Meta-Analysis of Quantitative Trait Loci Associated with Seedling-Stage Salt Tolerance in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Results
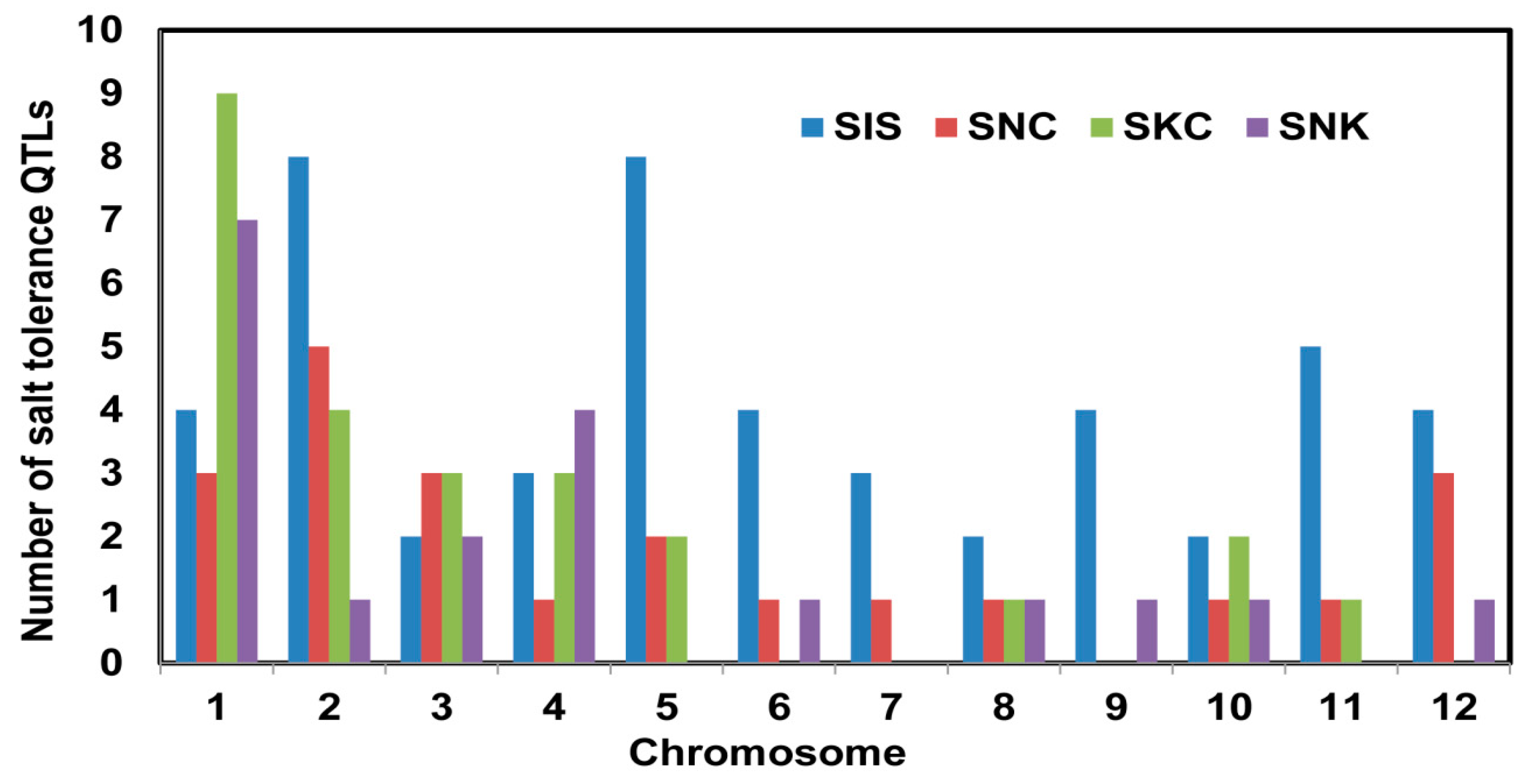
2.1. QTL Analysis of Traits Associated with Seedling-Stage Salt Tolerance
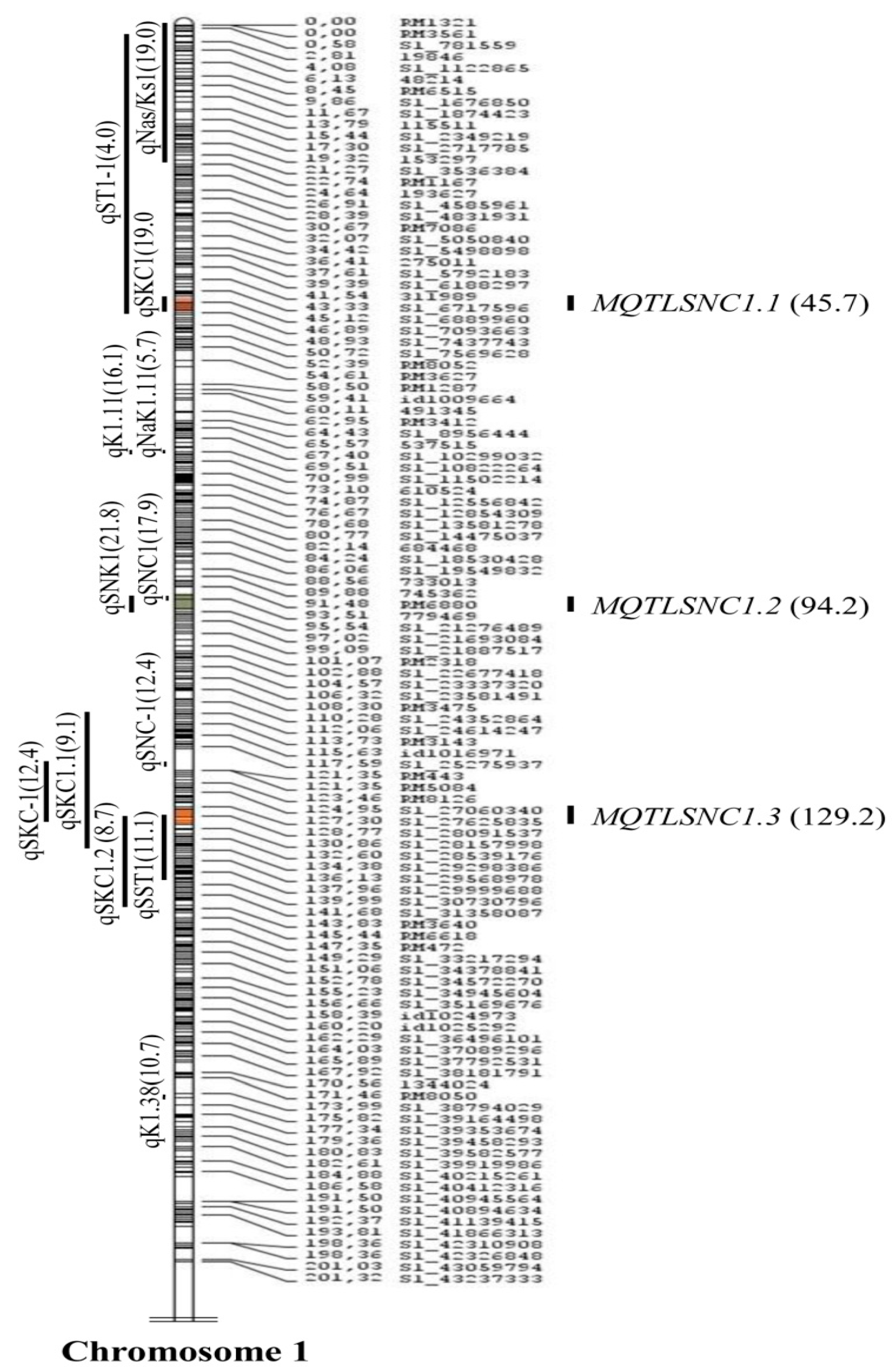
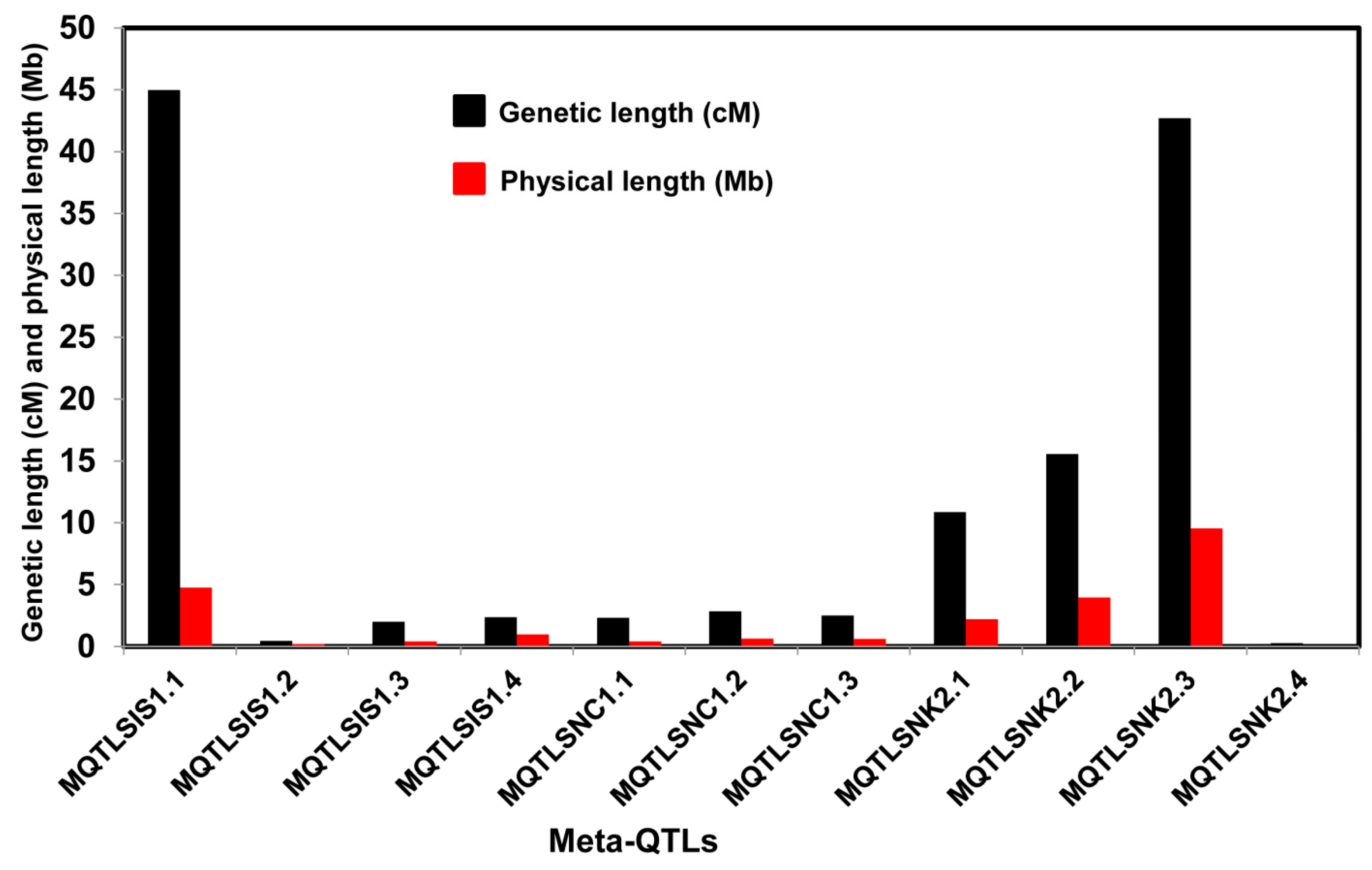
2.2. Meta-Analysis of the QTLs
2.3. Identification of Genes in the Meta-QTL Regions
2.4. Gene Ontology (GO) Analyses
2.5. Phenotyping and Genotyping of Rice Genotypes Using Meta-QTL-Linked Markers
3. Discussion
3.1. Meta-QTL Analysis of Salt Tolerance in Rice
3.2. Gene Content in the Meta-QTL Regions
3.3. Selection of Salt-Tolerant Germplasm Using Meta-QTL Linked Markers
3.4. Future Perspectives
4. Materials and Methods
4.1. Data Collection and Input File Preparation
4.2. Construction of Consensus Map and Projection of QTLs
4.3. Meta-Analysis of QTLs
4.4. Identification of Genes within the Meta-QTL Regions
4.5. Gene Ontology (GO) Enrichment Analysis
4.6. Salt Tolerance Screening and Marker Profiling in Rice Genotypes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Crops/Regions/World list/Production Quantity/Rice (Paddy) in 2014. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 11 December 2017).
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice 2016, 9, 52. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef]
- Bizimana, J.B.; Luzi-Kihupi, A.; Murori, R.W.; Singh, R.K. Identification of quantitative trait loci for salinity tolerance in rice (Oryza sativa L.) using IR29/Hasawi mapping population. J. Genet. 2017, 96, 571–582. [Google Scholar] [CrossRef]
- Dahanayaka, B.A.; Gimhani, D.; Kottearachchi, N.; Samarasighe, W.L.G. QTL mapping for salinity tolerance using an elite rice (Oryza sativa) breeding population. SABRAO J. Breed. Genet. 2017, 49, 123–134. [Google Scholar]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace ‘Pokkali’. PLoS ONE 2017, 12, e0175361. [Google Scholar] [CrossRef]
- Gimhani, D.R.; Gregorio, G.B.; Kottearachchi, N.S.; Samarasinghe, W.L.G. SNP-based discovery of salinity-tolerant QTLs in a bi-parental population of rice (Oryza sativa). Mol. Genet. Genom. 2016, 291, 2081–2099. [Google Scholar] [CrossRef]
- Puram, V.R.R.; Ontoy, J.; Linscombe, S.; Subudhi, P.K. Genetic dissection of seedling stage salinity tolerance in rice using introgression lines of a salt tolerant landrace Nona Bokra. J. Hered. 2017, 108, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Yuan, Z.; Liu, H.; Xiang, X.; Yang, L.; He, W.; Du, B.; Ye, G.; Xu, J.; Xing, D. Identification of salt tolerance-improving quantitative trait loci alleles from a salt-susceptible rice breeding line by introgression breeding. Plant Breed. 2015, 134, 653–660. [Google Scholar] [CrossRef]
- Thomson, M.J.; de Ocampo, M.; Egdane, J.; Rahman, M.A.; Sajise, A.G.; Adorada, D.L.; Tumimbang-Raiz, E.; Blumwald, E.; Seraj, Z.I.; Singh, R.K.; et al. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 2010, 3, 148–160. [Google Scholar] [CrossRef]
- Thomson, M.J.; Tai, T.H.; McClung, A.M.; Lai, X.H.; Hinga, M.E.; Lobos, K.B.; Xu, Y.; Martinez, C.P.; McCouch, S.R. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 2003, 107, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cao, M.; Ma, X.; Chen, W.; Zhao, J.; Sun, C.; Tan, L.; Liu, F. Integrated RNA sequencing and QTL mapping to identify candidate genes from Oryza rufipogon associated with salt tolerance at the seedling stage. Front. Plant Sci. 2017, 8, 1427. [Google Scholar] [CrossRef]
- Zheng, H.; Zhao, H.; Liu, H.; Wang, J.; Zou, D. QTL analysis of Na+ and K+ concentrations in shoots and roots under NaCl stress based on linkage and association analysis in japonica rice. Euphytica 2015, 201, 109–121. [Google Scholar] [CrossRef]
- Bonilla, P.; Dvorak, J.; Mackill, D.; Deal, K.; Gregorio, G. RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp. Agric. Sci. 2002, 85, 68–76. [Google Scholar]
- Gregorio, G.B. Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP). Ph.D. Thesis, University of the Philippines, Los Baños, CA, USA, 1997. [Google Scholar]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.C.; Tardieu, F.; Tuberosa, R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiol. 2008, 147, 469–486. [Google Scholar] [CrossRef]
- Swamy, B.P.M.; Vikram, P.; Dixit, S.; Ahmed, H.U.; Kumar, A. Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus. BMC Genom. 2011, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Goffinet, B.; Gerber, S. Quantitative trait loci: A meta-analysis. Genetics 2000, 155, 463–473. [Google Scholar]
- Veyrieras, J.B.; Goffinet, B.; Charcosset, A. MetaQTL: A package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinform. 2007, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Sosnowski, O.; Charcosset, A.; Johann Joets, J. BioMercator V3: An upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 2012, 28, 2082–2083. [Google Scholar] [CrossRef] [PubMed]
- Swamy, B.P.M.; Sarla, N. Meta-analysis of yield QTLs derived from inter-specific crosses of rice reveals consensus regions and candidate genes. Plant Mol. Biol. Rep. 2011, 29, 663–680. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, M.; Tao, X.; Guo, T.; Chen, Z.; Xiao, W. Quantitative trait loci identification and meta-analysis for rice panicle-related traits. Mol. Genet. Genom. 2016, 291, 1927–1940. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, J.; Mohammad, F. Identification and mapping of quantitative trait loci associated with salinity tolerance in rice (Oryza sativa) using SSR markers. Iran. J. Biotechnol. 2011, 9, 21–30. [Google Scholar]
- Puram, V.R.R.; Ontoy, J.; Subudhi, P.K. Identification of QTLs for salt tolerance traits and prebreeding lines with enhanced salt tolerance in an introgression line population of rice. Plant Mol. Biol. Rep. 2018. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome analysis of salt stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.). PLoS ONE 2016, 11, e0146242. [Google Scholar] [CrossRef] [PubMed]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Alam, R.; Sazzadur Rahman, M.; Seraj, Z.I.; Thomson, M.J.; Ismail, A.M.; Tumimbang-Raiz, E.; Gregorio, G.B. Investigation of seedling-stage salinity tolerance QTLs using backcross lines derived from Oryza sativa L. Pokkali. Plant Breed. 2011, 130, 430–437. [Google Scholar] [CrossRef]
- Lin, H.X.; Zhu, M.Z.; Yano, M.; Gao, J.P.; Liang, Z.W.; Su, W.A.; Hu, X.H.; Ren, Z.H.; Chao, D.Y. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [CrossRef]
- Sabouri, H.; Rezai, A.M.; Moumeni, A.; Kavousi, A.; Katouzi, M.; Sabouri, A. QTLs mapping of physiological traits related to salt tolerance in young rice seedlings. Biol. Plant. 2009, 53, 657–662. [Google Scholar] [CrossRef]
- Price, A.H. Believe it or not, QTLs are accurate! Trends Plant Sci. 2006, 11, 213–216. [Google Scholar] [CrossRef]
- Arcade, A.; Labourdette, A.; Falque, M.; Mangin, B.; Chardon, F.; Charcosset, A.; Joets, J. BioMercator: Integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 2004, 20, 2324–2326. [Google Scholar] [CrossRef]
- Courtois, B.; Ahmadi, N.; Khowaja, F.; Price, A.H.; Rami, J.F.; Frouin, J.; Hamelin, C.; Ruiz, M. Rice root genetic architecture: Meta-analysis from a drought QTL database. Rice 2009, 2, 115–128. [Google Scholar] [CrossRef]
- Griffiths, S.; Simmonds, J.; Leverington, M.; Wang, Y.; Fish, L.; Sayers, L.; Alibert, L.; Orford, S.; Wingen, L.; Herry, L.; et al. Meta-QTL analysis of the genetic control of ear emergence in elite European winter wheat germplasm. Theor. Appl. Genet. 2009, 119, 383–395. [Google Scholar] [CrossRef]
- Li, W.T.; Liu, C.; Liu, Y.X.; Pu, Z.E.; Dai, S.F.; Wang, J.R.; Lan, X.J.; Zheng, Y.L.; Wei, Y.M. Meta-analysis of QTL associated with tolerance to abiotic stresses in barley. Euphytica 2013, 189, 31–49. [Google Scholar] [CrossRef]
- Shinozuka, H.; Cogan, N.O.I.; Spangenberg, G.C.; Forster, J.W. Quantitative trait locus (QTL) meta-analysis and comparative genomics for candidate gene prediction in perennial ryegrass (Lolium perenne L.). BMC Genet. 2012, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Uddin, M.S.; Zou, C.; Xie, C.; Xu, Y.; Li, W.X. Meta-analysis and candidate gene mining of low-phosphorus tolerance in maize. J. Integr. Plant Biol. 2014, 56, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Agrama, H.A.S.; Moussa, M.E. Mapping QTLs in breeding for drought tolerance in maize (Zea mays L.). Euphytica 1996, 91, 89–97. [Google Scholar] [CrossRef]
- Wu, X.L.; Hu, Z.L. Meta-analysis of QTL mapping experiments. In Quantitative Trait Loci (QTL): Methods and Protocols; Rifkin, S.A., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 145–171. [Google Scholar]
- Khan, A.; Li, R.J.; Sun, J.T.; Ma, F.; Zhang, H.X.; Jin, J.H.; Ali, M.; Haq, S.U.; Wang, J.E.; Gong, Z.H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, N.; Negrão, S.; Katsantonis, D.; Frouin, J.; Ploux, J.; Letourmy, P.; Droc, G.; Babo, P.; Trindade, H.; Bruschi, G.; et al. Targeted association analysis identified japonica rice varieties achieving Na+/K+ homeostasis without the allelic make-up of the salt tolerant indica variety Nona Bokra. Theor. Appl. Genet. 2011, 123, 881–895. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Intl. J. Genom. 2014. [Google Scholar] [CrossRef]
- Li, W.T.; He, M.; Wang, J.; Wang, Y.P. Zinc finger protein (ZFP) in plants-a review. Plant Omics 2013, 6, 474–480. [Google Scholar]
- Hossain, M.R.; Bassel, G.W.; Pritchard, J.; Sharma, G.P.; Ford-Lloyd, B.V. Trait specific expression profiling of salt stress responsive genes in diverse rice genotypes as determined by modified significance analysis of microarrays. Front. J. Plant Sci. 2016, 7, 567. [Google Scholar] [CrossRef]
- Diédhiou, C.J.; Popova, O.V.; Dietz, K.J.; Golldack, D. The SNF1-type serine-threonine protein kinase SAPK4regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008, 8, 49. [Google Scholar] [CrossRef]
- Ouyang, S.Q.; Liu, Y.F.; Liu, P.; Lei, G.; He, S.J.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010, 62, 316–329. [Google Scholar] [CrossRef]
- Tanaka, H.; Osakabe, Y.; Katsura, S.; Mizuno, S.; Maruyama, K.; Kusakabe, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 2012, 70, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Santiago, P.; Lagunas-Gómez, D.; Barkla, B.J.; Vera-Estrella, R.; Lalonde, S.; Jones, A.; Frommer, W.B.; Zimmermannova, O.; Sychrova, H.; Pantoja, O. Identification of rice cornichon as a possible cargo receptor for the Golgi-localized sodium transporter OsHKT1;3. J. Exp. Bot. 2015, 66, 2733–2748. [Google Scholar] [CrossRef]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Darvasi, A.; Soller, M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 1997, 27, 125–132. [Google Scholar] [CrossRef]
- IRGSP. The map-based sequence of the rice genome. Nature 2005, 436, 793. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, G.; Senadhira, D.; Mendoza, R. Screening Rice for Salinity Tolerance; IRRI Discussion Paper Series; International Rice Research Institute: Manila, Philippines, 1997; Available online: http://irri.org/resources/publications/serials/discussion-papers-series/item/screening-rice-for-salinity-tolerance (accessed on 15 December 2018).
- Jones, J.B., Jr.; Case, V.W. Sampling, handling, and analyzing plant tissue samples. In Soil Testing and Plant Analysis, 3rd ed.; Westerman, R.L., Ed.; Book Series No. 3; Soil Science Society of America: Madison, WI, USA, 1990; pp. 389–427. [Google Scholar]
- Chen, D.H.; Ronald, P.C. A rapid DNA minipreparation method suitable for AFLP and other applications. Plant Mol. Biol. Rep. 1999, 17, 53–57. [Google Scholar] [CrossRef]


| Mapping Population | Markers Used | QTL Identified $ | References | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Parents | Type | Size | Type | Number | SIS | SNC | SKC | SNK | |
| Teqing × Tarome-Molaei | BC2F5 | 62 | SSR | 114 | - | - | 2 | 1 | [24] |
| IR29 × Hasawi | RIL | 142 | SNP | 194 | 5 | - | - | - | [4] |
| Bg352 × At354 | RIL | 100 | SSR/InDel | 158 | 2 | - | - | 2 | [5] |
| Bengal × Pokkali | RIL | 187 | SNP | 9303 | 7 | 3 | 2 | 2 | [2] |
| Bg352 × At354 | RIL | 94 | SNP | 1135 | 8 | 7 | 5 | 7 | [7] |
| Jupiter × Nona Bokra | IL | 138 | SSR | 126 | 4 | 4 | 4 | 2 | [8] |
| Cheniere × Nona Bokra | IL | 112 | SSR | 116 | 5 | 1 | 3 | 3 | [25] |
| Ce258 × IR75862 | BC1F10 | 200 | SSR | 128 | 4 | 1 | 3 | - | [9] |
| Zhongguangxiang1 × IR75862 | BC1F10 | 200 | SSR | 133 | 2 | 1 | 2 | - | [9] |
| IR29 × Pokkali (Experiment 1) | RIL | 140 | SSR | 100 | 2 | 1 | 1 | 2 | [10] |
| 93-11 × O. rufipogon | IL | 285 | SSR | 142 | 8 | - | - | - | [12] |
| Dongnong425 × Changbai10 | BC2F2 | 190 | SSR | 137 | 2 | 4 | 3 | - | [13] |
| Total | 11,786 | 49 | 22 | 25 | 19 | ||||
| Trait | Meta-QTLs | Ch | AIC Value | QTL Model | Marker Interval | Meta-QTL Peak Position (cM) | Physical Position (Mb) | No of Initial QTL | Mean Phenotypic Variance of the QTL (%) $ | Mean Initial CI (cM) | Meta-QTL CI (95%) (cM) | Physical Length of Meta-QTL (Mb) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIS | MQTLSIS1.1 | 1 | 72.6 | 4 | RM1321-RM1167 | 0 | 0.003–4.75 | 2 | 11.5 | 45.75 | 44.99 | 4.749 |
| MQTLSIS1.2 | S1_11502301-S1_11584932 | 71.5 | 14.07–14.27 | 2 | 10.9 | 0.64 | 0.45 | 0.198 | ||||
| MQTLSIS1.3 | 774607-RM5853 | 94.17 | 22.33–22.75 | 2 | 19.9 | 2.86 | 2.01 | 0.415 | ||||
| MQTLSIS1.4 | RM443-S1_26769954 | 122.57 | 28.34–29.32 | 3 | 11.3 | 13.96 | 2.37 | 0.983 | ||||
| SNC | MQTLSNC1.1 | 1 | 109.2 | 3 | S1_6824646-S1_7093663 | 45.68 | 8.52–8.94 | 2 | 11.5 | 24.4 | 2.32 | 0.415 |
| MQTLSNC1.2 | RM6880-S1_21352851 | 94.17 | 22.25–22.88 | 2 | 19.9 | 2.86 | 2.85 | 0.631 | ||||
| MQTLSNC1.3 | S1_27841959-S1_28157998 | 129.15 | 30.95–31.56 | 5 | 10.7 | 12.04 | 2.51 | 0.604 | ||||
| SNK | MQTLSNK2.1 | 2 | 92.1 | 4 | S2_2747069-S2_3978527 | 17.45 | 2.44–4.66 | 3 | 9.7 | 26.04 | 10.87 | 2.207 |
| MQTLSNK2.2 | S2_4889160-S2_7221617 | 38.74 | 5.90–9.88 | 4 | 10.1 | 23.42 | 15.57 | 3.973 | ||||
| MQTLSNK2.3 | RM3178-S2_22090860 | 76.68 | 14.99–24.53 | 3 | 9.0 | 55.32 | 42.7 | 9.541 | ||||
| MQTLSNK2.4 | RM3302-S2_29841039 | 141.96 | 32.89–32.93 | 4 | 23.7 | 2.22 | 0.28 | 0.037 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.S.; Ontoy, J.; Subudhi, P.K. Meta-Analysis of Quantitative Trait Loci Associated with Seedling-Stage Salt Tolerance in Rice (Oryza sativa L.). Plants 2019, 8, 33. https://doi.org/10.3390/plants8020033
Islam MS, Ontoy J, Subudhi PK. Meta-Analysis of Quantitative Trait Loci Associated with Seedling-Stage Salt Tolerance in Rice (Oryza sativa L.). Plants. 2019; 8(2):33. https://doi.org/10.3390/plants8020033
Chicago/Turabian StyleIslam, Md. Shofiqul, John Ontoy, and Prasanta K. Subudhi. 2019. "Meta-Analysis of Quantitative Trait Loci Associated with Seedling-Stage Salt Tolerance in Rice (Oryza sativa L.)" Plants 8, no. 2: 33. https://doi.org/10.3390/plants8020033
APA StyleIslam, M. S., Ontoy, J., & Subudhi, P. K. (2019). Meta-Analysis of Quantitative Trait Loci Associated with Seedling-Stage Salt Tolerance in Rice (Oryza sativa L.). Plants, 8(2), 33. https://doi.org/10.3390/plants8020033






