AtSIBP1, a Novel BTB Domain-Containing Protein, Positively Regulates Salt Signaling in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
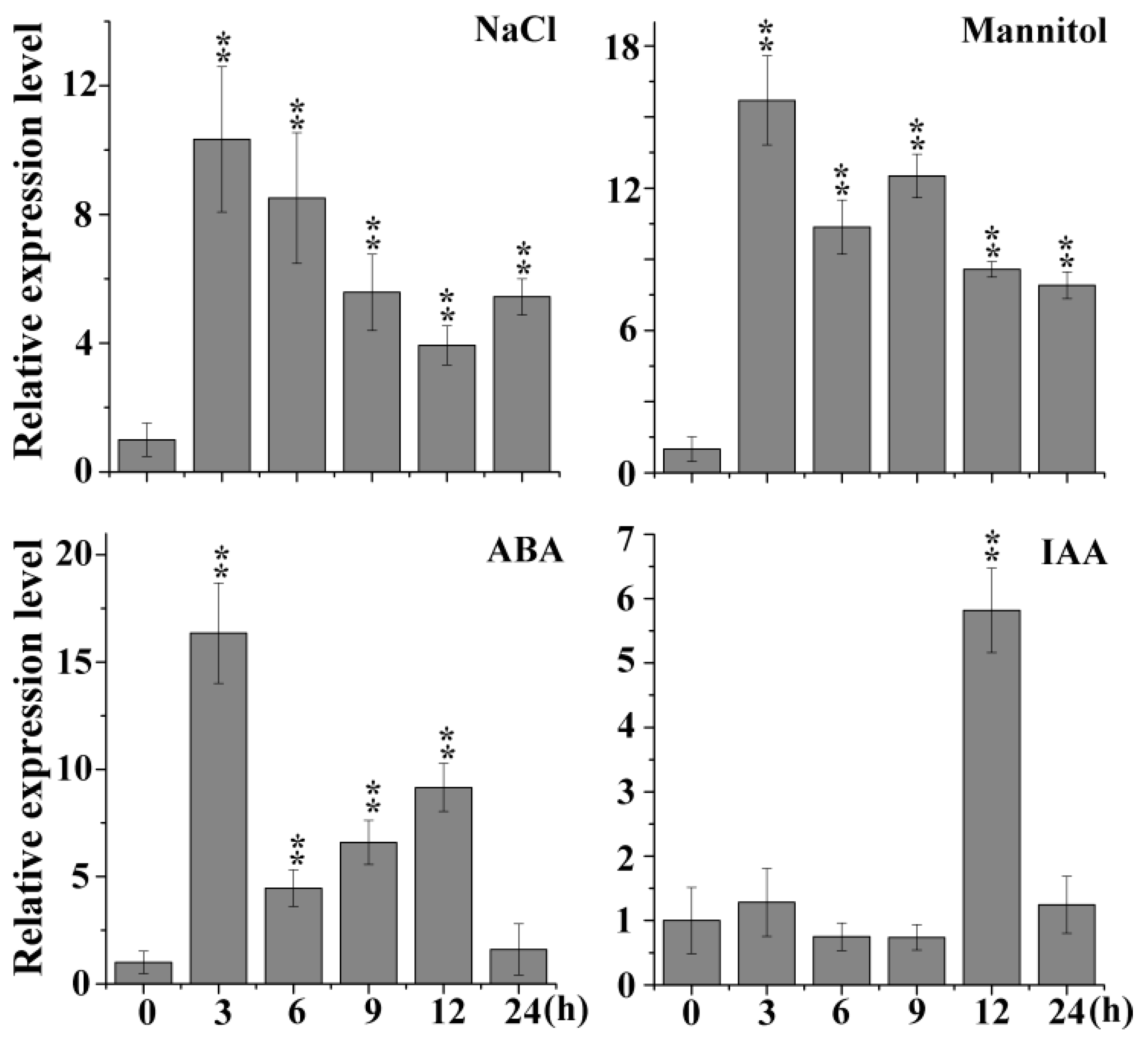
2.1. Expression Pattern of AtSIBP1
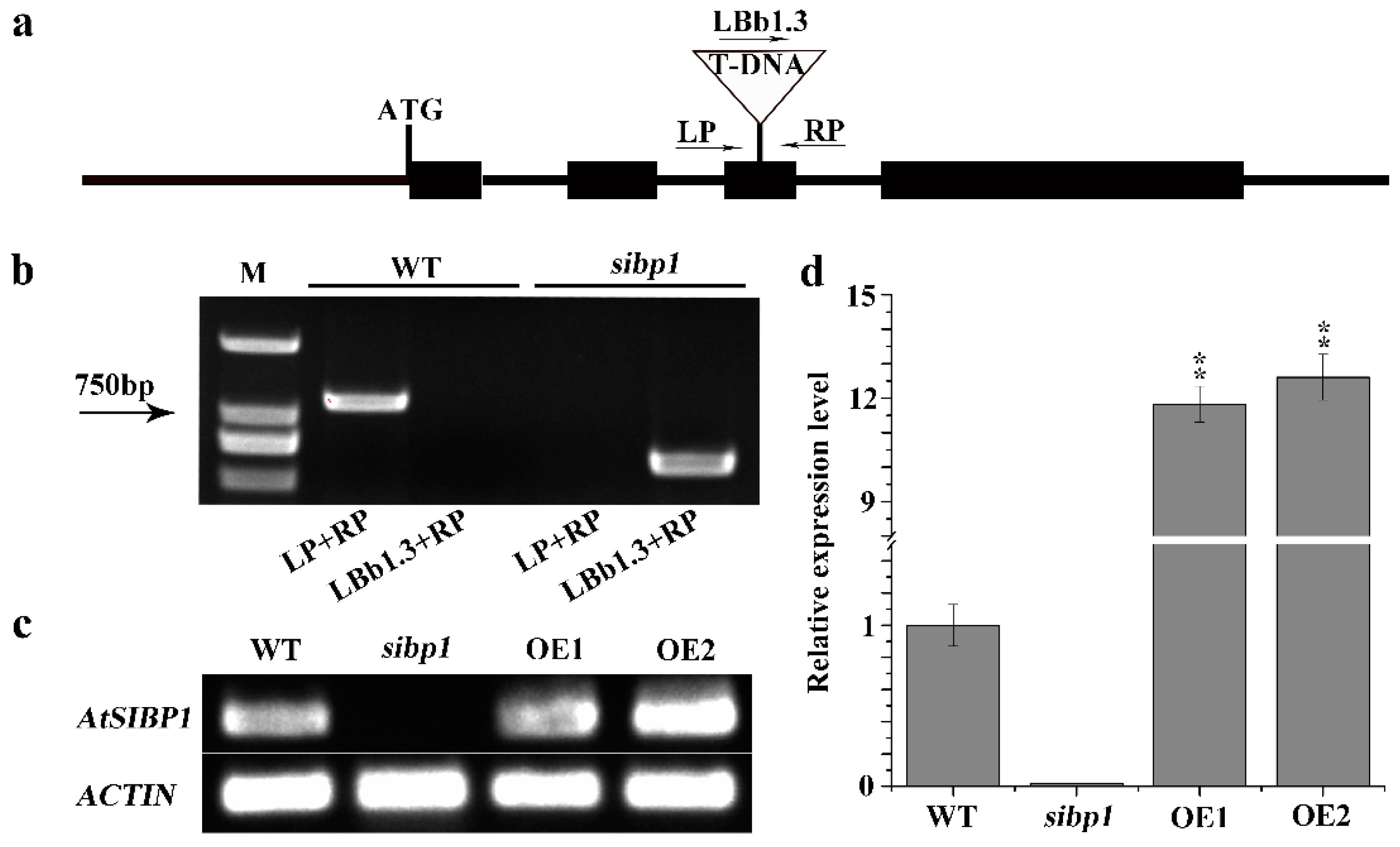
2.2. Identification of Arabidopsis sibp1 Mutant and AtSIBP1 Overexpression Lines
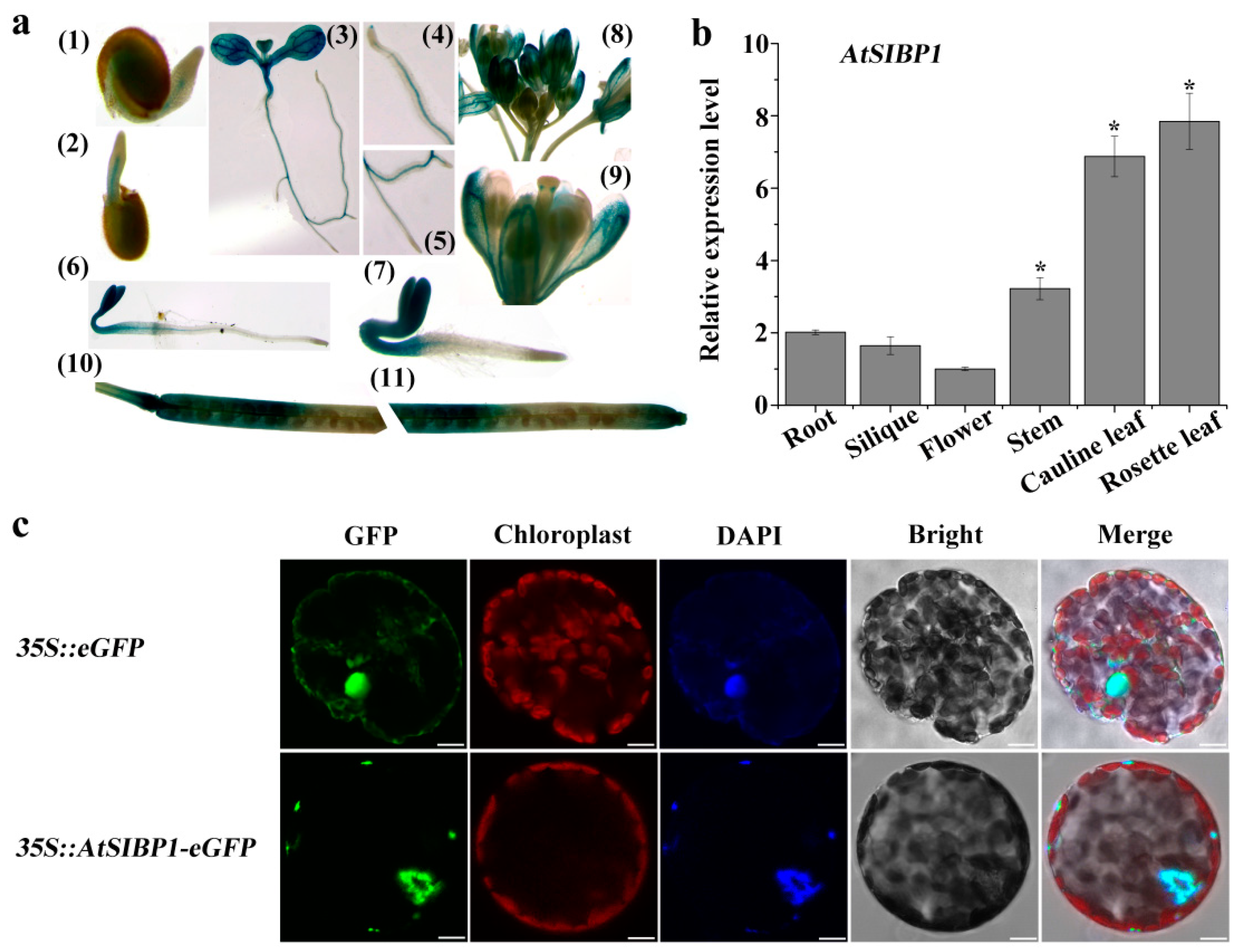
2.3. Tissue Expression Pattern of AtSIBP1 and Subcellular Localization of AtSIBP1-eGFP Fusion Protein
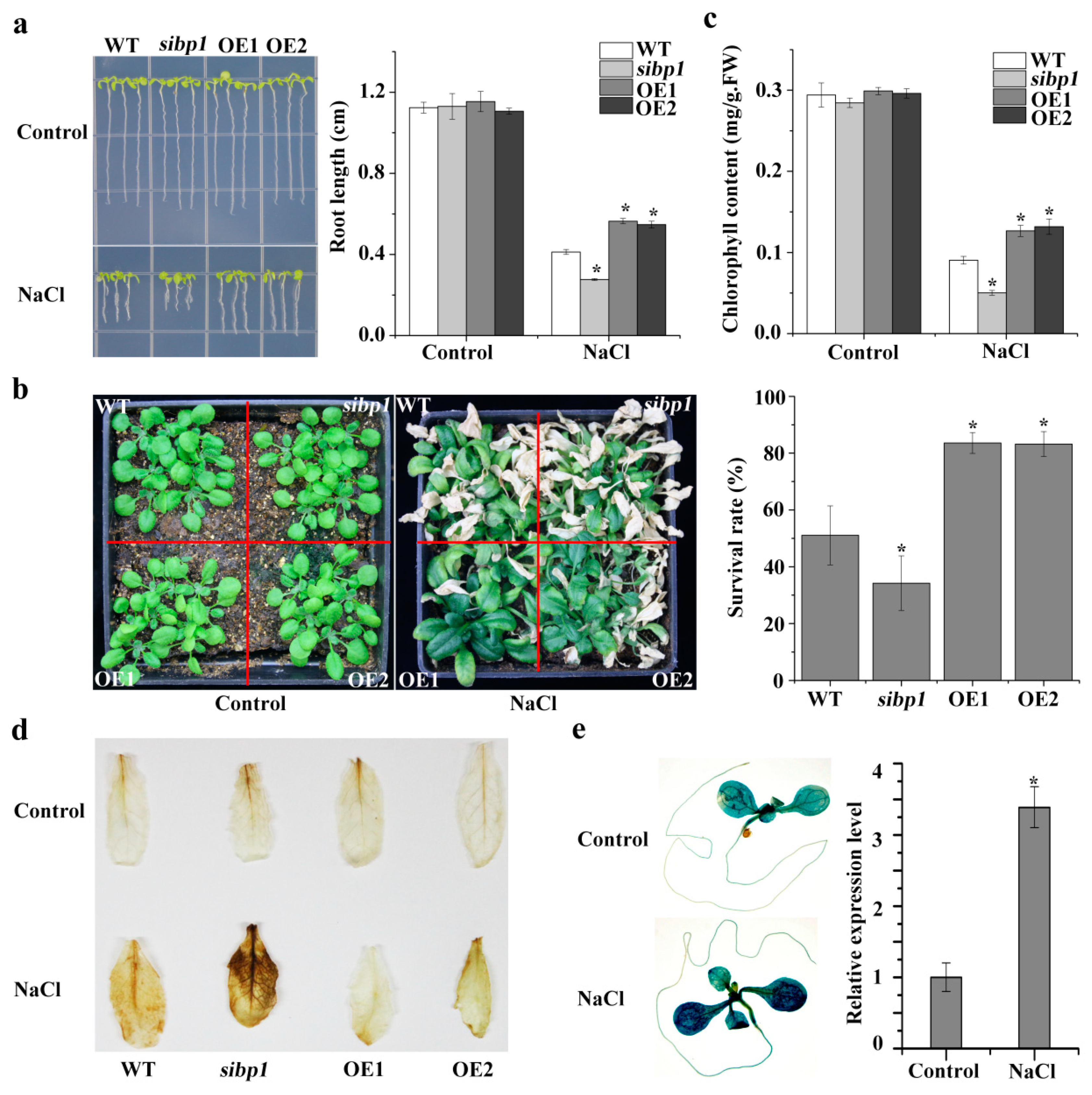
2.4. AtSIBP1 Acts as a Positive Regulator in Response to Salt Stress
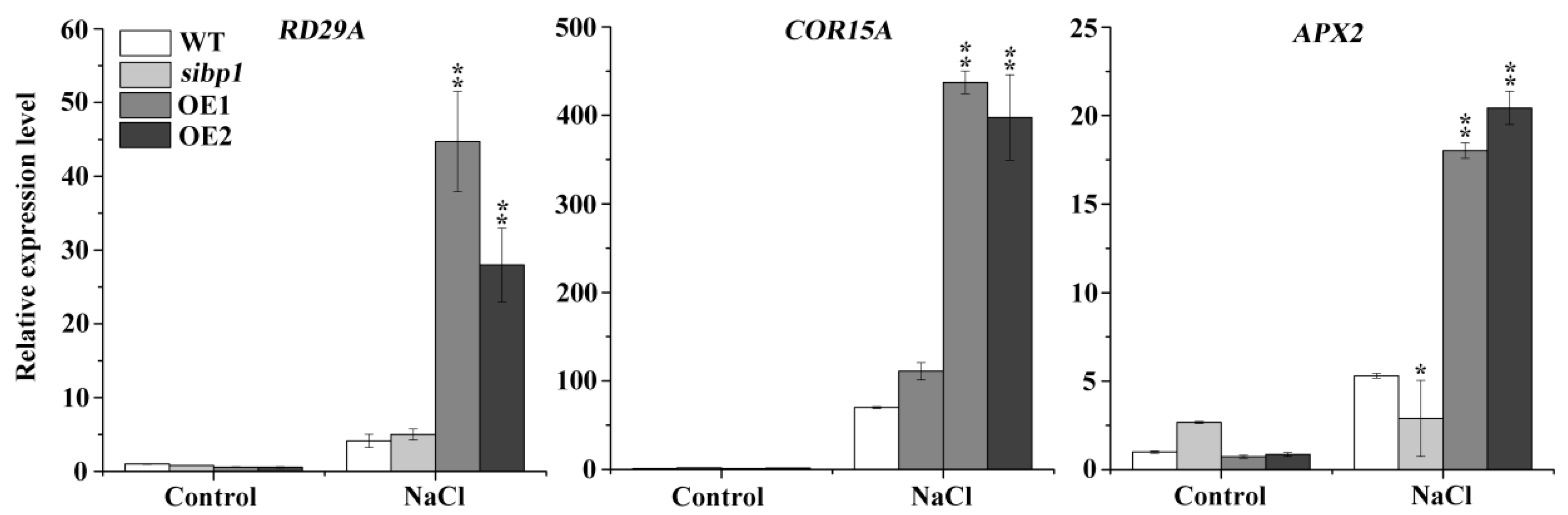
2.5. Disruption or Overexpressing of AtSIBP1 Altered the Expression of Stress Induced Genes
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Generation of Transgenic Plants
4.3. Subcellular Localization Assay
4.4. Phenotype Analysis
4.5. GUS and DAB Staining
4.6. Analysis of Gene Expression
4.7. Sequence Analysis of AtSIBP1
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Tada, Y. The HKT Transporter Gene from Arabidopsis, AtHKT1; 1, Is Dominantly Expressed in Shoot Vascular Tissue and Root Tips and Is Mild Salt Stress-Responsive. Plants 2019, 8, 204. [Google Scholar] [CrossRef]
- Zhu, J.; Fu, X.; Koo, Y.D.; Zhu, J.K.; Jenney, F.E.; Adams, M.W.; Zhu, Y.; Shi, H.; Yun, D.-J.; Hasegawa, P.M.; et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol. Cell. Biol. 2007, 27, 5214–5224. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003, 8, 135–142. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, W.T. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 2011, 31, 201–208. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Lechner, E.; Leonhardt, N.; Eisler, H.; Parmentier, Y.; Alioua, M.; Jacquet, H.; Leung, J.; Genschik, P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 2011, 21, 1116–1128. [Google Scholar] [CrossRef]
- Weber, H.; Bernhardt, A.; Dieterle, M.; Hano, P.; Mutlu, A.; Estelle, M.; Genschik, P.; Hellmann, H. Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 2005, 137, 83–93. [Google Scholar] [CrossRef]
- Thomann, A.; Dieterle, M.; Genschik, P. Plant CULLIN-based E3s: Phytohormones come first. FEBS Lett. 2005, 579, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.; Nandra, S.K.; Privé, G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef] [PubMed]
- Dow, M.R.; Mains, P.E. Genetic and Molecular Characterization of the Caenorhabditis elegans Gene, mel-26, a Postmeiotic Negative Regulator of MEI-1, a Meiotic-Specific Spindle Component. Genetics 1998, 150, 119–128. [Google Scholar] [PubMed]
- Bomont, P.; Cavalier, L.; Blondeau, F.; Hamida, C.B.; Belal, S.; Tazir, M.; Demir, E.; Topaloglu, H.; Korinthenberg, R.; Tüysüz, B.; et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 2000, 26, 370. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Stone, J.R.; Williams, A.J. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001, 21, 3609–3615. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. NPR1 Modulates Cross-Talk between Salicylate- and Jasmonate-Dependent Defense Pathways through a Novel Function in the Cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef]
- Dong, X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004, 7, 547–552. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van Loon, L.C. NPR1: The spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 2004, 7, 456–464. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.H.; Seo, D.H.; Chung, S.; Kim, S.W.; Lee, J.S.; Kim, W.T.; Lee, J.-H. ABA-HYPERSENSITIVE BTB/POZ PROTEIN 1 functions as a negative regulator in ABA-mediated inhibition of germination in Arabidopsis. Plant Mol. Biol. 2016, 90, 303–315. [Google Scholar] [CrossRef]
- Huang, K.; Peng, L.; Liu, Y.; Yao, R.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef]
- Cao, H.G.J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Larbat, R.; Le Bot, J.; Bourgaud, F.; Robin, C.; Adamowicz, S. Organ-specific responses of tomato growth and phenolic metabolism to nitrate limitation. Plant Biol. 2012, 14, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Huang, B. Root proteomic responses to heat stress in two Agrostis grass species contrasting in heat tolerance. J. Exp. Bot. 2008, 59, 4183–4194. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Jin, Q.; Borejsza-Wysocka, E.E.; He, S.Y.; Aldwinckle, H.S. Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Mol. Plant-Microbe Interact. 2007, 12, 1568–1580. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Qu, S.C.; Qiao, Y.S.; Zhang, Z.; Guo, Z.R. Overexpression of the Malus hupehensis MhNPR1 gene increased tolerance to salt and osmotic stress in transgenic tobacco. Mol. Biol. Rep. 2014, 41, 1553–1561. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Canet, J.V.; Dobón, A.; Fajmonová, J.; Tornero, P. The BLADE-ON-PETIOLE genes of Arabidopsis are essential for resistance induced by methyl jasmonate. BMC Plant Biol. 2012, 12, 199. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, L.; Wang, H.; Zhang, L.; Wang, F.; Yu, D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. Cell Mol. Biol. 2013, 74, 730–745. [Google Scholar] [CrossRef]
- Wang, X.; Hou, C.; Zheng, K.; Li, Q.; Chen, S.; Wang, S. Overexpression of ERF96, a small ethylene response factor gene, enhances salt tolerance in Arabidopsis. Biol. Plant. 2017, 61, 693–701. [Google Scholar] [CrossRef]
- Wang, X.; Huang, W.; Yang, Z.; Liu, J.; Huang, B. Transcriptional regulation of heat shock proteins and ascorbate peroxidase by CtHsfA2b from African bermudagrass conferring heat tolerance in Arabidopsis. Sci. Rep. 2016, 6, 28021. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Byun, M.Y.; Lee, J.; Cui, L.H.; Kang, Y.; Oh, T.K.; Park, H.; Lee, H.; Kim, W.T. Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. Int. J. Exp. Plant Biol. 2015, 236, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1987; p. 48. [Google Scholar]
- Liu, D.; Gong, Q.; Ma, Y.; Li, P.; Li, J.; Yang, S.; Yuan, L.; Yu, Y.; Pan, D.; Xu, F.; et al. cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J. Exp. Bot. 2010, 61, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, X.; Peng, L.; Xiong, J.; Li, X.; Wang, J.; Li, X.; Yang, Y. AtSIBP1, a Novel BTB Domain-Containing Protein, Positively Regulates Salt Signaling in Arabidopsis thaliana. Plants 2019, 8, 573. https://doi.org/10.3390/plants8120573
Wan X, Peng L, Xiong J, Li X, Wang J, Li X, Yang Y. AtSIBP1, a Novel BTB Domain-Containing Protein, Positively Regulates Salt Signaling in Arabidopsis thaliana. Plants. 2019; 8(12):573. https://doi.org/10.3390/plants8120573
Chicago/Turabian StyleWan, Xia, Lu Peng, Jie Xiong, Xiaoyi Li, Jianmei Wang, Xufeng Li, and Yi Yang. 2019. "AtSIBP1, a Novel BTB Domain-Containing Protein, Positively Regulates Salt Signaling in Arabidopsis thaliana" Plants 8, no. 12: 573. https://doi.org/10.3390/plants8120573
APA StyleWan, X., Peng, L., Xiong, J., Li, X., Wang, J., Li, X., & Yang, Y. (2019). AtSIBP1, a Novel BTB Domain-Containing Protein, Positively Regulates Salt Signaling in Arabidopsis thaliana. Plants, 8(12), 573. https://doi.org/10.3390/plants8120573




