Inheritance Pattern and Molecular Markers for Resistance to Blackleg Disease in Cabbage
Abstract
1. Introduction
2. Results
2.1. Inheritance of Blackleg Resistance in Cabbage
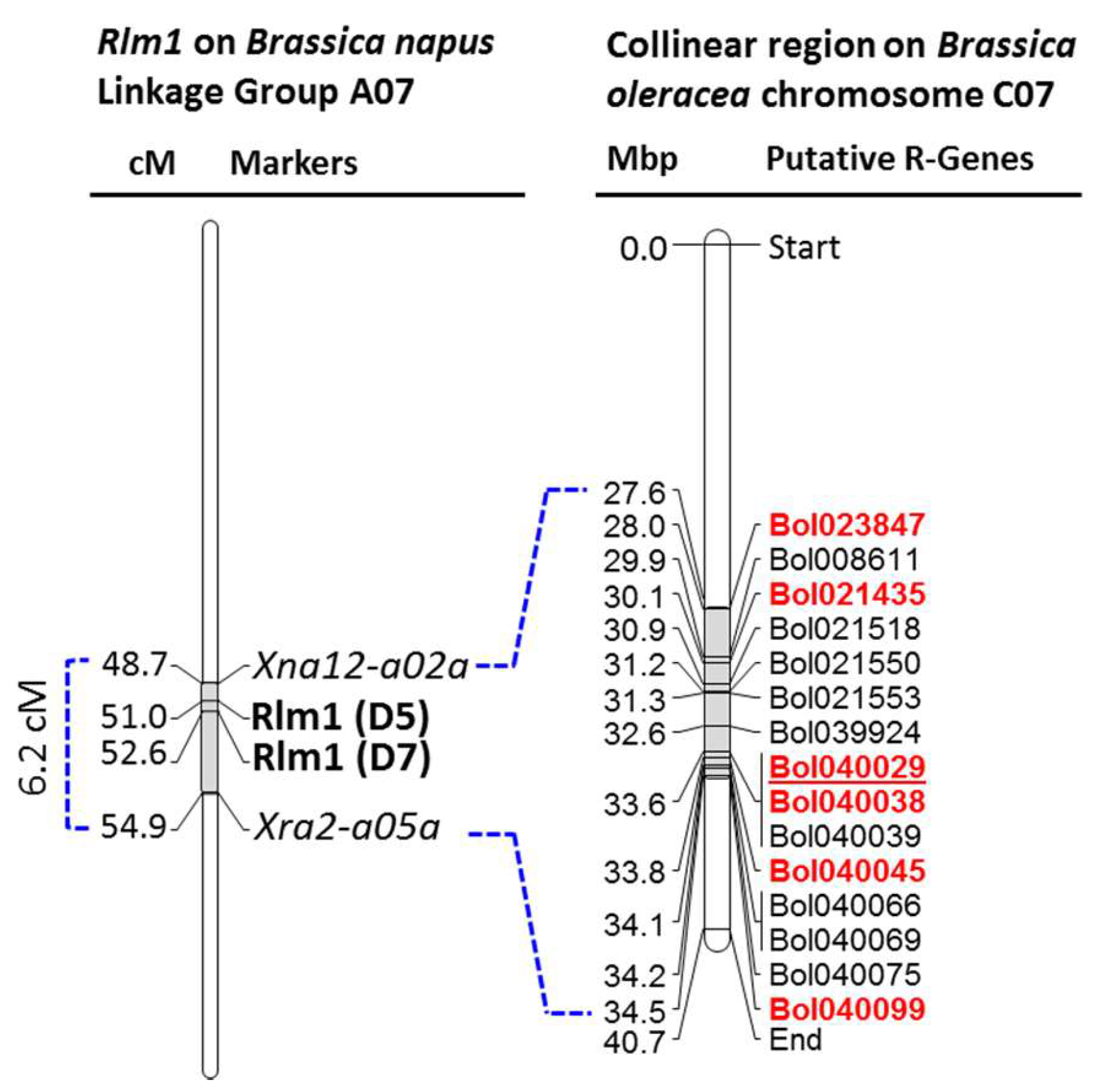
2.2. Selection of Genes and Detection of Length Polymorphism
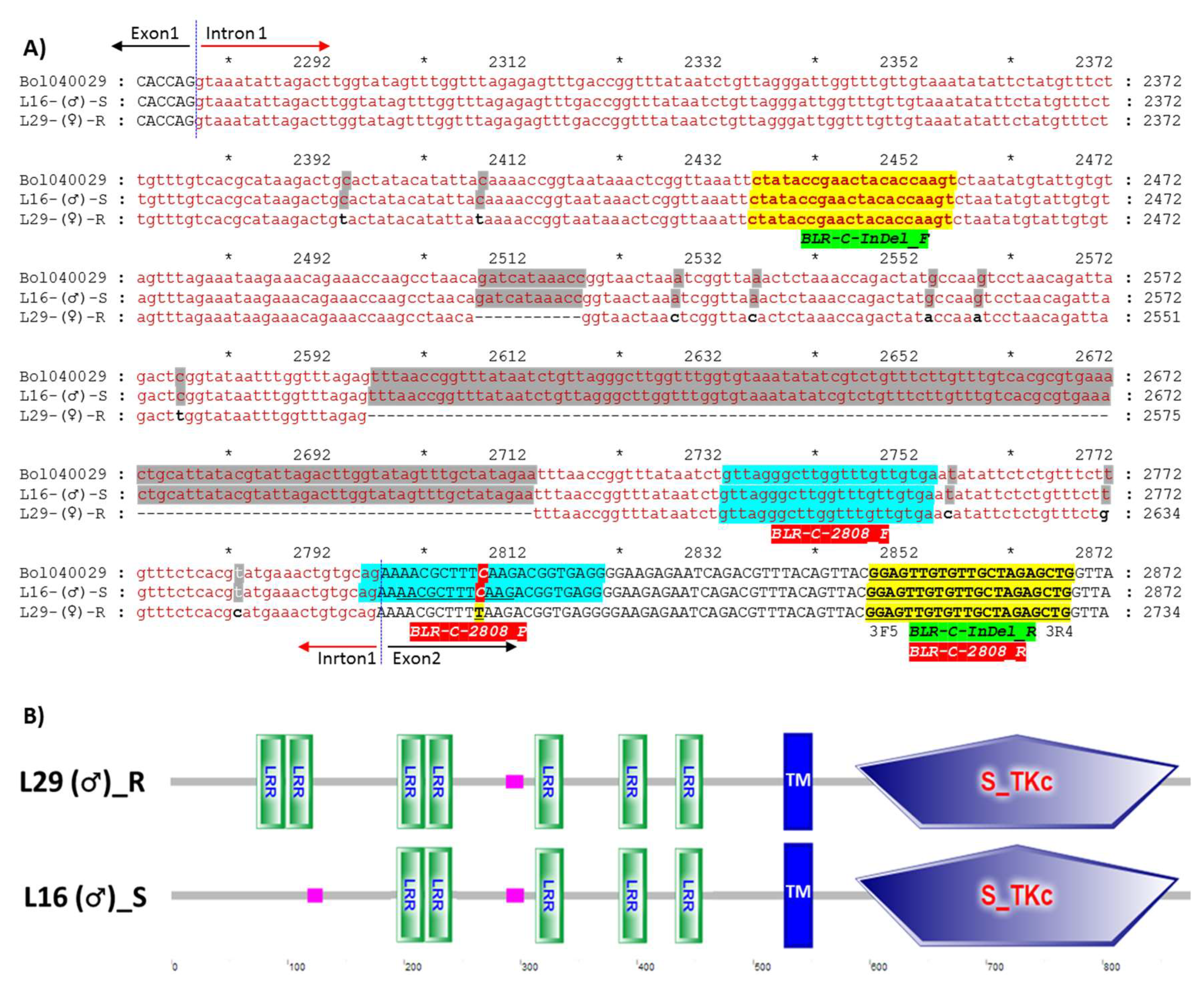
2.3. Cloning, Sequencing and Characterization of Polymorphism
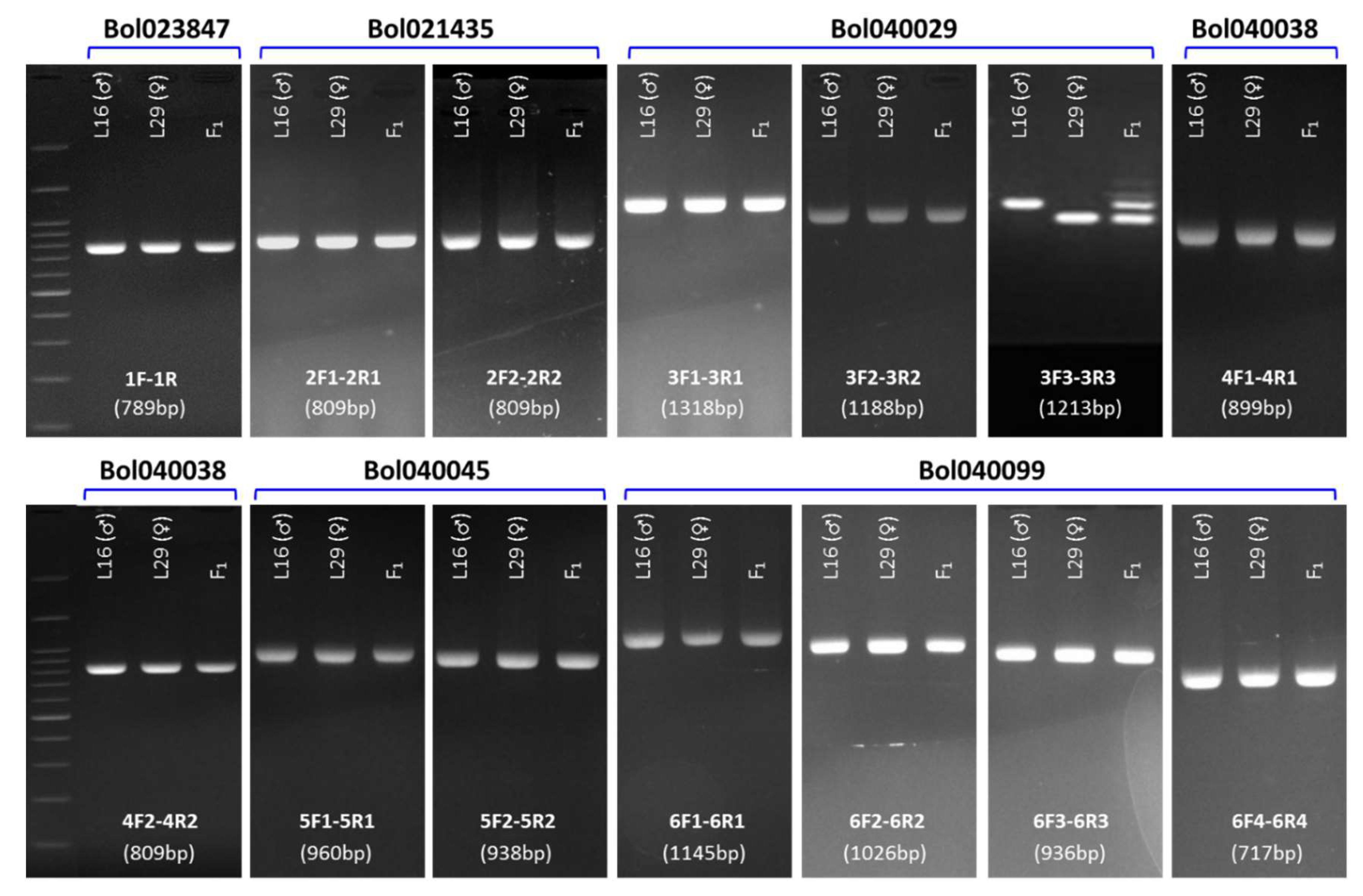
2.4. Development of Markers Linked with Blackleg Resistance
2.5. Validation of the Developed Markers
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Population Development
4.2. L. maculans Isolate: Culture, Inoculation, and Disease Scoring
4.3. Primer Design
4.4. DNA Extraction and PCR Based Detection of Length Polymorphism
4.5. Cloning and Sequencing of the Polymorphic Gene
4.6. High Resolution Melting (HRM) Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Robin, A.H.K.; Larkan, N.J.; Laila, R.; Park, J.I.; Ahmed, N.U.; Borhan, H.; Parkin, I.A.P.; Nou, I.S. Korean Brassica oleracea germplasm offers a novel source of qualitative resistance to blackleg disease. Eur. J. Plant Pathol. 2017, 149, 611–623. [Google Scholar] [CrossRef]
- West, J.S.; Kharbanda, P.D.; Barbetti, M.J.; Fitt, B.D.L. Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol. 2001, 50, 10–27. [Google Scholar] [CrossRef]
- Howlett, B.J. Current knowledge of the interaction between Brassica napus and Leptosphaeria maculans. Can. J. Plant Pathol. 2004, 26, 245–252. [Google Scholar] [CrossRef]
- Fitt, B.D.L.; Brun, H.; Barbetti, M.J.; Rimmer, S.R. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 2006, 114, 3–15. [Google Scholar] [CrossRef]
- Zhang, X.; Fernando, W.G.D. Insights into fighting against blackleg disease of Brassica napus in Canada. Crop Pasture Sci. 2018, 69, 40–47. [Google Scholar] [CrossRef]
- Mendes-Pereira, E.; Balesdent, M.-H.; Brun, H.; Rouxel, T. Molecular phylogeny of the Leptosphaeria maculans—L. biglobosa species complex. Mycol. Res. 2003, 107, 1287–1304. [Google Scholar] [CrossRef] [PubMed]
- Howlett, B.J.; Idnurm, A.; Pedras, M.S.C. Leptosphaeria maculans, the causal agent of blackleg disease of Brassicas. Fungal Genet. Biol. 2001, 33, 1–14. [Google Scholar] [CrossRef]
- Henderson, M.P. The black-leg disease of cabbage caused by Phoma lingam (Tode) Desmaz. Phytopathology 1918, 8, 379–431. [Google Scholar]
- Williams, P.H. Biology of Leptosphaeria maculans. Can. J. Plant Pathol. 1992, 14, 30–35. [Google Scholar] [CrossRef]
- Li, H.; Kuo, J.; Barbetti, M.J.; Sivasithamparam, K. Differences in the responses of stem tissues of spring-type Brassica napus cultivars with polygenic resistance and single dominant gene-based resistance to inoculation with Leptosphaeria maculans. Botany 2007, 85, 191–203. [Google Scholar] [CrossRef]
- Ghanbarnia, K.; Dilantha Fernando, W.G.; Crow, G. Developing rainfall-and temperature-based models to describe infection of canola under field conditions caused by pycnidiospores of Leptosphaeria maculans. Phytopathology 2009, 99, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fitt, B.D.L.; Welham, S.J.; Gladders, P.; Sansford, C.E.; West, J.S. Effects of severity and timing of stem canker (Leptosphaeria maculans) symptoms on yield of winter oilseed rape (Brassica napus) in the UK. Eur. J. Plant Pathol. 1999, 105, 715–728. [Google Scholar] [CrossRef]
- Raman, H.; Raman, R.; Larkan, N. Genetic Dissection of Blackleg Resistance Loci in Rapeseed (Brassica napus L.). In Plant Breeding from Laboratories to Fields; InTech: London, UK, 2013; pp. 85–120. [Google Scholar] [CrossRef]
- Sprague, S.J.; Balesdent, M.-H.; Brun, H.; Hayden, H.L.; Marcroft, S.J.; Pinochet, X.; Rouxel, T.; Howlett, B.J. Major gene resistance in Brassica napus (oilseed rape) is overcome by changes in virulence of populations of Leptosphaeria maculans in France and Australia. In Sustainable Strategies for Managing Brassica Napus (Oilseed Rape) Resistance to Leptosphaeria; Fitt, B.D.L., Evans, N., Howlett, B.J., Cooke, B.M., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 33–40. ISBN 978-1-4020-4525-7. [Google Scholar] [CrossRef]
- Brun, H.; Levivier, S.; Somda, I.; Ruer, D.; Renard, M.; Chèvre, A.M. A Field Method for Evaluating the Potential Durability of New Resistance Sources: Application to the Leptosphaeria maculans-Brassica napus Pathosystem. Phytopathology 2000, 90, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Barret, P.; Guérif, J.; Reynoird, J.P.; Delourme, R.; Eber, F.; Renard, M.; Chèvre, A.M. Selection of stable Brassica napus-Brassica juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). A ‘to and fro’ strategy to localise and characterise interspecific introgressions on the B. napus genome. Theor. Appl. Genet. 1998, 96, 1097–1103. [Google Scholar] [CrossRef]
- Rouxel, T.; Penaud, A.; Pinochet, X.; Brun, H.; Gout, L.; Delourme, R.; Schmit, J.; Balesdent, M.-H. A 10-year Survey of Populations of Leptosphaeria maculans in France Indicates a Rapid Adaptation Towards the Rlm1 Resistance Gene of Oilseed Rape. Eur. J. Plant Pathol. 2003, 109, 871–881. [Google Scholar] [CrossRef]
- Yu, F.; Lydiate, D.J.; Rimmer, S.R. Identification and mapping of a third blackleg resistance locus in Brassica napus derived from B. rapa subsp. sylvestris. Genome 2008, 51, 64–72. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, M.; Wang, X.; Tong, C.; Huang, S.; Tehrim, S.; Liu, Y. Bolbase: A comprehensive genomics database for Brassica oleracea. BMC Genomics 2013, 14, 664. [Google Scholar] [CrossRef]
- Zhang, X.; White, R.P.; Demir, E.; Jedryczka, M.; Lange, R.M.; Islam, M.; Li, Z.Q.; Huang, Y.J.; Hall, A.M.; Zhou, G. Leptosphaeria spp., phoma stem canker and potential spread of L. maculans on oilseed rape crops in China. Plant Pathol. 2014, 63, 598–612. [Google Scholar] [CrossRef]
- Liu, Z.; Latunde-Dada, A.O.; Hall, A.M.; Fitt, B.D.L. Phoma stem canker disease on oilseed rape (Brassica napus) in China is caused by Leptosphaeria biglobosa ‘brassicae’. Eur. J. Plant Pathol. 2014, 140, 841–857. [Google Scholar] [CrossRef]
- Hong, S.-K.; Kim, W.-G.; Shin, D.-B.; Choi, H.-W.; Lee, Y.-K.; Lee, S.-Y. Occurrence of stem canker on rape caused by Leptosphaeria biglobosa in Korea. Plant Pathol. J. 2009, 25, 294–298. [Google Scholar] [CrossRef]
- Hao, L.; Song, P.; Huangfu, H.; Li, Z. Genetic diversity and differentiation of Leptosphaeria biglobosa on oilseed rape in China. Phytoparasitica 2015, 43, 253–263. [Google Scholar] [CrossRef]
- Cai, X.; Huang, Y.; Jiang, D. Evaluation of oilseed rape seed yield losses caused by Leptosphaeria biglobosa in central China. Eur. J. Plant Pathol. 2017, 150, 179–190. [Google Scholar] [CrossRef]
- Fitt, B.D.L.; Hu, B.C.; Li, Z.Q.; Liu, S.Y.; Lange, R.M.; Kharbanda, P.D.; Butterworth, M.H.; White, R.P. Strategies to prevent spread of Leptosphaeria maculans (phoma stem canker) onto oilseed rape crops in China; costs and benefits. Plant Pathol. 2008, 57, 652–664. [Google Scholar] [CrossRef]
- Liu, Z. Strategies for Managing Leptosphaeria Maculans and Leptosphaeria Biglobosa to Decrease Severity of Phoma Stem Canker Epidemics on Winter Oilseed Rape. Ph.D. Thesis, University of Hertfordshire, Hertfordshire, UK, 2008. [Google Scholar]
- Fitt, B.D.L.; Huang, Y.-J.; van den Bosch, F.; West, J.S. Coexistence of related pathogen species on arable crops in space and time. Annu. Rev. Phytopathol. 2006, 44, 163–182. [Google Scholar] [CrossRef] [PubMed]
- Ananga, A.O.; Cebert, E.; Soliman, K.; Kantety, R.; Pacumbaba, R.P.; Konan, K. RAPD markers associated with resistance to blackleg disease in Brassica species. J. Biotechnol. 2006, 5, 2041–2048. [Google Scholar] [CrossRef]
- Badawy, H.M.A.; Hoppe, H.; Koch, E. Differential reactions between the genus Brassica and aggressive single spore isolates of Leptosphaeria maculans. J. Phytopathol. 1991, 131, 109–119. [Google Scholar] [CrossRef]
- Ferreira, M.E.; Dias, J.S.; Mengistu, A.; Williams, P.H. Screening of Portuguese cole landraces (Brassica oleracea L.) with Leptosphaeria maculans and Xanthomonas campestris pv. campestris. Euphytica 1992, 65, 219–227. [Google Scholar] [CrossRef]
- Leflon, M.; Brun, H.; Eber, F.; Delourme, R.; Lucas, M.O.; Vallée, P.; Ermel, M.; Balesdent, M.H.; Chèvre, A.M. Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus. Theor. Appl. Genet. 2007, 115, 897–906. [Google Scholar] [CrossRef]
- Mithen, R.F.; Lewis, B.G.; Heaney, R.K.; Fenwick, G.R. Resistance of leaves of Brassica species to Leptosphaeria maculans. Trans. Br. Mycol. Soc. 1987, 88, 525–531. [Google Scholar] [CrossRef]
- Bohman, S.; Wang, M.; Dixelius, C. Arabidopsis thaliana-derived resistance against Leptosphaeria maculans in a Brassica napus genomic background. Theor. Appl. Genet. 2002, 105, 498–504. [Google Scholar] [CrossRef]
- Chèvre, A.M.; Barret, P.; Eber, F.; Dupuy, P.; Brun, H.; Tanguy, X.; Renard, M. Selection of stable Brassica napus-B. juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). Identification of molecular markers, chromosomal and genomic origin of the introgression. Theor. Appl. Genet. 1997, 95, 1104–1111. [Google Scholar] [CrossRef]
- Delourme, R.; Chevre, A.M.; Brun, H.; Rouxel, T.; Balesdent, M.H.; Dias, J.S.; Salisbury, P.; Biop, U.M.R.; Cedex, L.R.; St-cyr, R. De Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus). Eur. J. Plant Pathol. 2006, 114, 41–52. [Google Scholar] [CrossRef]
- Marcroft, S.J.; Wratten, N.; Purwantara, A.; Salisbury, P.A.; Potter, T.D.; Barbetti, M.J.; Khangura, R.; Howlett, B.J. Reaction of a range of Brassica species under Australian conditions to the fungus, Leptosphaeria maculans, the causal agent of blackleg. Aust. J. Exp. Agric. 2002, 42, 587–594. [Google Scholar] [CrossRef]
- Pang, E.C.K.; Halloran, G.M. The genetics of blackleg [Leptosphaeria maculans (Desm.) Ces. et De Not.] resistance in rapeseed (Brassica napus L.). Theor. Appl. Genet. 1996, 93, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Rimmer, S.R.; van den Berg, C.G.J. Resistance of oilseed Brassica spp. to blackleg caused by Leptosphaeria maculans. Can. J. Plant Pathol. 1992, 14, 56–66. [Google Scholar] [CrossRef]
- Snowdon, R.J.; Winter, H.; Diestel, A.; Sacristán, M.D. Development and characterisation of Brassica napus-Sinapis arvensis addition lines exhibiting resistance to Leptosphaeria maculans. Theor. Appl. Genet. 2000, 101, 1008–1014. [Google Scholar] [CrossRef]
- Christianson, J.A.; Rimmer, S.R.; Good, A.G.; Lydiate, D.J. Mapping genes for resistance to Leptosphaeria maculans in Brassica juncea. Genome 2006, 41, 30–41. [Google Scholar] [CrossRef]
- Balesdent, M.H.; Attard, A.; Kühn, M.L.; Rouxel, T. New Avirulence Genes in the Phytopathogenic Fungus Leptosphaeria maculans. Genet. Resist. 2002, 92, 1122–1133. [Google Scholar] [CrossRef]
- Plieske, J.; Struss, D.; Röbbelen, G. Inheritance of resistance derived from the B-genome of Brassica against Phoma lingam in rapeseed and the development of molecular markers. Theor. Appl. Genet. 1998, 97, 929–936. [Google Scholar] [CrossRef]
- Delourme, R.; Pilet-Nayel, M.L.; Archipiano, M.; Horvais, R.; Tanguy, X.; Rouxel, T.; Brun, H.; Renard, M.; Balesdent, M.H. A Cluster of Major Specific Resistance Genes to Leptosphaeria maculans in Brassica napus. Phytopathology 2004, 94, 578–583. [Google Scholar] [CrossRef]
- Yu, F.; Lydiate, D.J.; Rimmer, S.R. Identification of two novel genes for blackleg resistance in Brassica napus. Theor. Appl. Genet. 2005, 110, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Saal, B.; Brun, H.; Glais, I.; Struss, D. Identification of a Brassica juncea-derived recessive gene conferring resistance to Leptosphaeria maculans in oilseed rape. Plant Breed. 2004, 123, 505–511. [Google Scholar] [CrossRef]
- Yu, F.; Gugel, R.K.; Kutcher, H.R.; Peng, G.; Rimmer, S.R. Identification and mapping of a novel blackleg resistance locus LepR4 in the progenies from Brassica napus × B. rapa subsp. sylvestris. Theor. Appl. Genet. 2013, 126, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Delourme, R.; Laperche, A.; Bouchet, A.; Jubault, M.; Paillard, S.; Nesi, N. Genes and Quantitative Trait Loci Mapping for Major Agronomic Traits in Brassica napus L. In The Brassica Napus Genome; Springer International Publishing: New York, NY, USA, 2018; pp. 41–85. ISBN 9783319436944. [Google Scholar] [CrossRef]
- Raman, R.; Taylor, B.; Marcroft, S.; Stiller, J.; Eckermann, P. Molecular mapping of qualitative and quantitative loci for resistance to Leptosphaeria maculans causing blackleg disease in canola (Brassica napus L.). Theor. Appl. Genet. 2012, 125, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Dion, Y.; Gugel, R.K.; Rakow, G.F.; Seguin-Swartz, G.; Landry, B.S. RFLP mapping of resistance to the blackleg disease [causal agent, Leptosphaeria maculans (Desm.) Ces. et de Not.] in canola (Brassica napus L.). Theor. Appl. Genet. 1995, 91, 1190–1194. [Google Scholar] [CrossRef]
- Ferreira, M.E.; Rimmer, S.R.; Williams, P.H.; Osborn, T.C. Mapping loci controlling Brassica napus resistance to Leptosphaeria maculans under different screening conditions. Phytopathology 1995, 85, 213–217. [Google Scholar] [CrossRef]
- Mayerhofer, R.; Good, A.G.; Bansal, V.K.; Thiagarajah, M.R.; Stringam, G.R. Molecular mapping of resistance to Leptosphaeria maculans in Australian cultivars of Brassica napus. Genome 1997, 40, 294–301. [Google Scholar] [CrossRef]
- Saal, B.; Struss, D. RGA-and RAPD-derived SCAR markers for a Brassica B-genome introgression conferring resistance to blackleg in oilseed rape. Theor. Appl. Genet. 2005, 111, 281–290. [Google Scholar] [CrossRef]
- Huang, Y.J.; Pirie, E.J.; Evans, N.; Delourme, R.; King, G.J.; Fitt, B.D.L. Quantitative resistance to symptomless growth of Leptosphaeria maculans (phoma stem canker) in Brassica napus (oilseed rape). Plant Pathol. 2009, 58, 314–323. [Google Scholar] [CrossRef]
- Raman, H.; Raman, R.; Diffey, S.; Qiu, Y.; McVittie, B.; Barbulescu, D.M.; Salisbury, P.A.; Marcroft, S.; Delourme, R. Stable Quantitative Resistance Loci to Blackleg Disease in Canola (Brassica napus L.) Over Continents. Front. Plant Sci. 2018, 9, 1622. [Google Scholar] [CrossRef]
- Pilet, M.L.; Delourme, R.; Foisset, N.; Renard, M. Identification of loci contributing to quantitative field resistance to blackleg disease, causal agent Leptosphaeria maculans (Desm.) Ces. et de Not., in Winter rapeseed (Brassica napus L.). Theor. Appl. Genet. 1998, 96, 23–30. [Google Scholar] [CrossRef]
- Huang, Y.; Qi, A.; King, G.J.; Fitt, B.D.L. Assessing Quantitative Resistance against Leptosphaeria maculans (Phoma Stem Canker) in Brassica napus (Oilseed Rape) in Young Plants. PLoS ONE 2014, 9, e84924. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Golicz, A.A.; Tirnaz, S.; Chan, C.K.K.; Edwards, D.; Batley, J. Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol. J. 2019, 17, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Goff, K.E.; Ramonell, K.M. The Role and Regulation of Receptor-Like Kinases in Plant Defense. Gene Regul. Syst. Biol. 2007, 1, 167–175. [Google Scholar] [CrossRef]
- Liu, P.L.; Du, L.; Huang, Y.; Gao, S.M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef]
- Sekhwal, M.K.; Li, P.; Lam, I.; Wang, X.; Cloutier, S.; You, F.M. Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 2015, 16, 19248–19290. [Google Scholar] [CrossRef]
- Saintenac, C.; Lee, W.-S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 2018, 50, 368. [Google Scholar] [CrossRef]
- Larkan, N.J.; Ma, L.; Borhan, M.H. The I-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 2015, 13, 983–992. [Google Scholar] [CrossRef]
- Larkan, N.J.; Lydiate, D.J.; Parkin, I.A.P.; Nelson, M.N.; Epp, D.J.; Cowling, W.A.; Rimmer, S.R.; Borhan, M.H. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013, 197, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Maniatis, T.; Reed, R. An extensive network of coupling among gene expression machines. Nature 2002, 416, 499. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-F.; Zhu, T.; Niu, D.-K. Association of intron loss with high mutation rate in Arabidopsis: Implications for genome size evolution. Genome Biol. Evol. 2013, 5, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, A.; Cooper, T.A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011, 12, 715. [Google Scholar] [CrossRef] [PubMed]
- Gorlova, O.; Fedorov, A.; Logothetis, C.; Amos, C.; Gorlov, I. Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol. Biol. 2014, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.-Y.; Lin, S.-L. Intronic microRNAs. Biochem. Biophys. Res. Commun. 2005, 326, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.B. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 2004, 40, 744–751. [Google Scholar] [CrossRef]
- Rose, A.B.; Emami, S.; Bradnam, K.; Korf, I. Evidence for a DNA-based mechanism of intron-mediated enhancement. Front. Plant Sci. 2011, 2, 98. [Google Scholar] [CrossRef]
- Chorev, M.; Carmel, L. The function of introns. Front Genet. 2012, 3, 55. [Google Scholar] [CrossRef]
- Abuyusuf, M.; Nath, U.K.; Kim, H.; Islam, R.; Park, J.; Nou, I. Molecular markers based on sequence variation in BoFLC1. C9 for characterizing early- and late-flowering cabbage genotypes. BMC Genet. 2019, 20, 42. [Google Scholar] [CrossRef]
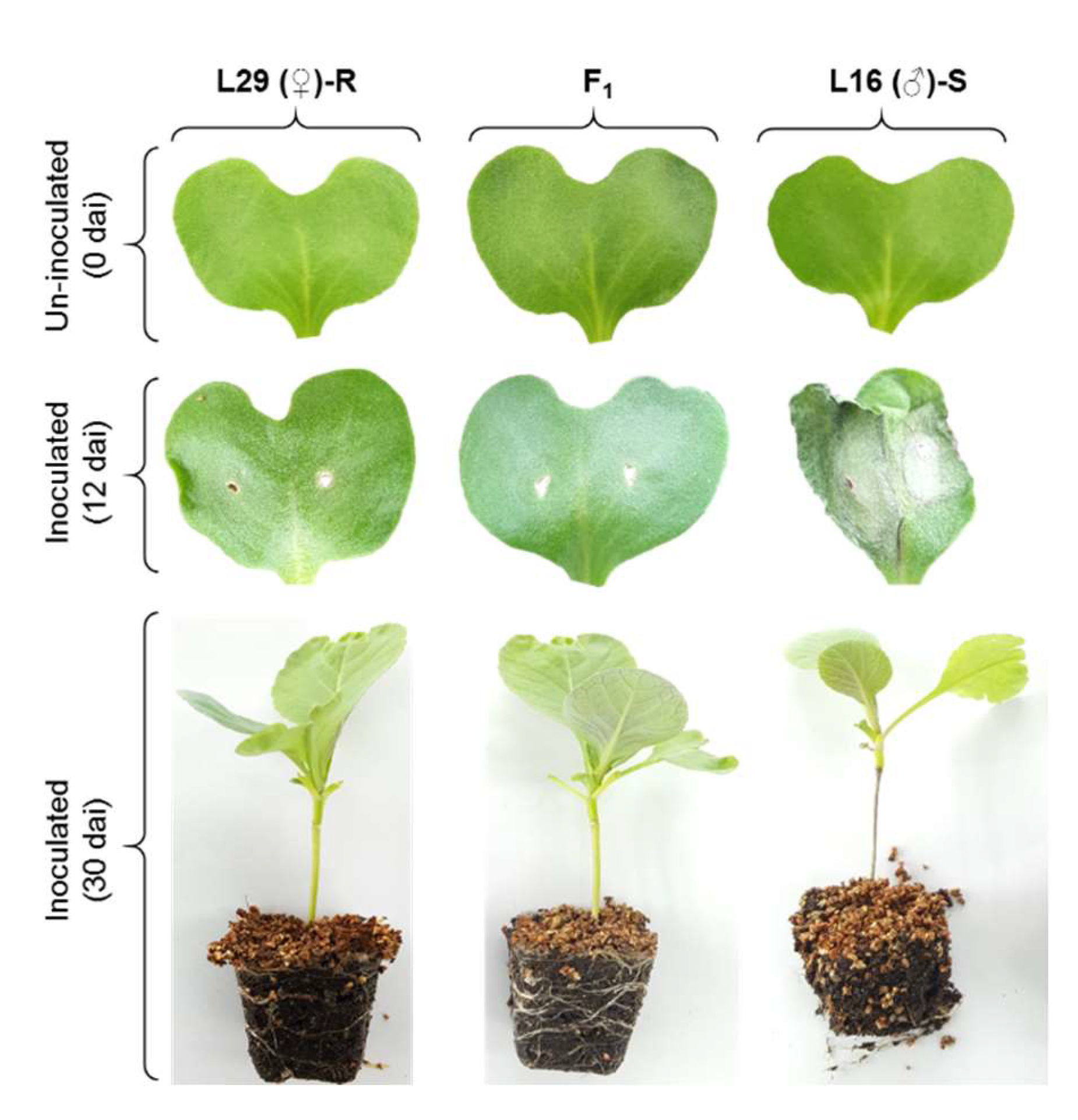
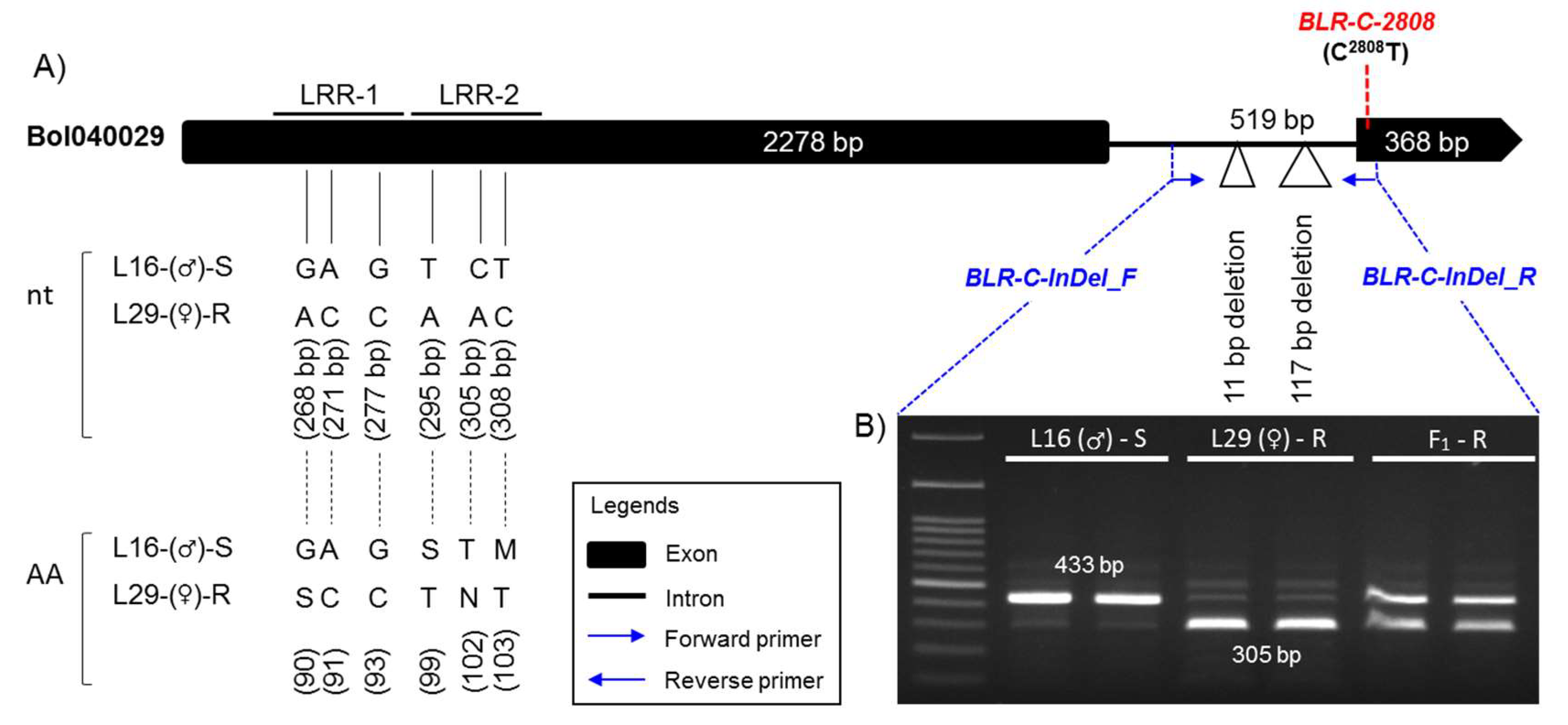
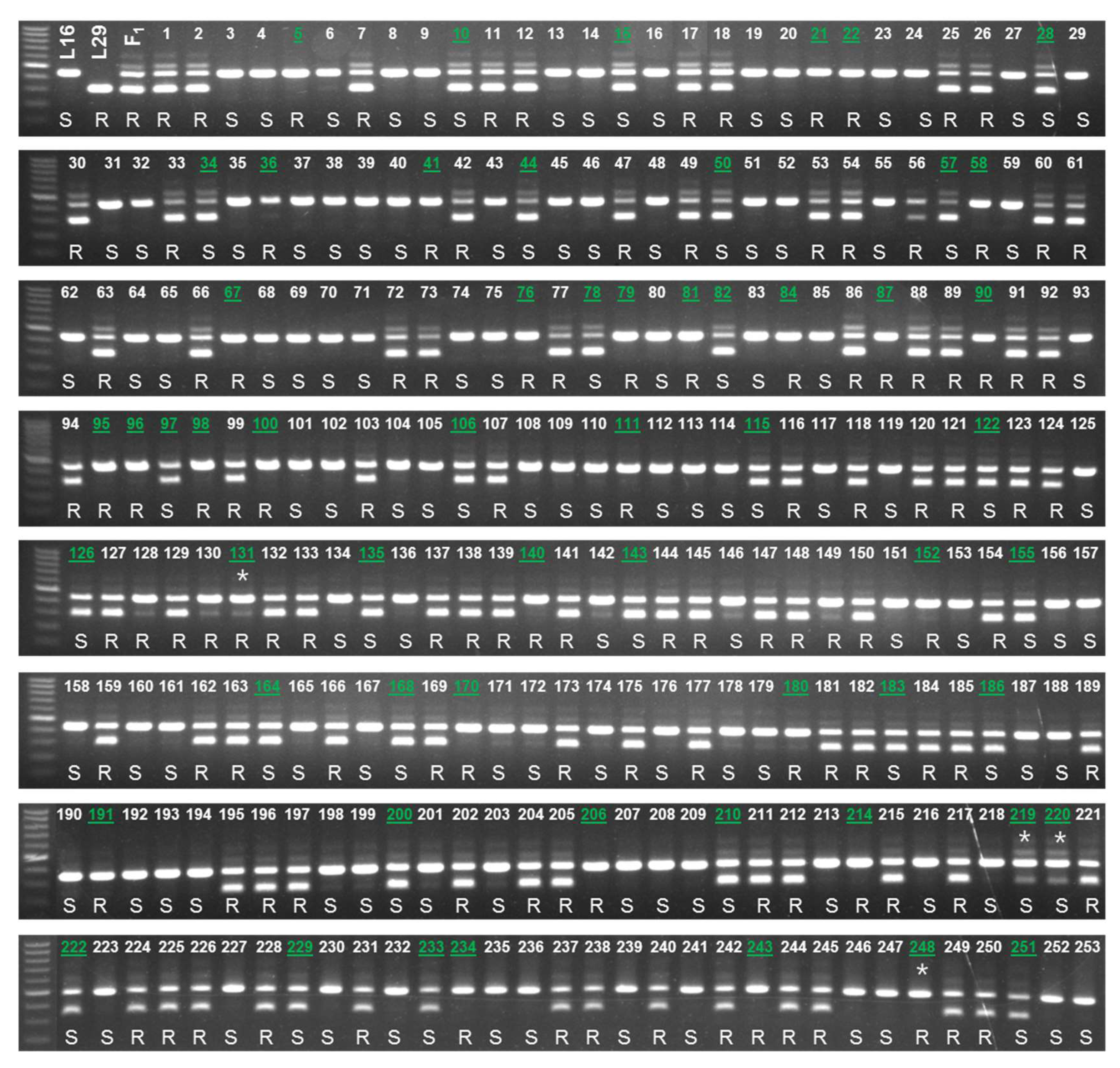

| Parent/Cross | Resistant (Score 0–5) | Susceptible (Score 6–9) | Phenotypic Ratio (R:S) | Chi-Square (χ2) | P-Value |
|---|---|---|---|---|---|
| L29 (♀) | 15 | 0 | |||
| L16 (♂) | 0 | 15 | |||
| F1 [L29 (♀)× L16 (♂)] | 15 | 0 | |||
| BC1 [F1 (♀) × L16 (♂)] | 122 | 131 | 1:1 | 0.32 | 0.572 |
| Sl | Gene ID | Ensemble ID | Brassica rapa Homolog | Arabidopsis Hit | Swissprot ID | Description | Key Domains |
|---|---|---|---|---|---|---|---|
| 1 | Bol023847 * | Bo6g077080 | Bra003415 | AT3G60490 | Q9M210 | Ethylene-responsive transcription factor ERF035 | APETALA2; EREBPs |
| 2 | Bol008611 | Bo6g067950 | Bra003178 | AT3G54320 | Q6X5Y6 | Ethylene-responsive transcription factor WRI1 | APETALA2; EREBPs |
| 3 | Bol021435 * | Bo6g080150 | Bra003549 | AT1G80080 | Q9SSD1 | Protein TOO MANY MOUTHS_TMM | LRR |
| 4 | Bol021518 | Bo6g081090 | Bra003614 | AT1G79280 | P12270 | TPR/MLP1/MLP2-like protein | CC, TPR_MLP1_2, SMC |
| 5 | Bol021550 | Bo6g081540 | Bra003638 | AT1G78750 | Q9ZV93 | F-box/FBD/LRR-repeat protein | LRR, FBD |
| 6 | Bol021553 | Bo6g081570 | Bra003641 | AT1G78430 | Q9M9F9 | Interactor of constitutive active ROPs 4 | CC |
| 7 | Bol039924 | Bo6g086740 | Bra003780 | AT1G74930 | Q9S7L5 | Ethylene-responsive transcription factor ERF018 | APETALA2; EREBPs |
| 8 | Bol040029 * | Bo6g088090 | Bra003858 | AT1G73080 | P93194 | Receptor-like protein kinase | LRR-RLK, STKc |
| 9 | Bol040038 * | Bo6g089160 | Bra003864 | AT1G72890 | O82500 | disease resistance protein (TIR-NBS class), putative | TIR, NB-ARC |
| 10 | Bol040039 | Bo6g116350 | Bra016027 | AT1G72890 | Q40392 | disease resistance protein (TIR-NBS class), putative | TIR, NB-ARC |
| 11 | Bol040045 * | Bo6g089290 | Bra003866 | AT1G72850 | O82500 | disease resistance protein (TIR-NBS class), putative | TIR, NB-ARC |
| 12 | Bol040066 | Bo6g091510 | Bra003880 | AT1G72460 | C0LGU0 | LRR receptor-like serine/threonine-protein kinase | LRR-RLK, STKc |
| 13 | Bol040069 | Bo6g091540 | Bra003883 | AT1G72360 | Q8H0T5 | Ethylene-responsive transcription factor ERF073 | APETALA2; EREBPs |
| 14 | Bol040075 | Bo6g092630 | Bra003889 | AT1G72180 | Q9SGP2 | Receptor-like protein kinase | LRR-RLK, STKc |
| 15 | Bol040099 * | Bo6g093010 | Bra003911 | AT1G71830 | Q94AG2 | Somatic embryogenesis receptor kinase 1 | LRR-RLK, STKc |
| SL | Gene ID | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Tm (°C) | Product Size (bp) | Primer Position | Detection of Polymorphism |
|---|---|---|---|---|---|---|---|
| 1 | Bol023847 | 1F GCAGACCACTTCAACTTGTAACC | 1R GGGTACTTTAGTCATCTAGCC | 59 | 789 | Promoter - 3′ UTR | - |
| 2 | Bol021435 | 2F1 TGCCATATGCTCCTTGTGTT | 2R1 CCGTTTGACTGGTTCGATTC | 59 | 809 | Exon-1 - Exon-3 | - |
| 2F2 CCGTTTGACTGGTTCGATTC | 2R2 ACGCGAAATTGAACACAACA | 60 | 809 | Promoter - 3′-UTR | - | ||
| 3 | Bol040029 | 3F1 GGTTGGTTCTTTGCCTGAGA | 3R1 CTTATCCGGAAGCTCACCTG | 60 | 1318 | Promoter - Exon-1 | - |
| 3F2 GCGTTTTGACGTTGGGTTTA | 3R2 GCCAAACCAAAGTCACCAAT | 60 | 1188 | Exon-1 - Exon-1 | - | ||
| 3F3 CTTGAGTGGTCTGCACGGTA | 3R3 GCCCATTATAGGCCGAGTTA | 60 | 1213 | Exon-1 - 3′ UTR | + | ||
| 4 | Bol040038 | 4F1 TGAGCACGATGTTGGAAAAA | 4R1 GGTTATTACCATTGCTTAGTGT | 58 | 899 | Exon-1 - Exon-2 | - |
| 4F2 TCAGAGATGTTGTCCACGGT | 4R2 TCCAAAGGAGGGCGTAATC | 60 | 809 | Exon-2 - 3′ UTR | - | ||
| 5 | Bol040045 | 5F1 GGACTTTTCCTCTGCTCGAA | 5R1 GGATGGACTGATCGGCTTAT | 61 | 960 | Promoter - Exon-2 | - |
| 5F2 CGATGCAAGATTTTCATTCAC | 5R2 ACATCATGACAACCGCATAAA | 60 | 938 | Intron-1 - 3′ UTR | - | ||
| 6 | Bol040099 | 6F1 TGGGTTGATTAGGGATTTGA | 6R1 GCTCACCAAGTTCGTCAGGT | 59 | 1145 | Promoter - Exon-4 | - |
| 6F2 CCCTCTCGTTTCACTTTAAATC | 6R2 GAAAAGCAAAGCAGCACCTG | 60 | 1026 | Intron-2 - Exon-8 | - | ||
| 6F3 TCCACCCCGAGTAAGTTGTC | 6R3 CGCTGTTGTCACGTGAGTGT | 61 | 936 | Intron-7 - Exon-9 | - | ||
| 6F4 GGCTCAGCTCGTGGTTTATC | 6R4 TGGGACCAGACAACTCAACA | 58 | 717 | Exon-9 - Exon-9 | - |
| Marker Type | Target Polymorphism * | Primer Name | Primer Sequence (5′-3′) | Primer Position | Amplicon Size/SNP Allele |
|---|---|---|---|---|---|
| InDel | 2508–2518 bp and 2597–2713 bp deletion (total 128 bp) in the R line | BLR-C-InDel_F | CTATACCGAACTACACCAAGT | 1st Intron | S line (433 bp) R line (305 bp) |
| BLR-C-InDel_R | CAGCTCTAGCAACACAACTCC | 2nd Exon | |||
| HRM | C2808T SNP in S and R line | BLR-C-2808_F | GTTAGGGCTTGGTTTGTTGTGA | 1st Intron | R line (T2808) S line (C2808) |
| BLR-C-2808_R | CAGCTCTAGCAACACAACTC | 2nd Exon | |||
| BLR-C-2808_P | AGAAAACGCTTTCAAGACGGTGAGG | 2nd Exon |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferdous, M.J.; Hossain, M.R.; Park, J.-I.; Robin, A.H.K.; Jesse, D.M.I.; Jung, H.-J.; Kim, H.-T.; Nou, I.-S. Inheritance Pattern and Molecular Markers for Resistance to Blackleg Disease in Cabbage. Plants 2019, 8, 583. https://doi.org/10.3390/plants8120583
Ferdous MJ, Hossain MR, Park J-I, Robin AHK, Jesse DMI, Jung H-J, Kim H-T, Nou I-S. Inheritance Pattern and Molecular Markers for Resistance to Blackleg Disease in Cabbage. Plants. 2019; 8(12):583. https://doi.org/10.3390/plants8120583
Chicago/Turabian StyleFerdous, Mostari Jahan, Mohammad Rashed Hossain, Jong-In Park, Arif Hasan Khan Robin, Denison Michael Immanuel Jesse, Hee-Jeong Jung, Hoy-Taek Kim, and Ill-Sup Nou. 2019. "Inheritance Pattern and Molecular Markers for Resistance to Blackleg Disease in Cabbage" Plants 8, no. 12: 583. https://doi.org/10.3390/plants8120583
APA StyleFerdous, M. J., Hossain, M. R., Park, J.-I., Robin, A. H. K., Jesse, D. M. I., Jung, H.-J., Kim, H.-T., & Nou, I.-S. (2019). Inheritance Pattern and Molecular Markers for Resistance to Blackleg Disease in Cabbage. Plants, 8(12), 583. https://doi.org/10.3390/plants8120583






