Genome-Wide Investigation of the Role of MicroRNAs in Desiccation Tolerance in the Resurrection Grass Tripogon loliiformis
Abstract
1. Introduction
2. Results
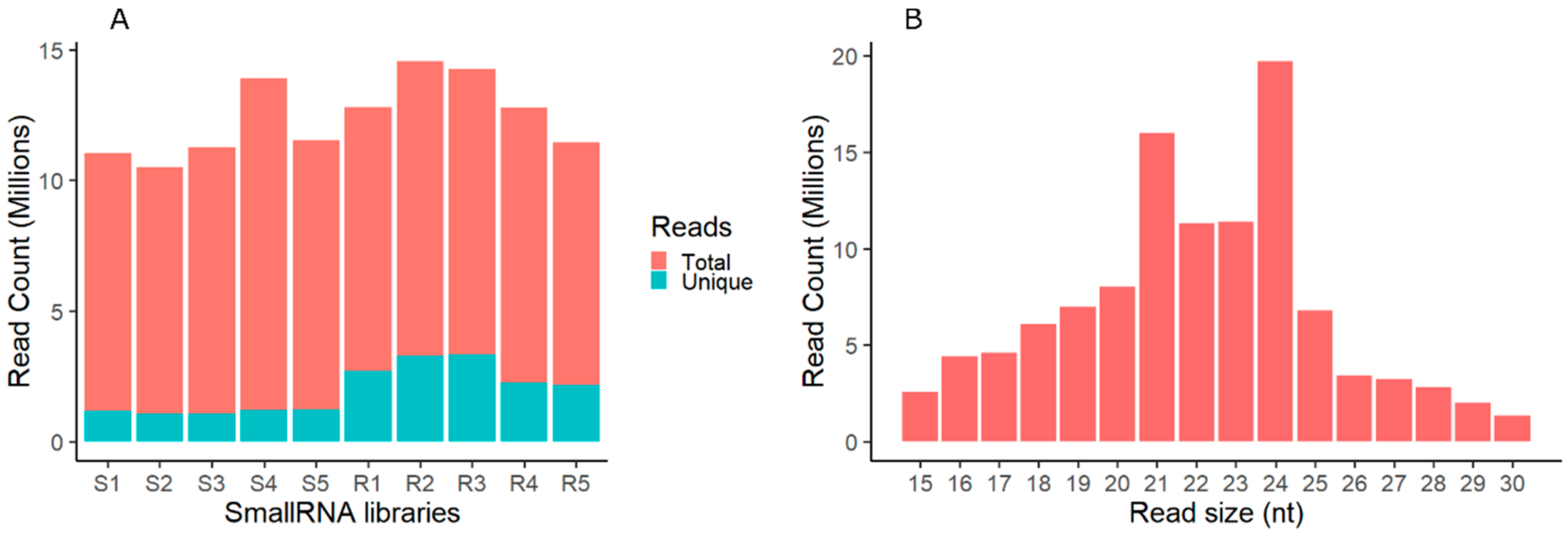
2.1. Analysis of Small RNAs in Shoot and Root of Tripogon loliiformis
2.2. Evolutionary Conservation of MiRNAs in Tripogon loliiformis
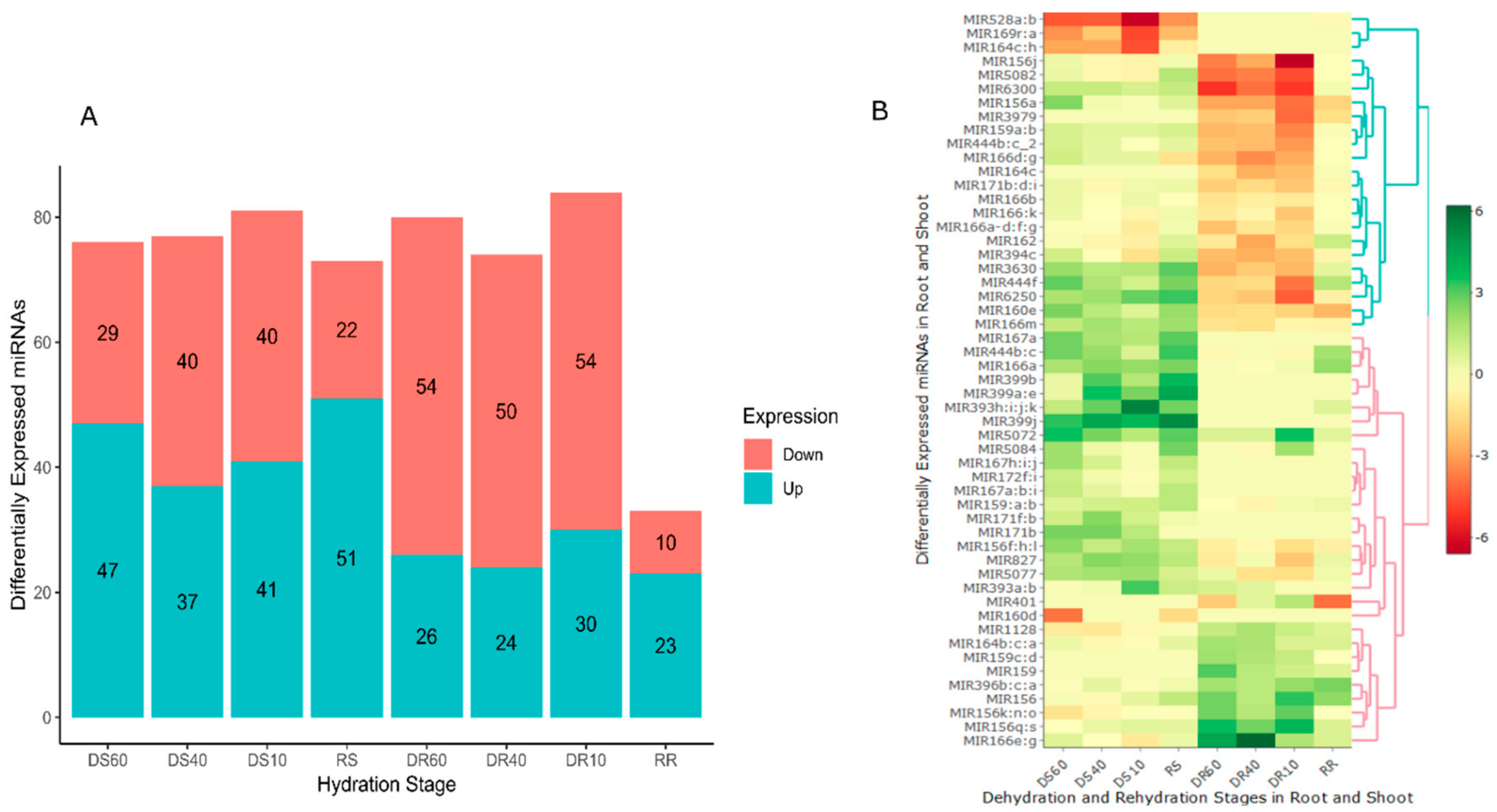
2.3. Spatiotemporal Expression of MiRNAs in Tripogon loliiformis Tissues
2.4. Tripogon loliiformis Stress-Associated MiRNAs Targets
2.5. Functional Roles of MiRNA Targets
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Total RNA Extraction and High Throughput Sequencing
4.3. Small RNAs Data Analysis and MiRNAs Identification
4.4. MiRNAs Expression Analysis
4.5. MiRNAs Targets and Their Functional Enrichment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Höfler, K.; Migsch, H.; Rottenburg, W. Über die Austrocknungresistenz landwirtschaftlicher Kulturpflanzen. Forschungsdienst 1941, 12, 50–61. [Google Scholar]
- Rascio, N.; Rocca, N.L. Resurrection Plants: The Puzzle of Surviving Extreme Vegetative Desiccation. Crit. Rev. Plant Sci. 2005, 24, 209–225. [Google Scholar] [CrossRef]
- Scott, P. Resurrection Plants and the Secrets of Eternal Leaf. Ann. Bot. 2000, 85, 159–166. [Google Scholar] [CrossRef]
- Bernacchia, G.; Salamini, F.; Bartels, D. Molecular characterization of the rehydration process in the resurrection plant Craterostigma plantagineum. Plant Physiol. 1996, 111, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.M. A comparison of mechanisms of desicassion tolerance among 3 angiosperms resurrection plant species. Plant Ecol. 2000, 151, 29–39. [Google Scholar] [CrossRef]
- Moore, J.P.; Farrant, J.M. A Systems-Based Molecular Biology Analysis of Resurrection Plants for Crop and Forage Improvement in Arid Environments. In Improving Crop Resistance to Abiotic Stress; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2012; pp. 399–418. [Google Scholar] [CrossRef]
- Peters, S.; Mundree, S.G.; Thomson, J.A.; Farrant, J.M.; Keller, F. Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): Both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. J. Exp. Bot. 2007, 58, 1947–1956. [Google Scholar] [CrossRef] [PubMed]
- Asami, P.; Mundree, S.; Williams, B. Saving for a rainy day: Control of energy needs in resurrection plants. Plant Sci. 2018, 271, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Karbaschi, M.R.; Williams, B.; Taji, A.; Mundree, S.G. Tripogon loliiformis elicits a rapid physiological and structural response to dehydration for desiccation tolerance. Funct. Plant Biol. 2016. [Google Scholar] [CrossRef]
- Orchard, E.; Wilson, A. Flora of Australia. In Poaceae Australian Biological Resources Study; K Mallett, T.M., Ed.; CSIRO: Canberra, Australia, 2005. [Google Scholar]
- Gaff, D.; Pate, J.S. The Biology of Resurrection Plants; Pate, J.S., McComb, A.J., Eds.; The Biology of Australian Plants, University Western Australia Press: Nedlands, Australia, 1981; pp. 114–146. [Google Scholar]
- Williams, B.; Njaci, I.; Moghaddam, L.; Long, H.; Dickman, M.B.; Zhang, X.; Mundree, S. Trehalose Accumulation Triggers Autophagy during Plant Desiccation. PLoS Genet. 2015, 11, e1005705. [Google Scholar] [CrossRef] [PubMed]
- Gechev, T.S.; Benina, M.; Obata, T.; Tohge, T.; Sujeeth, N.; Minkov, I.; Hille, J.; Temanni, M.-R.; Marriott, A.S.; Bergström, E. Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cell. Mol. Life Sci. 2013, 70, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.C.S.; Edsgard, D.; Hussain, S.S.; Alquezar, D.; Rasmussen, M.; Gilbert, T.; Nielsen, B.H.; Bartels, D.; Mundy, J. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 2010, 63, 212–228. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, X.; Jiao, Y.; Qin, Y.; Liu, X.; He, K.; Chen, C.; Ma, L.; Wang, J.; Xiong, L. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol. Biol. 2007, 63, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R. MicroRNAs with macro-effects on plant stress responses. Semin. Cell Dev. Biol. 2010, 21, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Li, J.; Song, R.; Messing, J.; Chen, X. CARPEL FACTORY, a Dicer Homolog, and HEN1, a Novel Protein, Act in microRNA Metabolism in Arabidopsis thaliana. Curr. Biol. 2002, 12, 1484–1495. [Google Scholar] [CrossRef]
- Zhao, T.; Li, G.; Mi, S.; Li, S.; Hannon, G.J.; Wang, X.-J.; Qi, Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007, 21, 1190–1203. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.W.; Xue, L.G.; An, L.Z. Functional diversity of miRNA in plants. Plant Sci. 2007, 172, 423–432. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, Y.Q. Insights into the mechanism of plant development: Interactions of miRNAs pathway with phytohormone response. Biochem. Biophys. Res. Commun. 2009, 384, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Shukla, L.I.; Chinnusamy, V.; Sunkar, R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2008, 1779, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Frazier, T.P.; Sun, G.; Burklew, C.E.; Zhang, B. Salt and Drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 2011, 49, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qin, Y.; Duan, H.; Yin, W.; Xia, X. Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 2011, 62, 3765–3779. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Guo, G.; Du, J.; Guo, W.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol. 2014, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zheng, Y.; Jagadeeswaran, G.; Li, Y.; Gowdu, K.; Sunkar, R. Identification and temporal expression analysis of conserved and novel microRNAs in Sorghum. Genomics 2011, 98, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Matts, J.; Jagadeeswaran, G.; Roe, B.A.; Sunkar, R. Identification of microRNAs and their targets in switchgrass, a model biofuel plant species. J. Plant Physiol. 2010, 167, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Akpinar, A. Dehydration stress-responsive miRNA in Brachypodium distachyon: Evident by genome-wide screening of microRNAs expression. OMICS 2011, 15, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Liang, R.; Ge, L.; Li, W.; Xiao, H.; Lin, H.; Ruan, K.; Jin, Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006, 46, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; Van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Guo, Z.; Li, L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 2013, 380, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Oliver, M.J.; Guo, L.; Alexander, D.C.; Ryals, J.A.; Wone, B.W.; Cushman, J.C. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell Online 2011, 23, 1231–1248. [Google Scholar] [CrossRef] [PubMed]
- Gechev, T.; Dinakar, C.; Benina, M.; Toneva, V.; Bartels, D. Molecular mechanisms of desiccation tolerance in resurrection plants. Cell. Mol. Life Sci. 2012, 69, 3175–3186. [Google Scholar] [CrossRef] [PubMed]
- Sra, N. Bioproject: Transcriptome Analysis of Tripogon Loliiformis throughout Dehydration, Desiccation and Rehydration. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA288839 (accessed on 22 April 2018).
- Whittaker, A.; Bochicchio, A.; Vazzana, C.; Lindsey, G.; Farrant, J. Changes in leaf hexokinase activity and metabolite levels in response to drying in the desiccation-tolerant species Sporobolus stapfianus and Xerophyta viscosa. J. Exp. Bot. 2001, 52, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef] [PubMed]
- Alpert, P.; Oliver, M.J. 1 Drying Without Dying. In Desiccation and Survival in Plants: Drying without Dying; CABI: Wallingford, UK, 2002; p. 3. [Google Scholar]
- Farrant, J.; Cooper, K.; Hilgart, A.; Abdalla, K.; Bentley, J.; Thomson, J.; Dace, H.W.; Peton, N.; Mundree, S.; Rafudeen, M. A molecular physiological review of vegetative desiccation tolerance in the resurrection plant Xerophyta viscosa (Baker). Planta 2015, 242, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, J.; Hussain, S.S.; Shi, B.J. Role of microRNAs in plant drought tolerance. Plant Biotechnol. J. 2015, 13, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Geng, S.; Wang, D.; Feng, N.; Zhang, D.; Wu, L.; Hao, C.; Zhang, X.; Li, A.; Mao, L. Characterization of Squamosa Promoter Binding Protein-LIKE genes in wheat. J. Plant Biol. 2015, 58, 220–229. [Google Scholar] [CrossRef]
- Wang, C.; Deng, P.; Chen, L.; Wang, X.; Ma, H.; Hu, W.; Yao, N.; Feng, Y.; Chai, R.; Yang, G.; et al. A Wheat WRKY Transcription Factor TaWRKY10 Confers Tolerance to Multiple Abiotic Stresses in Transgenic Tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Besseau, S.; Törönen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Ruberti, C.; Kim, S.-J.; Stefano, G.; Brandizzi, F. Unfolded protein response in plants: One master, many questions. Curr. Opin. Plant Biol. 2015, 27, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Verchot, J.; Dickman, M. When Supply Does Not Meet Demand-ER Stress and Plant Programmed Cell Death. Front. Plant Sci. 2014, 5, 211. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Tuteja, N. Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal. Behav. 2011, 6, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Sarwat, M.; Naqvi, A.R. Heterologous expression of rice calnexin (OsCNX) confers drought tolerance in Nicotiana tabacum. Mol. Biol. Rep. 2013, 40, 5451–5464. [Google Scholar] [CrossRef] [PubMed]
- Ghasempour, H.; Gaff, D.; Williams, R.; Gianello, R. Contents of sugars in leaves of drying desiccation tolerant flowering plants, particularly grasses. Plant Growth Regul. 1998, 24, 185–191. [Google Scholar] [CrossRef]
- Farrant, J.M.; Moore, J.P. Programming desiccation-tolerance: From plants to seeds to resurrection plants. Curr. Opin. Plant Biol. 2011, 14, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Illing, N.; Denby, K.J.; Collett, H.; Shen, A.; Farrant, J.M. The signature of seeds in resurrection plants: A molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integr. Comp. Biol. 2005, 45, 771–787. [Google Scholar] [CrossRef] [PubMed]
- Ried, J.L.; Walker-Simmons, M. Group 3 late embryogenesis abundant proteins in desiccation-tolerant seedlings of wheat (Triticum aestivum L.). Plant Physiol. 1993, 102, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Bender, K.W.; Snedden, W.A. Calmodulin-Related Proteins Step Out from the Shadow of Their Namesake. Plant Physiol. 2013, 163, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Hu, X.; Cai, W.; Huang, W.; Zhou, X.; Luo, Q.; Yang, H.; Wang, J.; Huang, J. Arabidopsis miR171-Targeted Scarecrow-Like Proteins Bind to GT cis-Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions. PLOS Genet. 2014, 10, e1004519. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.T. Molecular and Functional Characterisation of an Osmotin Gene from the Resurrection Plant Tripogon Loliiformis. Ph.D. Thesis, Queensland University of Technology, Brisbane, Australia, 2018. Available online: https://eprints.qut.edu.au/115835 (accessed on 30 August 2018).
- Barrs, H.; Weatherley, P. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef]
- Qiagen. CLC Genomics Workbench. Available online: https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/ (accessed on 22 April 2018).
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
| Name | Contig Number | Accession Number | Target Description |
|---|---|---|---|
| ABIOTIC STRESS RESPONSE | |||
| MIR408 | Contig_1263 | XP_004951273.1 | BAG family molecular chaperone regulator 6 |
| MIR172j | Contig_4654 | EMS50802.1 | DnaJ homolog subfamily C member 7 |
| MIR3630 | Contig_195 | CAA47948.2 | heat shock protein 70 |
| MIR6250 | Contig_29066 | XP_003578630.1 | hydrophobic protein OSR8-like |
| MIR166d | Contig_10495 | XP_003557818.1 | probable methyltransferase PMT2-like |
| CARBOHYDRATE METABOLISM | |||
| MIR319a | Contig_91996 | XP_003568782.1 | hexokinase-7-like |
| MIR172h | Contig_914 | XP_004984086.1 | sucrose synthase 2-like |
| MIR167c | Contig_12310 | XP_004965756.1 | sucrose-phosphate synthase 3-like |
| MIR394c | Contig_1 | XP_003566746.1 | callose synthase 7-like |
| MIR166b | Contig_1239 | XP_004984579.1 | galactinol synthase 2-like |
| MIR167e | Contig_3855 | XP_004958079.1 | xylose isomerase-like |
| DEVELOPMENT ASSOCIATED | |||
| MIR5021 | Contig_16313 | XP_004966491.1 | senescence-associated protein DIN1-like |
| MIR172b | Contig_2082 | XP_004962343.1 | serine/threonine-protein kinase TOR-like |
| MIR528 | Contig_16524 | XP_004951509.1 | serine-threonine kinase receptor protein |
| MIR156c | Contig_10275 | XP_004957197.1 | squamosa promoter-binding-like protein 17-like |
| MIR156 | Contig_10275 | XP_004957197.1 | squamosa promoter-binding-like protein 17-like |
| HORMONE METABOLISM | |||
| MIR160d | Contig_39233 | EMT29346.1 | 1-aminocyclopropane-1-carboxylate synthase 7 |
| MIR167h | Contig_16664 | XP_004961973.1 | ABSCISIC ACID-INSENSITIVE 5-like protein |
| MIR164a | Contig_14909 | XP_004985140.1 | Allene oxide synthase 2-like |
| MIR172b | Contig_975 | XP_004956401.1 | auxin transport protein BIG-like |
| MIR171e | Contig_10826 | XP_004976258.1 | IAA-amino acid hydrolase ILR1-like 5-like |
| DNA REPAIR/SYNTHESIS/CHROMATIN STRUCTURE | |||
| MIR166h | Contig_3556 | XP_003576514.1 | DNA mismatch repair protein Msh6-1-like |
| MIR167a | Contig_548 | XP_004975516.1 | DNA repair helicase XPB1-like |
| MIR172a | Contig_49068 | NP_001151792.1 | DNA binding protein |
| MIR319g:l | Contig_5793 | NP_001065884.1 | HUA enhancer 2 |
| MIR399a:e | Contig_961 | XP_004961835.1 | probable histone H2A.4-like |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Njaci, I.; Williams, B.; Castillo-González, C.; Dickman, M.B.; Zhang, X.; Mundree, S. Genome-Wide Investigation of the Role of MicroRNAs in Desiccation Tolerance in the Resurrection Grass Tripogon loliiformis. Plants 2018, 7, 68. https://doi.org/10.3390/plants7030068
Njaci I, Williams B, Castillo-González C, Dickman MB, Zhang X, Mundree S. Genome-Wide Investigation of the Role of MicroRNAs in Desiccation Tolerance in the Resurrection Grass Tripogon loliiformis. Plants. 2018; 7(3):68. https://doi.org/10.3390/plants7030068
Chicago/Turabian StyleNjaci, Isaac, Brett Williams, Claudia Castillo-González, Martin B. Dickman, Xiuren Zhang, and Sagadevan Mundree. 2018. "Genome-Wide Investigation of the Role of MicroRNAs in Desiccation Tolerance in the Resurrection Grass Tripogon loliiformis" Plants 7, no. 3: 68. https://doi.org/10.3390/plants7030068
APA StyleNjaci, I., Williams, B., Castillo-González, C., Dickman, M. B., Zhang, X., & Mundree, S. (2018). Genome-Wide Investigation of the Role of MicroRNAs in Desiccation Tolerance in the Resurrection Grass Tripogon loliiformis. Plants, 7(3), 68. https://doi.org/10.3390/plants7030068





