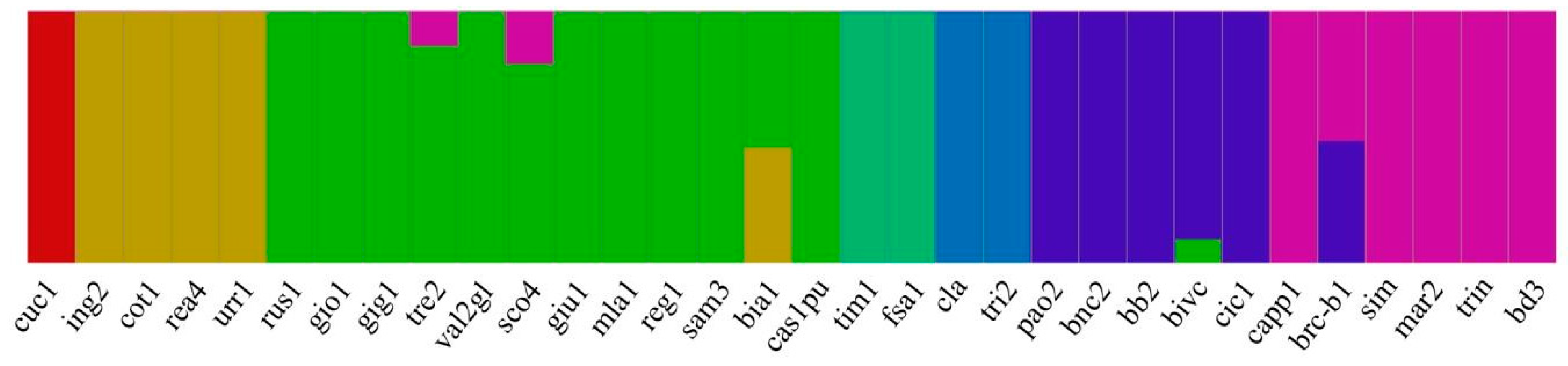
| ID | Test Weight (kg/hL) | Thousand Kernels Weight (g) | Starchy Kernels (%) | Protein Content (% d.m.) | Wet Gluten Content (%) | Dry Gluten Content (%) | Water Binding in Wet Gluten | Gluten Index (0–100) | Brown Index (100−L*) | Red Index (a*) | Yellow Index (b*) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| bia1 | 79.2 ± 0.14e | 41.9 ± 0.14q | 2.5 ± 0.71ln | 15.5 ± 0.07a | 36.00 ± 0.57eg | 11.10 ± 0.2eg | 24.90 ± 0.28e | 48.60 ± 0.81J | 17.75 ± 0.0b | 1.44 ± 0.03bc | 14.80 ± 0.11lm |
| bd3 | 77.6 ± 0.14gh | 39.2 ± 0.14r | 5.5 ± 0.71il | 17.1 ± 0.00a | 42.45 ± 0.64 b | 14.70 ± 0.28a | 27.75 ± 0.35c | 56.65 ± 0.65eg | 14.84 ± 0.12mn | 0.59 ± 0.09ef | 15.57 ± 0.01ij |
| bivc | 80.2 ± 0.28d | 54.9 ± 0.07d | 43.5 ± 2.12cd | 14.1 ± 0.07e | 32.25 ± 0.92 il | 9.65 ± 0.21jk | 22.60 ± 0.71gh | 36.10 ± 1.82mn | 15.64 ± 0.18ij | 0.60 ± 0.01ef | 14.15 ± 0.20no |
| bb2 | 71.8 ± 0.14q | 45.3 ± 0.14no | 15.5 ± 2.12f | 14.9 ± 0.07cd | 31.45 ± 0.92jl | 10.10 ± 0.28hj | 21.35 ± 0.64gi | 54.51 ± 1.33fh | 16.63 ± 0.24de | 0.70 ± 0.08de | 14.06 ± 0.0o |
| bnc2 | 73.9 ± 0.07no | 43.2 ± 0.14p | 9.5 ± 2.12gi | 14.1 ± 0.00e | 34.40 ± 1.13gi | 9.95 ± 0.35hj | 24.45 ± 1.48ef | 37.18 ± 2.07mn | 15.72 ± 0.01hi | 1.44 ± 0.06bc | 13.36 ± 0.12p |
| brc-b1 | 75.6 ± 0.14l | 49.2 ± 0.21ij | 28.5 ± 2.12e | 14.1 ± 0.07e | 42.25 ± 0.50 b | 11.80 ± 0.14ce | 30.45 ± 0.35b | 48.40 ± 0.60 j | 14.50 ± 0.06n | 1.01 ± 0.06d | 15.50 ± 0.09ik |
| capp1 | 81.4 ± 0.07b | 49.6 ± 0.58i | 9.0 ± 1.41gi | 15.1 ± 0.00ab | 38.10 ± 0.85 ce | 12.00 ± 0.14cde | 26.10 ± 0.71cde | 19.67 ± 1.79q | 13.87 ± 0.35o | 0.89 ± 0.0d | 18.24 ± 0.69c |
| cas1pu | 76.8 ± 0.07ik | 49.7 ± 0.21i | 1.0 ± 0.00no | 14.8 ± 0.07d | 32.00 ± 1.27 il | 10.00 ± 0.1hj | 22.00 ± 1.13gi | 29.01 ± 2.82op | 19.15 ± 0.35a | 1.43 ± 0.03bc | 16.06 ± 0.33gi |
| cic1 | 78.4 ± 0.07f | 42.8 ± 0.21 p | 0.5 ± 0.71op | 16.7 ± 0.00a | 40.10 ± 0.85bc | 12.55 ± 0.21bc | 27.55 ± 0.64ce | 35.40 ± 1.37n | 17.87 ± 0.06b | 1.26 ± 0.08cd | 14.95 ± 0.3km |
| cla | 82.6 ± 0.28a | 48.4 ± 0.35k | 11 ± 1.41fh | 13.95 ± 0.07e | 30.79 ± 0.72jl | 9.85 ± 0.78kl | 21.74 ± 0.65gi | 83.07 ± 1.39b | 17.68 ± 0.00bc | 0.59 ± 0.08ef | 19.45 ± 0.18b |
| cot1 | 77.8 ± 0.00gh | 48.8 ± 0.28jk | 46.5 ± 3.54c | 14.0 ± 0.07e | 32.09 ± 1.56hk | 9.85 ± 0.78ik | 23.05 ± 0.78fg | 59.53 ± 1.91e | 16.16 ± 0.07eh | 0.41 ± 0.01fg | 14.70 ± 0.09mn |
| cuc1 | 73.6 ± 0.14op | 47.7 ± 0.21l | 11.5 ± 2.12fh | 15.4 ± 0.00a | 33.55 ± 0.78gj | 11.10 ± 0.42eg | 22.45 ± 0.35gi | 66.31 ± 0.78d | 11.40 ± 0.14p | -0.92 ± 0.02k | 8.74 ± 0.04r |
| fsa1 | 78.5 ± 0.00f | 50.6 ± 0.28h | 2.5 ± 0.71ln | 12.5 ± 0.07h | 20.70 ± 0.42n | 7.00 ± 0.42n | 13.70 ± 0.00l | 18.82 ± 1.66q | 15.14 ± 0.13km | 1.51 ± 0.06ab | 17.98 ± 0.20c |
| gig1 | 74.1 ± 0.00no | 47.5 ± 0.28lm | 1.0 ± 0.00no | 16.2 ± 0.00a | 40.50 ± 0.99bc | 13.15 ± 0.07b | 27.35 ± 0.92ce | 44.43 ± 1.36k | 16.60 ± 0.16df | 1.75 ± 0.05a | 16.68 ± 0.13ef |
| gio1 | 75.5 ± 0.21lm | 53.5 ± 0.14e | 1.5 ± 0.71mo | 14.7 ± 0.07d | 30.20 ± 0.71 l | 9.65 ± 0.21jk | 20.55 ± 0.49l | 26.47 ± 1.72p | 15.86 ± 0.12gi | 0.61 ± 0.05df | 17.41 ± 0.13d |
| giu1 | 76.5 ± 0.21jk | 45.0 ± 0.07o | 2.5 ± 0.71ln | 15.5 ± 0.07a | 35.25 ± 0.64fh | 10.80 ± 0.28fh | 24.45 ± 0.35ef | 34.17 ± 1.19n | 16.19 ± 0.23eh | 0.22 ± 0.09gh | 16.55 ± 0.22eg |
| ing2 | 77.2 ± 0.14hi | 49.3 ± 0.07ij | 3.5 ± 0.7jn | 16.1 ± 0.00a | 39.45 ± 0.35 cd | 12.30 ± 0.14bd | 27.15 ± 0.21cd | 43.47 ± 0.51k | 16.32 ± 0.09eg | 0.12 ± 0.06 | 15.65 ± 0.08ij |
| mar2 | 75.4 ± 0.07lm | 51.5 ± 0.07g | 14.0 ± 1.4fg | 12.9 ± 0.00g | 20.20 ± 1.70 n | 7.75 ± 0.35no | 12.45 ± 1.34lm | 78.64 ± 1.79c | 15.77 ± 0.10hi | 1.39 ± 0.0bc | 17.08 ± 0.13de |
| mla1 | 75.1 ± 0.14m | 46.9 ± 0.14m | 7.0 ± 1.4hij | 12.9 ± 0.00g | 18.05 ± 1.061 o | 6.75 ± 0.35m | 11.30 ± 0.71mn | 91.12 ± 0.52a | 15.60 ± 0.06ik | 1.21 ± 0.04cd | 16.93 ± 0.02de |
| pao2 | 76.9 ± 0.07ik | 57.4 ± 0.14b | 66.5 ± 3.54b | 15.0 ± 0.00bc | 45.40 ± 0.71a | 12.50 ± 0.42bc | 32.90 ± 0.28a | 50.88 ± 0.77ij | 14.98 ± 0.06lm | 1.15 ± 0.01d | 11.64 ± 0.05q |
| rea4 | 73.7 ± 0.14no | 41.4 ± 0.42q | 9.0 ± 1.41gi | 15.0 ± 0.07bc | 31.75 ± 0.35jl | 10.80 ± 0.42fh | 20.95 ± 0.07hi | 52.12 ± 0.53hi | 16.12 ± 0.13fh | 1.01 ± 0.03d | 18.03 ± 0.11c |
| reg1 | 76.4 ± 0.21k | 56.6 ± 0.28c | 4.0 ± 1.41jm | 16.3 ± 0.00a | 39.00 ± 0.28cd | 11.35 ± 0.07dg | 27.65 ± 0.21c | 42.31 ± 0.42kl | 16.52 ± 0.05df | 0.96 ± 0.01d | 15.18 ± 0.03im |
| rus1 | 77.1 ± 0.07ik | 57.4 ± 0.21b | 10 ± 1.41fi | 13.3 ± 0.07f | 24.90 ± 0.28 m | 8.80 ± 0.28l | 16.10 ± 0.00k | 57.83 ± 0.48ef | 15.41 ± 0.01il | 0.83 ± 0.06de | 16.26 ± 0.22fh |
| sam3 | 80.2 ± 0.14d | 51.2 ± 0.2gh | 6.5 ± 0.71hk | 14.9 ± 0.07ce | 38.80 ± 0.57 cd | 11.65 ± 0.07cf | 27.15 ± 0.49ce | 48.45 ± 0.75j | 16.50 ± 0.45df | 0.45 ± 0.03fg | 16.68 ± 0.20ef |
| sco4 | 73.3 ± 0.21p | 44.7 ± 0.14o | 0.0 ± 0.00p | 16.7 ± 0.07a | 38.45 ± 0.64ce | 12.10 ± 0.14cd | 26.35 ± 0.78ce | 54.22 ± 0.76gi | 17.29 ± 0.09c | 0.82 ± 0.03de | 18.37 ± 0.06c |
| sim | 79.1 ± 0.14e | 52.3 ± 0.28f | 3.0 ± 1.41kn | 14.65 ± 0.07d | 31.63 ± 0.56jl | 9,30 ± 0.14jl | 22.33 ± 0.42gi | 86.30 ± 0.25b | 17.29 ± 0.18c | 0.60 ± 0.04ef | 20.06 ± 0.29a |
| tim1 | 75.3 ± 0.28lm | 33.5 ± 0.00s | 9.0 ± 1.41gi | 13.5 ± 0.14f | 26.95 ± 0.50 m | 8.55 ± 0.07l | 18.40 ± 0.42j | 30.60 ± 1.27o | 19.29 ± 0.11a | 1.87 ± 0.01a | 14.81 ± 0.02lm |
| tre2 | 80.3 ± 0.14cd | 47.5 ± 0.21lm | 26.5 ± 2.12 e | 11.9 ± 0.07n | 14.00 ± 0.28 p | 5.30 ± 0.14p | 8.70 ± 0.14o | 67.14 ± 0.66d | 15.25 ± 0.01jm | -0.05 ± 0.02i | 16.28 ± 0.01fh |
| trin | 80.3 ± 0.14cd | 56.5 ± 0.21c | 7.0 ± 2.83hj | 14.80 ± 0.00ij | 32.35 ± 1.34 il | 10.70 ± 0.42gi | 21.65 ± 0.92gi | 83.60 ± 0.68b | 16.28 ± 0.29eg | 0.70 ± 0.16j | 15.74 ± 0.17hj |
| tri2 | 82.3 ± 0.07a | 39.3 ± 0.07r | 96.0 ± 1.41a | 8.5 ± 0.04k | 15.00 ± 0.14p | 5.40 ± 0.14p | 9.60 ± 0.28no | 77.33 ± 0.21c | 11.24 ± 0.04p | −0.69 ± 0.28j | 13.49 ± 0.11p |
| urr1 | 80.7 ± 0.00c | 45.7 ± 0.14n | 38.0 ± 1.41d | 11.5 ± 0.07j | 18.85 ± 0.07no | 6.20 ± 0.00o | 12.65 ± 0.07lm | 38.99 ± 0.23lm | 15.75 ± 0.18hi | 0.24 ± 0.0gh | 11.19 ± 0.16q |
| val2gl | 82.3 ± 0.28a | 59.2 ± 0.21a | 7.0 ± 1.41hj | 15.7 ± 0.00a | 37.15 ± 0.07df | 11.50 ± 0.14dg | 25.65 ± 0.07de | 34.05 ± 0.13n | 16.84 ± 0.01d | 0.84 ± 0.0de | 15.34 ± 0.31jl |
| Parameters | Values |
|---|---|
| N † | 32 |
| Total number of loci | 81,587 |
| No. of failed loci | 5,594 |
| No. of monomorphic loci | 41,926 |
| No. of used loci | 18,170 |
| No. of polymorphic loci | 13,528 |
| MAF # | 0.232 |
| ID | Accession | Origin |
|---|---|---|
| bb2 | Bufala bianca | In collection at the ESS* and sampled in ‘99-’04 on farm in Randazzo (Catania) |
| bd3 | Bidì | IPK TRI 26213, in collection at the ESS* since 2004 (selection from Tunisian landrace Bidì, line AP4) |
| bia1 | Biancuccia | In collection at the ESS* and sampled in ‘99-’04 on farm in Salemi (Trapani) |
| bivc | Casedda Bivona | In collection at the ESS* and sampled in ‘99-’04 on farm in S. Stefano Quisqinia (Agrigento) |
| bnc2 | Bufala nera corta | IPK3517, in collection at the ESS* since 2004 |
| brc-b1 | Bufala rossa corta b | In collection at the ESS* and sampled in ‘99-’04 on farm in Randazzo (Catania) |
| capp1 | Cappelli | Historical variety - Old collection of ESS* (selection from Tunisian landrace Jenah Rhetifah) |
| cas1pu | Castiglione pubescente | CGN 8213, in collection at the ESS* since 2004 |
| cic1 | Ciciredda | In collection at the ESS* and sampled in ‘99-’04 on farm in Maletto (Catania) |
| cot1 | Cotrone | USDA 157975, in collection at the ESS* since 2004 |
| cuc1 | Cuccitta | In collection at the ESS* and sampled in ‘99-’04 on farm in Fiumedinisi (Messina) |
| fsa1 | Francesa | Old collection of ESS* |
| gig1 | Gigante | CGN 8206, in collection at the ESS* since 2004 |
| gio1 | Gioia | IPK 3851, in collection at the ESS* since 2004 |
| giu1 | Giustalisa | USDA 278354, in collection at the ESS* since 2004 |
| ing2 | Inglesa | IPK 3519, in collection at the ESS* since 2004 |
| mar2 | Margherito | In collection at the ESS* and sampled in ‘99-’04 on farm in Chiaramonte Gulfi (Ragusa) (selection from Tunisian landrace Mahmoudi) |
| mla1 | Martinella | USDA 157971, in collection at the ESS* since 2004 |
| pao2 | Paola | In collection at the ESS* and sampled in ‘99-’04 on farm in Randazzo (Catania) |
| rea4 | Realforte | IPK TRI 28452, in collection at the ESS* since 2004 |
| reg1 | Regina | Old collection of ESS* |
| rus1 | Russello | In collection at the ESS* and sampled in ‘99-’04 on farm in Randazzo (Catania) |
| sam3 | Sammartinara | USDA 157958, in collection at the ESS* since 2004 |
| sco4 | Scorsonera | In collection at the ESS* and sampled in ‘99-’04 on farm in Santa Croce Camerina (Ragusa) |
| tim1 | Timilia | In collection at the ESS* and sampled in ‘99-’04 on farm in Maletto (Catania) |
| tre2 | Trentino | USDA 157965, in collection at the ESS* since 2004 |
| tri2 | Tripolino | Old collection of ESS* |
| urr1 | Urria | In collection at the ESS* and sampled in ‘99-’04 on farm in Santa Croce Camerina (Ragusa) |
| val2gl | Vallelunga glabra | USDA 157979, in collection at the ESS since 2004 |
| trin | Trinakria | Historical variety - Old collection of ESS*(B14 x Capeiti 8) |
| sim | Simeto | Modern variety (Capeiti 8 × Valnova) |
| cla | Claudio | Modern variety (Cimmyt selection × Durango) × (IS1938 × Grazia) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiore, M.C.; Mercati, F.; Spina, A.; Blangiforti, S.; Venora, G.; Dell’Acqua, M.; Lupini, A.; Preiti, G.; Monti, M.; Pè, M.E.; et al. High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily. Plants 2019, 8, 116. https://doi.org/10.3390/plants8050116
Fiore MC, Mercati F, Spina A, Blangiforti S, Venora G, Dell’Acqua M, Lupini A, Preiti G, Monti M, Pè ME, et al. High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily. Plants. 2019; 8(5):116. https://doi.org/10.3390/plants8050116
Chicago/Turabian StyleFiore, Maria Carola, Francesco Mercati, Alfio Spina, Sebastiano Blangiforti, Gianfranco Venora, Matteo Dell’Acqua, Antonio Lupini, Giovanni Preiti, Michele Monti, Mario Enrico Pè, and et al. 2019. "High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily" Plants 8, no. 5: 116. https://doi.org/10.3390/plants8050116
APA StyleFiore, M. C., Mercati, F., Spina, A., Blangiforti, S., Venora, G., Dell’Acqua, M., Lupini, A., Preiti, G., Monti, M., Pè, M. E., & Sunseri, F. (2019). High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily. Plants, 8(5), 116. https://doi.org/10.3390/plants8050116








