Gene Regulation Mediated by microRNA-Triggered Secondary Small RNAs in Plants
Abstract
1. Introduction
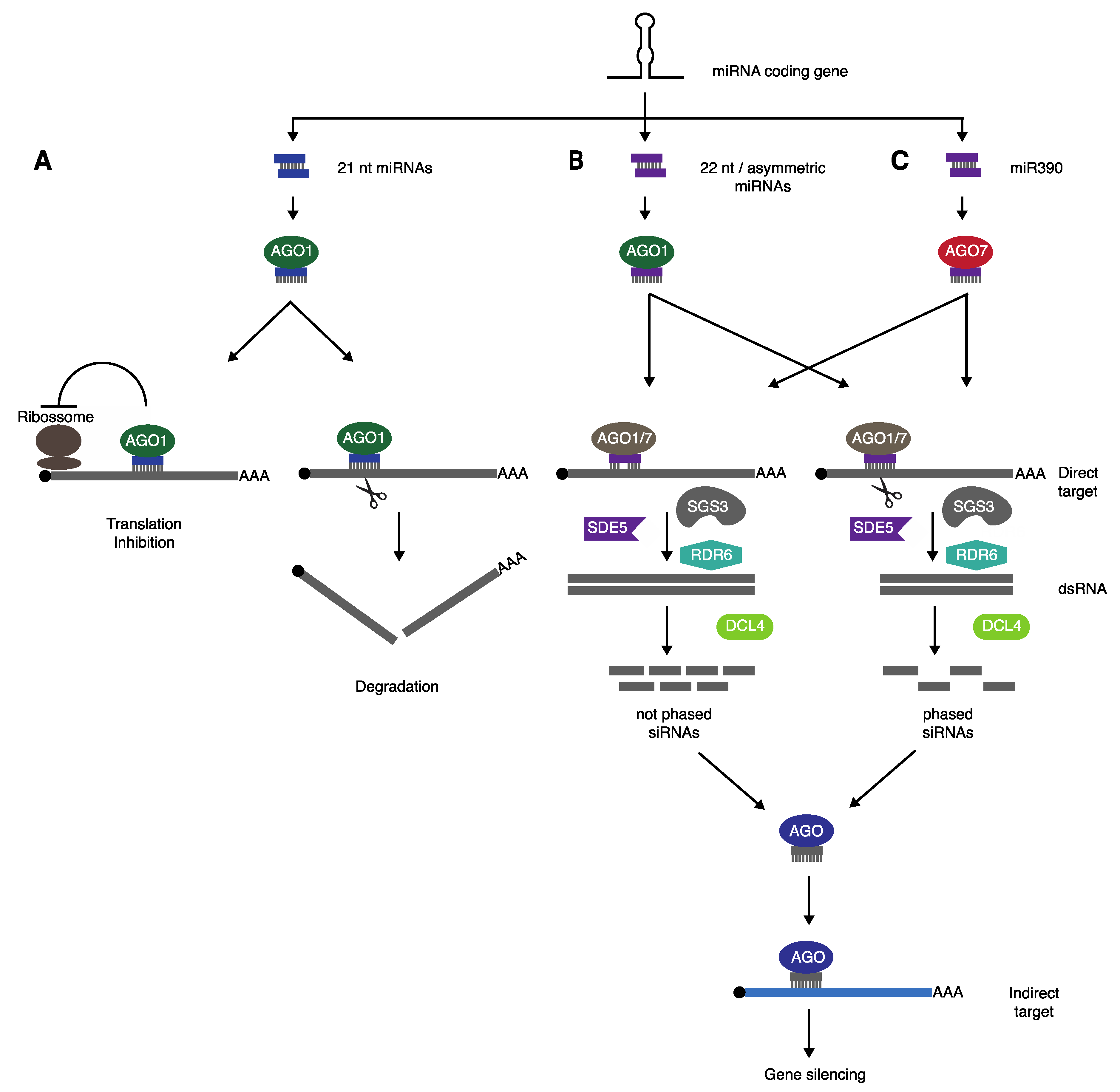
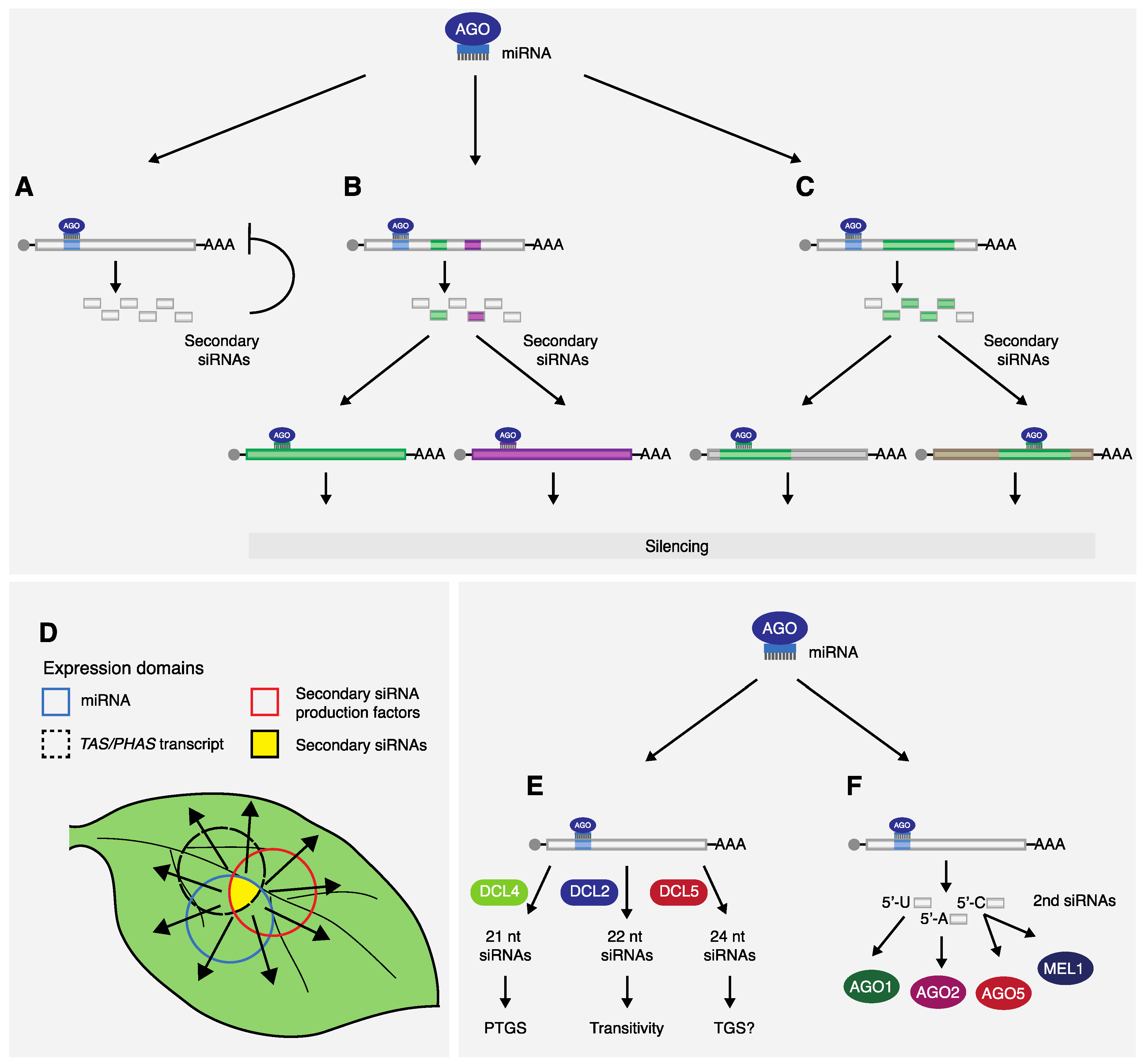
2. Biogenesis of miRNA-Triggered Secondary siRNAs
2.1. “Two-Hit” Model
2.2. “One-Hit” Model
2.3. A Unified “One-Hit” Model
2.4. Other Elements Involved in Secondary siRNA Production
3. Features and Advantages of miRNA-Triggered Secondary siRNA Gene Regulation
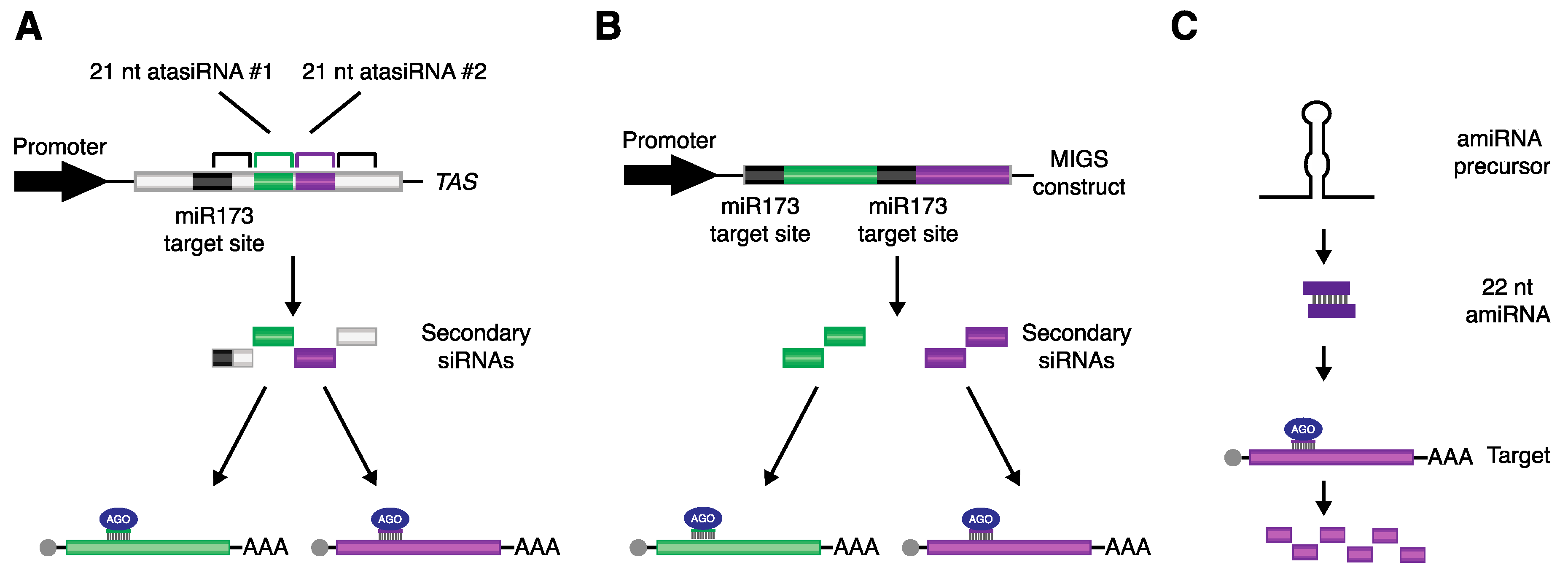
4. Utilizing miRNA-Triggered Secondary siRNAs to Promote Directed Gene Silencing
5. Conclusion and Final Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small rnas in arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Castillo-González, C.; Yu, B.; Zhang, X. The functions of plant small rnas in development and in stress responses. Plant J. 2017, 90, 654–670. [Google Scholar] [PubMed]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. Sgs3 and sgs2/sde1/rdr6 are required for juvenile development and the production of trans-acting sirnas in arabidopsis. Genes Dev. 2004, 18, 2368–2379. [Google Scholar] [CrossRef]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.C.; Hilbert, J.-L.; Bartel, D.P.; Crété, P. Endogenous trans-acting sirnas regulate the accumulation of arabidopsis mrnas. Mol. Cell 2004, 16, 69–79. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. Microrna-directed phasing during trans-acting sirna biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Carles, C.C.; Osmont, K.S.; Fletcher, J.C. A database analysis method identifies an endogenous trans-acting short-interfering rna that targets the arabidopsis arf2, arf3, and arf4 genes. Proc. Natl. Acad. Sci. USA 2005, 102, 9703–9708. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting sirnas in arabidopsis. Genes Dev. 2005, 19, 2164–2175. [Google Scholar] [CrossRef]
- Hernandez-Pinzon, I.; Yelina, N.E.; Schwach, F.; Studholme, D.J.; Baulcombe, D.; Dalmay, T. Sde5, the putative homologue of a human mrna export factor, is required for transgene silencing and accumulation of trans-acting endogenous sirna. Plant J. 2007, 50, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Jauvion, V.; Elmayan, T.; Vaucheret, H. The conserved rna trafficking proteins hpr1 and tex1 are involved in the production of endogenous and exogenous small interfering rna in arabidopsis. Plant Cell 2010, 22, 2697–2709. [Google Scholar] [CrossRef] [PubMed]
- Gasciolli, V.; Mallory, A.C.; Bartel, D.P.; Vaucheret, H. Partially redundant functions of arabidopsis dicer-like enzymes and a role for dcl4 in producing trans-acting sirnas. Curr. Biol. 2005, 15, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Allen, E.; Wilken, A.; Carrington, J.C. Dicer-like 4 functions in trans-acting small interfering rna biogenesis and vegetative phase change in arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 12984–12989. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Kasprzewska, A.; Tennessen, K.; Fernandes, J.; Nan, G.-L.; Walbot, V.; Sundaresan, V.; Vance, V.; Bowman, L.H. Clusters and superclusters of phased small rnas in the developing inflorescence of rice. Genome Res. 2009, 19, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering rnas in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Vazquez, F.; Hohn, T. Biogenesis and biological activity of secondary sirnas in plants. Scientifica 2013, 2013, 783253. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of micrornas in arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef]
- Arif, M.A.; Fattash, I.; Ma, Z.; Cho, S.H.; Beike, A.K.; Reski, R.; Axtell, M.J.; Frank, W. Dicer-like3 activity in physcomitrella patens dicer-like4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 2012, 5, 1281–1294. [Google Scholar] [CrossRef]
- Li, F.; Orban, R.; Baker, B. Somart: A web server for plant mirna, tasirna and target gene analysis. Plant J. 2012, 70, 891–901. [Google Scholar] [CrossRef]
- Zhang, C.; Li, G.; Wang, J.; Fang, J. Identification of trans-acting sirnas and their regulatory cascades in grapevine. Bioinformatics 2012, 28, 2561–2568. [Google Scholar] [CrossRef]
- Zuo, J.; Wang, Q.; Han, C.; Ju, Z.; Cao, D.; Zhu, B.; Luo, Y.; Gao, L. Srnaome and degradome sequencing analysis reveals specific regulation of srna in response to chilling injury in tomato fruit. Physiol. Plant 2017, 160, 142–154. [Google Scholar] [CrossRef]
- Zhai, J.; Jeong, D.-H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. Micrornas as master regulators of the plant nb-lrr defense gene family via the production of phased, trans-acting sirnas. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef]
- Deng, P.; Muhammad, S.; Cao, M.; Wu, L. Biogenesis and regulatory hierarchy of phased small interfering rnas in plants. Plant Biotechnol. J. 2018, 16, 965–975. [Google Scholar] [CrossRef]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A two-hit trigger for sirna biogenesis in plants. Cell 2006, 127, 565–577. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Yoo, S.J.; Fahlgren, N.; Gilbert, S.D.; Howell, M.D.; Sullivan, C.M.; Alexander, A.; Nguyen, G.; Allen, E.; Ahn, J.H.; et al. Ago1-mir173 complex initiates phased sirna formation in plants. Proc. Natl. Acad. Sci. USA 2008, 105, 20055–20062. [Google Scholar] [CrossRef]
- Felippes, F.F.; Weigel, D. Triggering the formation of tasirnas in arabidopsis thaliana: The role of microrna mir173. EMBO Rep. 2009, 10, 264–270. [Google Scholar] [CrossRef]
- Chen, H.-M.; Chen, L.-T.; Patel, K.; Li, Y.-H.; Baulcombe, D.C.; Wu, S.-H. 22-nucleotide rnas trigger secondary sirna biogenesis in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 15269–15274. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Burke, R.T.; Takeda, A.; Sullivan, C.M.; Gilbert, S.D.; Montgomery, T.A.; Carrington, J.C. Unique functionality of 22-nt mirnas in triggering rdr6-dependent sirna biogenesis from target transcripts in arabidopsis. Nat. Struct. Mol. Biol. 2010, 17, 997–1003. [Google Scholar] [CrossRef]
- Manavella, P.A.; Koenig, D.; Weigel, D. Plant secondary sirna production determined by microrna-duplex structure. Proc. Natl. Acad. Sci. USA 2012, 109, 2461–2466. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Yu, Y.; Liu, L.; Zhang, Y.; Baldrich, P.; Dai, Q.; Chen, X.; Meyers, B.C. Biogenesis of a 22-nt microrna in phaseoleae species by precursor-programmed uridylation. Proc. Natl. Acad. Sci. USA 2018, 115, 8037–8042. [Google Scholar] [CrossRef]
- de Felippes, F.F.; Marchais, A.; Sarazin, A.; Oberlin, S.; Voinnet, O. A single mir390 targeting event is sufficient for triggering tas3-tasirna biogenesis in arabidopsis. Nucleic Acids Res. 2017, 45, 5539–5554. [Google Scholar] [CrossRef] [PubMed]
- Krasnikova, M.S.; Milyutina, I.A.; Bobrova, V.K.; Ozerova, L.V.; Troitsky, A.V.; Solovyev, A.G.; Morozov, S.Y. Novel mir390-dependent transacting sirna precursors in plants revealed by a pcr-based experimental approach and database analysis. J. Biomed. Biotechnol. 2009, 2009, 952304–952309. [Google Scholar] [CrossRef]
- Xia, R.; Zhu, H.; An, Y.-Q.; Beers, E.P.; Liu, Z. Apple mirnas and tasirnas with novel regulatory networks. Genome Biol. 2012, 13, R47. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Arikit, S.; Meyers, B.C. Extensive families of mirnas and phas loci in norway spruce demonstrate the origins of complex phasirna networks in seed plants. Mol. Biol. Evol. 2015, 32, 2905–2918. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of argonaute7-mir390 interaction and dual functionality in tas3 trans-acting sirna formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Arribas-Hernández, L.; Marchais, A.; Poulsen, C.; Haase, B.; Hauptmann, J.; Benes, V.; Meister, G.; Brodersen, P. The slicer activity of argonaute1 is required specifically for the phasing, not production, of trans-acting short interfering rnas in arabidopsis. Plant Cell 2016, 28, 1563–1580. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small rnas into arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef]
- Takeda, A.; Iwasaki, S.; Watanabe, T.; Utsumi, M.; Watanabe, Y. The mechanism selecting the guide strand from small rna duplexes is different among argonaute proteins. Plant Cell Physiol. 2008, 49, 493–500. [Google Scholar] [CrossRef]
- Endo, Y.; Iwakawa, H.O.; Tomari, Y. Arabidopsis argonaute7 selects mir390 through multiple checkpoints during risc assembly. EMBO Rep 2013, 14, 652–658. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature 2008, 456, 209–213. [Google Scholar] [CrossRef]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with host sgs3 is required for suppression of rna silencing by tomato yellow leaf curl virus v2 protein. Proc. Natl. Acad. Sci. USA 2008, 105, 157–161. [Google Scholar] [CrossRef]
- Elmayan, T.; Adenot, X.; Gissot, L.; Lauressergues, D.; Gy, I.; Vaucheret, H. A neomorphic sgs3 allele stabilizing mirna cleavage products reveals that sgs3 acts as a homodimer. FEBS J. 2009, 276, 835–844. [Google Scholar] [CrossRef]
- Kumakura, N.; Takeda, A.; Fujioka, Y.; Motose, H.; Takano, R.; Watanabe, Y. Sgs3 and rdr6 interact and colocalize in cytoplasmic sgs3/rdr6-bodies. FEBS Lett. 2009, 583, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Jouannet, V.; Moreno, A.B.; Elmayan, T.; Vaucheret, H.; Crespi, M.D.; Maizel, A. Cytoplasmic arabidopsis ago7 accumulates in membrane-associated sirna bodies and is required for ta-sirna biogenesis. EMBO J. 2012, 31, 1704–1713. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Sarazin, A.; Jullien, P.E.; Bologna, N.G.; Oberlin, S.; Voinnet, O. DNA methylation influences the expression of dicer-like4 isoforms, which encode proteins of alternative localization and function. Plant Cell 2016, 28, 2786–2804. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Le, B.; Ma, X.; Li, S.; You, C.; Yu, Y.; Zhang, B.; Liu, L.; Gao, L.; Shi, T.; et al. Biogenesis of phased sirnas on membrane-bound polysomes in arabidopsis. elife 2016, 5, e22750. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Iki, T.; Tsutsui, Y.; Miyashita, K.; Poethig, R.S.; Habu, Y.; Ishikawa, M. 3′ fragment of mir173-programmed risc-cleaved rna is protected from degradation in a complex with risc and sgs3. Proc. Natl. Acad. Sci. USA 2013, 110, 4117–4122. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Iki, T.; Numa, H.; Miyashita, K.; Meshi, T.; Ishikawa, M. A short open reading frame encompassing the microrna173 target site plays a role in trans-acting small interfering rna biogenesis. Plant Physiol. 2016, 171, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Yelina, N.E.; Smith, L.M.; Jones, A.M.; Patel, K.; Kelly, K.A.; Baulcombe, D.C. Putative arabidopsis tho/trex mrna export complex is involved in transgene and endogenous sirna biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 13948–13953. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.Y.; Lee, W.C.; Chou, H.C.; Chen, A.P.; Chou, S.J.; Chen, H.M. Global analysis of truncated rna ends reveals new insights into ribosome stalling in plants. Plant Cell 2016, 28, 2398–2416. [Google Scholar] [CrossRef]
- Bazin, J.; Baerenfaller, K.; Gosai, S.J.; Gregory, B.D.; Crespi, M.; Bailey-Serres, J. Global analysis of ribosome-associated noncoding rnas unveils new modes of translational regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E10018–E10027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ng, D.W.; Lu, J.; Chen, Z.J. Roles of target site location and sequence complementarity in trans-acting sirna formation in arabidopsis. Plant J. 2012, 69, 217–226. [Google Scholar] [CrossRef]
- Adenot, X.; Elmayan, T.; Lauressergues, D.; Boutet, S.; Bouché, N.; Gasciolli, V.; Vaucheret, H. Drb4-dependent tas3 trans-acting sirnas control leaf morphology through ago7. Curr. Biol. 2006, 16, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of auxin response factor3 by tas3 ta-sirna affects developmental timing and patterning in arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.-K.; Lee, W.S. Auxin regulation of the microrna390-dependent transacting small interfering rna pathway in arabidopsis lateral root development. Nucleic Acids Res. 2009, 38, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. Mir390, arabidopsis tas3 tasirnas, and their auxin response factor targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Mizunashi, K.; Tanaka, M.; Kaminuma, E.; Nguyen, A.H.; Nakajima, M.; Kim, J.-M.; Nguyen, D.V.; Toyoda, T.; Seki, M. Tasirna-arf pathway moderates floral architecture in arabidopsis plants subjected to drought stress. BioMed Res. Int. 2014, 2014, 303451. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ke, L.; Wu, G.; Xu, Y.; Wu, X.; Xia, R.; Deng, X.; Xu, Q. Mir3954 is a trigger of phasirnas that affects flowering time in citrus. Plant J. 2017, 92, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ren, S.; Guo, J.; Wang, Q.; Zhang, X.; Liao, P.; Li, S.; Sunkar, R.; Zheng, Y. Genome-wide identification and comprehensive analysis of micrornas and phased small interfering rnas in watermelon. BMC Genomics 2018, 19, 111. [Google Scholar] [CrossRef]
- Jagadeeswaran, G.; Zheng, Y.; Li, Y.-F.; Shukla, L.I.; Matts, J.; Hoyt, P.; Macmil, S.L.; Wiley, G.B.; Roe, B.A.; Zhang, W.; et al. Cloning and characterization of small rnas from medicago truncatula reveals four novel legume-specific microrna families. New Phytol. 2009, 184, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Rajeswaran, R.; Aregger, M.; Zvereva, A.S.; Borah, B.K.; Gubaeva, E.G.; Pooggin, M.M. Sequencing of rdr6-dependent double-stranded rnas reveals novel features of plant sirna biogenesis. Nucleic Acids Res. 2012, 40, 6241–6254. [Google Scholar] [CrossRef]
- Guan, X.; Pang, M.; Nah, G.; Shi, X.; Ye, W.; Stelly, D.M.; Chen, Z.J. Mir828 and mir858 regulate homoeologous myb2 gene functions in arabidopsis trichome and cotton fibre development. Nat. Commun. 2014, 5, 3050. [Google Scholar] [CrossRef]
- Axtell, M.J.; Snyder, J.A.; Bartel, D.P. Common functions for diverse small rnas of land plants. Plant Cell 2007, 19, 1750–1769. [Google Scholar] [CrossRef]
- Xia, R.; Meyers, B.C.; Liu, Z.; Beers, E.P.; Ye, S.; Liu, Z.; Liu, Z. Microrna superfamilies descended from mir390 and their roles in secondary small interfering rna biogenesis in eudicots. Plant Cell 2013, 25, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Nogueira, F.T.S.; Howell, M.D.; Montgomery, T.A.; Carrington, J.C.; Timmermans, M.C.P. Pattern formation via small rna mobility. Genes Dev. 2009, 23, 549–554. [Google Scholar] [CrossRef]
- Schwab, R.; Maizel, A.; Ruiz-Ferrer, V.; Garcia, D.; Bayer, M.; Crespi, M.; Voinnet, O.; Martienssen, R.A. Endogenous tasirnas mediate non-cell autonomous effects on gene regulation in arabidopsis thaliana. PLoS ONE 2009, 4, e5980. [Google Scholar] [CrossRef]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate arabidopsis organ asymmetry via modulation of kanadi activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, P.; Zhai, J.; Zhou, M.; Ma, L.; Liu, B.; Jeong, D.-H.; Nakano, M.; Cao, S.; Liu, C.; et al. Roles of dcl4 and dcl3b in rice phased small rna biogenesis. Plant J. 2011, 69, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Zhang, H.; Arikit, S.; Huang, K.; Nan, G.L.; Walbot, V.; Meyers, B.C. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasirnas in maize anthers. Proc. Natl. Acad. Sci. USA 2015, 112, 3146–3151. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Chen, C.; Pokhrel, S.; Ma, W.; Huang, K.; Patel, P.; Wang, F.; Xu, J.; Liu, Z.; Li, J.; et al. 24-nt reproductive phasirnas are broadly present in angiosperms. Nat Commun 2019, 10, 627. [Google Scholar] [CrossRef]
- Komiya, R.; Ohyanagi, H.; Niihama, M.; Watanabe, T.; Nakano, M.; Kurata, N.; Nonomura, K.-I. Rice germline-specific argonaute mel1 protein binds to phasirnas generated from more than 700 lincrnas. Plant J. 2014, 78, 385–397. [Google Scholar] [CrossRef]
- Nonomura, K.I.; Morohoshi, A.; Nakano, M.; Eiguchi, M.; Miyao, A.; Hirochika, H.; Kurata, N. A germ cell specific gene of the argonaute family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 2007, 19, 2583–2594. [Google Scholar] [CrossRef] [PubMed]
- de Felippes, F.F.; Ott, F.; Weigel, D. Comparative analysis of non-autonomous effects of tasirnas and mirnas in arabidopsis thaliana. Nucleic Acids Res. 2011, 39, 2880–2889. [Google Scholar] [CrossRef] [PubMed]
- McHale, M.; Eamens, A.L.; Finnegan, E.J.; Waterhouse, P.M. A 22-nt artificial microrna mediates widespread rna silencing in arabidopsis. Plant J. 2013, 76, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Senthil-Kumar, M.; Mysore, K.S. Advances in plant gene silencing methods. In Plant Gene Silencing; Mysore, K.S., Senthil-Kumar, M., Eds.; Springer: New York, NY, USA, 2015; Volume 1287, pp. 3–23. [Google Scholar]
- de la Luz Gutiérrez-Nava, M.; Aukerman, M.J.; Sakai, H.; Tingey, S.V.; Williams, R.W. Artificial trans-acting sirnas confer consistent and effective gene silencing. Plant Physiol. 2008, 147, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Takeda, A.; Fahlgren, N.; Johnson, S.C.; Cuperus, J.T.; Carrington, J.C. New generation of artificial microrna and synthetic trans-acting small interfering rna vectors for efficient gene silencing in arabidopsis. Plant Physiol. 2014, 165, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Daròs, J.-A. Artificial micrornas and synthetic trans-acting small interfering rnas interfere with viroid infection. Mol. Plant Pathol. 2017, 18, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial micrornas in arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene silencing in plants using artificial micrornas and other small rnas. Plant J. 2008, 53, 674–690. [Google Scholar]
- de Felippes, F.F.; Wang, J.; Weigel, D. Migs: Mirna-induced gene silencing. Plant J. 2012, 541–547. [Google Scholar] [CrossRef]
- de Felippes, F.F. Downregulation of plant genes with mirna-induced gene silencing. In Sirna Design; Taxman, D., Ed.; Humana Press: Totowa, NJ, USA, 2013; Volume 942, pp. 379–387. [Google Scholar]
- Benstein, R.M.; Ludewig, K.; Wulfert, S.; Wittek, S.; Gigolashvili, T.; Frerigmann, H.; Gierth, M.; Flügge, U.-I.; Krueger, S. Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 2013, 25, 5011–5029. [Google Scholar] [CrossRef] [PubMed]
- Imin, N.; Mohd-Radzman, N.A.; Ogilvie, H.A.; Djordjevic, M.A. The peptide-encoding cep1 gene modulates lateral root and nodule numbers in medicago truncatula. J. Exp. Bot. 2013, 64, 5395–5409. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, B.; Qin, X.; Li, M.; Guo, Y. Investigation of a mirna-induced gene silencing technique in petunia reveals alterations in mir173 precursor processing and the accumulation of secondary sirnas from endogenous genes. PLoS ONE 2015, 10, e0144909–e0144916. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, T.B.; Lawler, N.J.; LaFayette, P.R.; Vodkin, L.O.; Parrott, W.A. Simple gene silencing using the trans-acting sirna pathway. Plant Biotechnol. J. 2015, 14, 117–127. [Google Scholar] [CrossRef]
- Zheng, X.; Yang, L.; Li, Q.; Ji, L.; Tang, A.; Zang, L.; Deng, K.; Zhou, J.; Zhang, Y. Migs as a simple and efficient method for gene silencing in rice. Front. Plant Sci. 2018, 9, 662. [Google Scholar] [CrossRef]
- Baykal, U.; Liu, H.; Chen, X.; Nguyen, H.T.; Zhang, Z.J. Novel constructs for efficient cloning of srna-encoding DNA and uniform silencing of plant genes employing artificial trans- acting small interfering rna. Plant Cell Rep. 2016, 35, 2137–2150. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Felippes, F.F. Gene Regulation Mediated by microRNA-Triggered Secondary Small RNAs in Plants. Plants 2019, 8, 112. https://doi.org/10.3390/plants8050112
de Felippes FF. Gene Regulation Mediated by microRNA-Triggered Secondary Small RNAs in Plants. Plants. 2019; 8(5):112. https://doi.org/10.3390/plants8050112
Chicago/Turabian Stylede Felippes, Felipe Fenselau. 2019. "Gene Regulation Mediated by microRNA-Triggered Secondary Small RNAs in Plants" Plants 8, no. 5: 112. https://doi.org/10.3390/plants8050112
APA Stylede Felippes, F. F. (2019). Gene Regulation Mediated by microRNA-Triggered Secondary Small RNAs in Plants. Plants, 8(5), 112. https://doi.org/10.3390/plants8050112




