N6-Methyladenosine (m6A) Methylation-Mediated Transcriptional Regulation in Maize Root Response to Salt Stress
Abstract
1. Introduction
2. Results
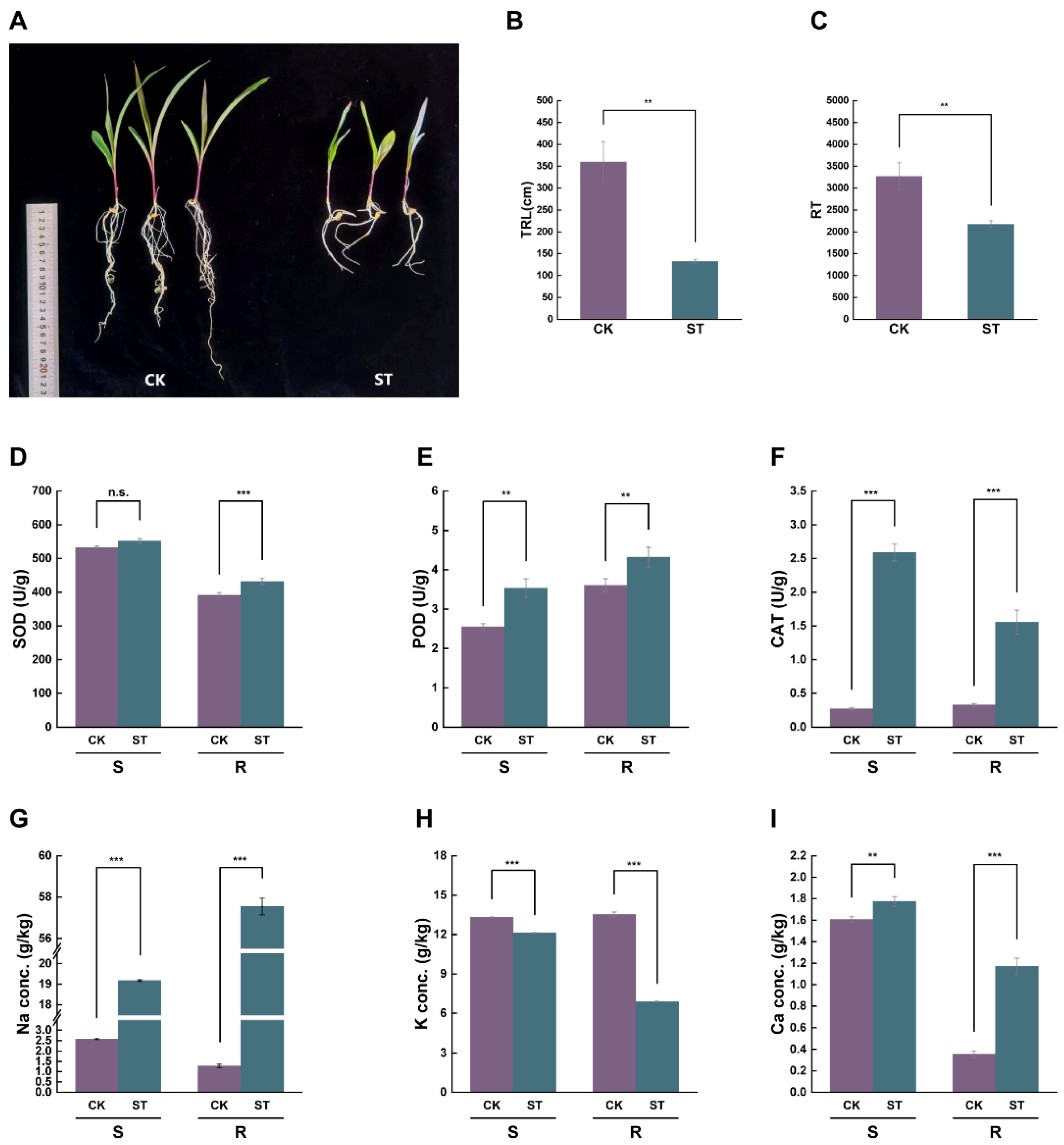
2.1. Impact of Salt Stress on Maize Seedling Growth, Antioxidant Enzyme Activities, and Ion Contents
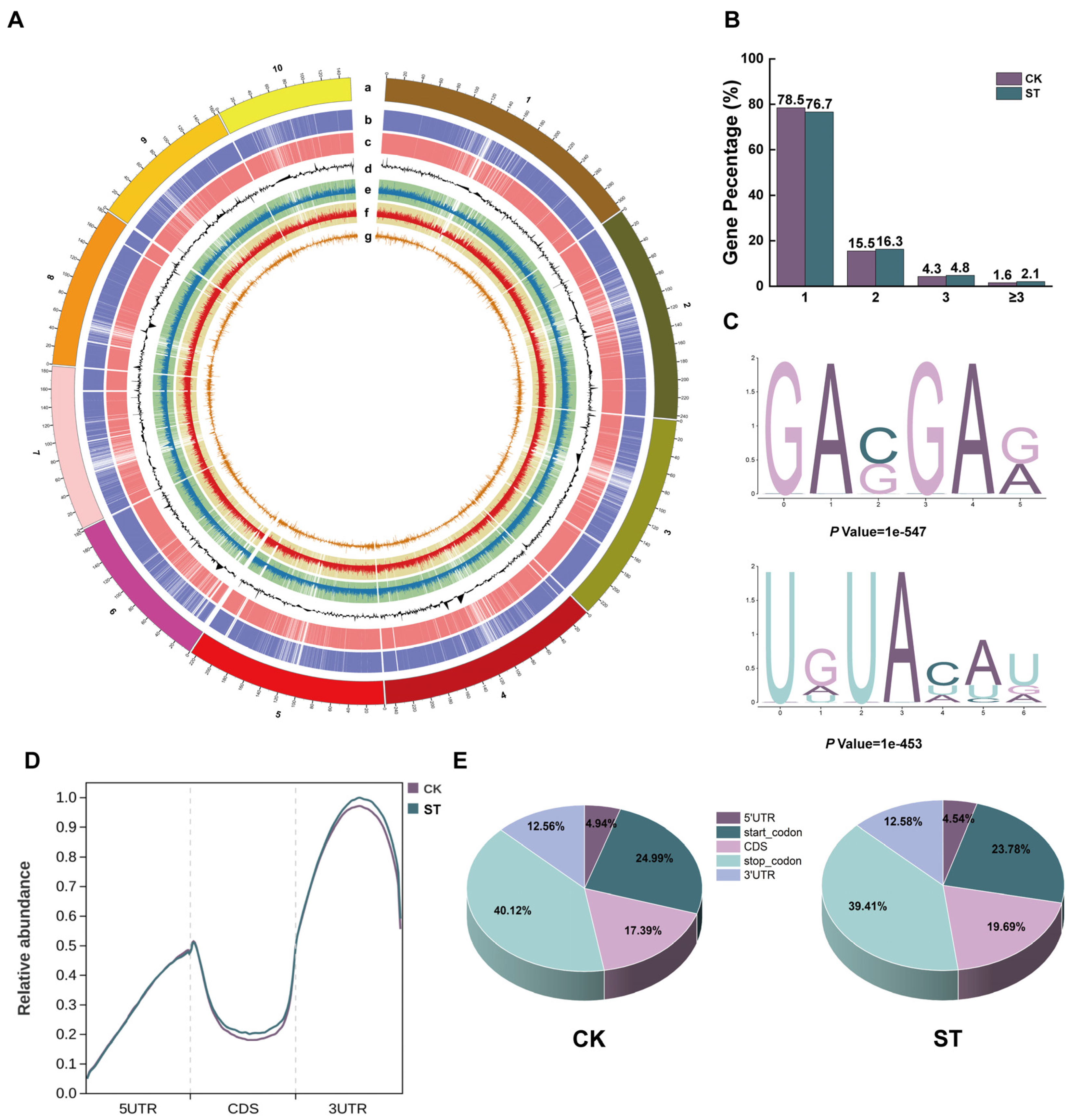
2.2. Genome-Wide Analysis of m6A Methylation Profile in Maize Roots Under Salt Stress
2.3. Identification and Functional Enrichment Analysis of DMPs in Maize Roots Under Salt Stress
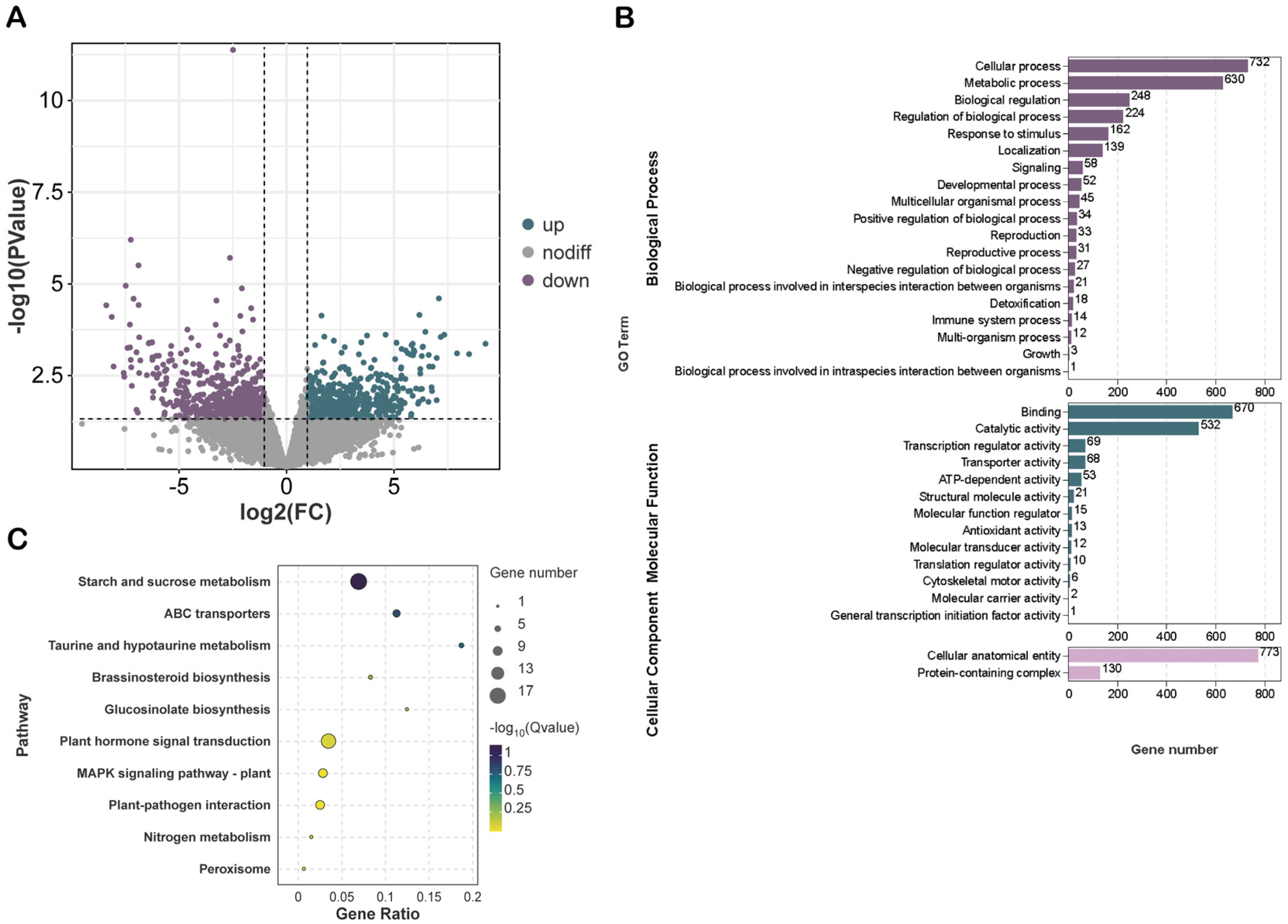
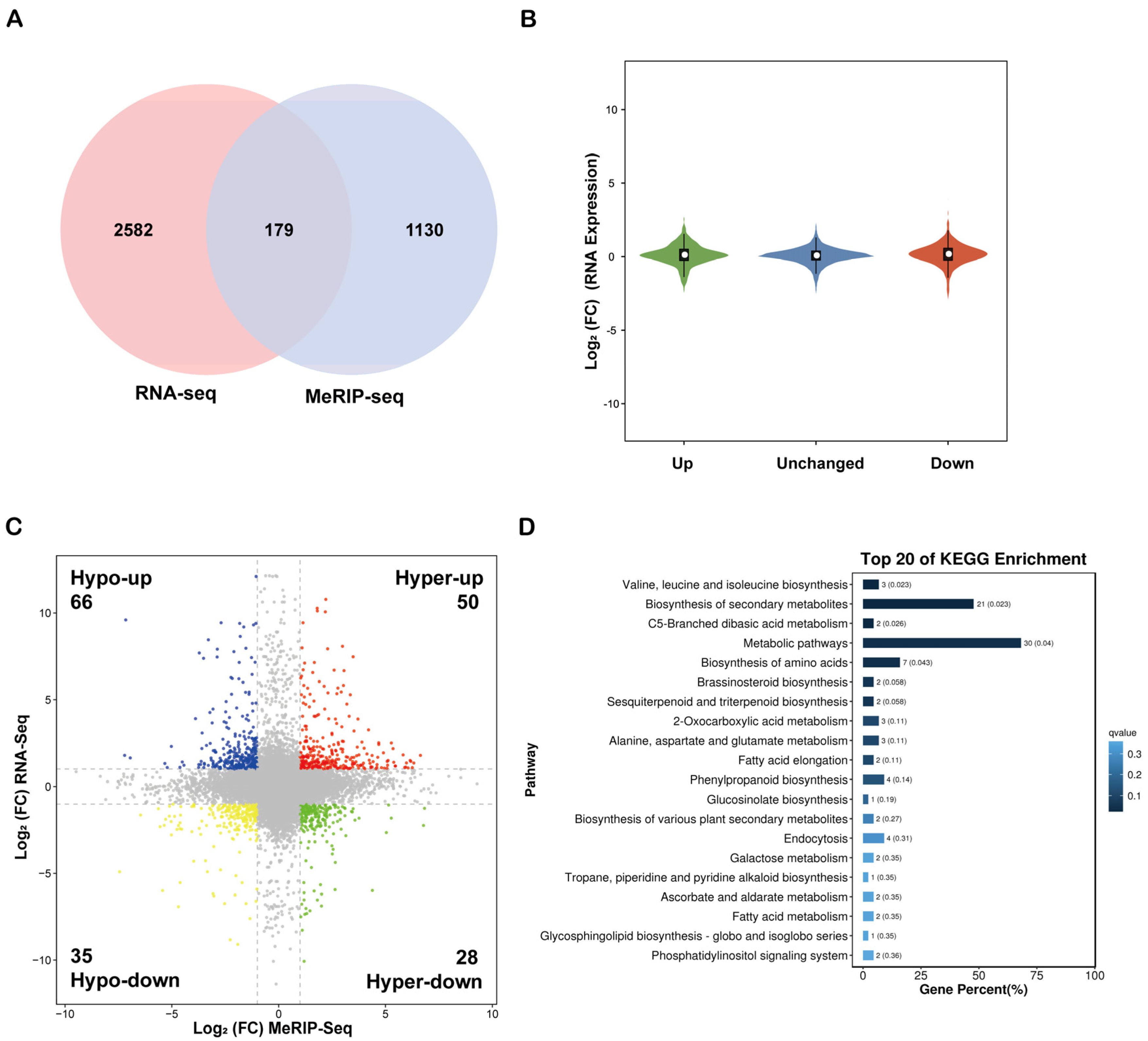
2.4. Global Analysis and Functional Enrichment of DEGs in Maize Root Transcriptome Under Salt Stress
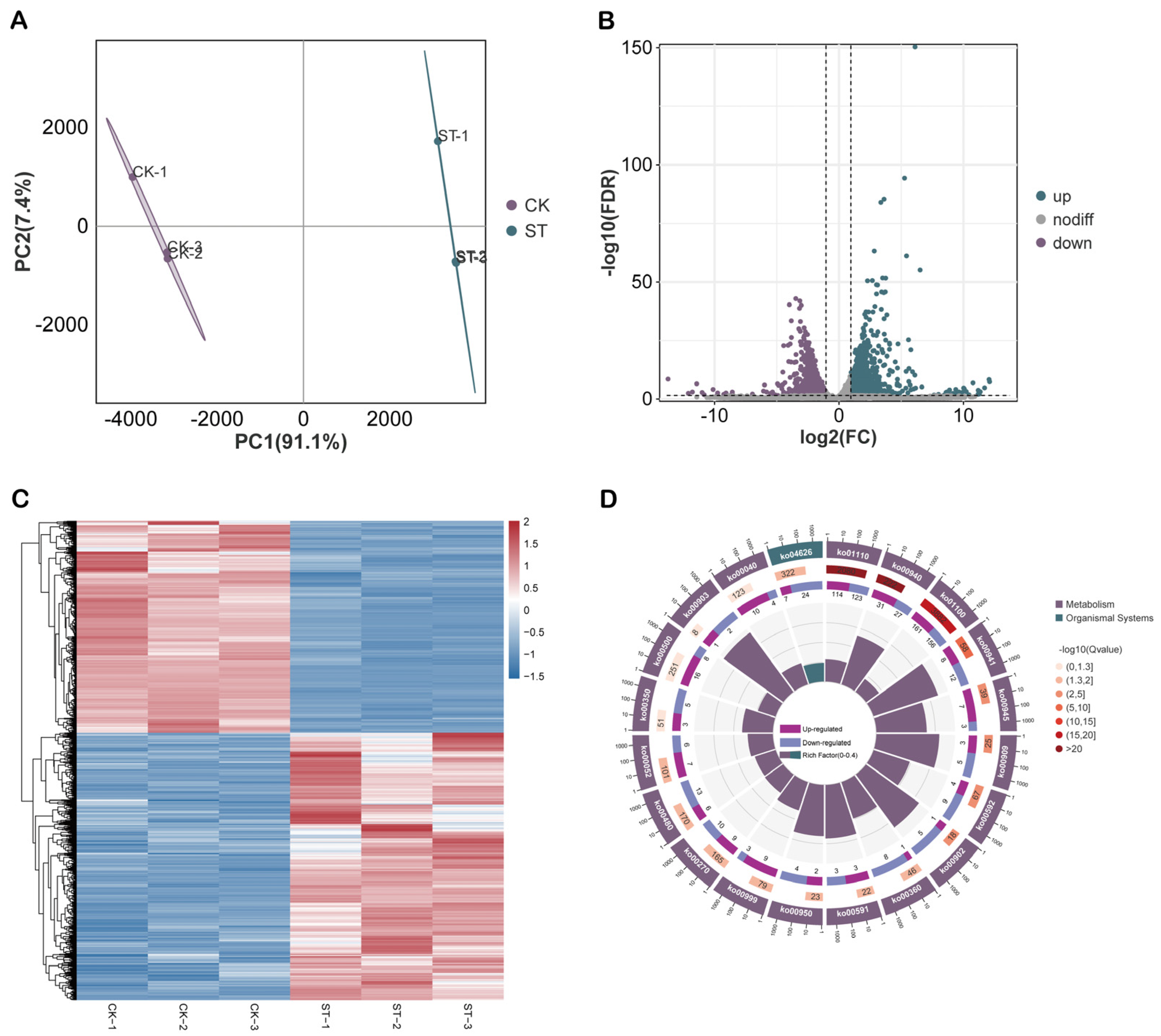
2.5. Association Analysis of m6A Methylation and Transcriptome
2.6. Response of m6A Methylation-Related Gene Writers, Readers, and Erasers to Salt Stress
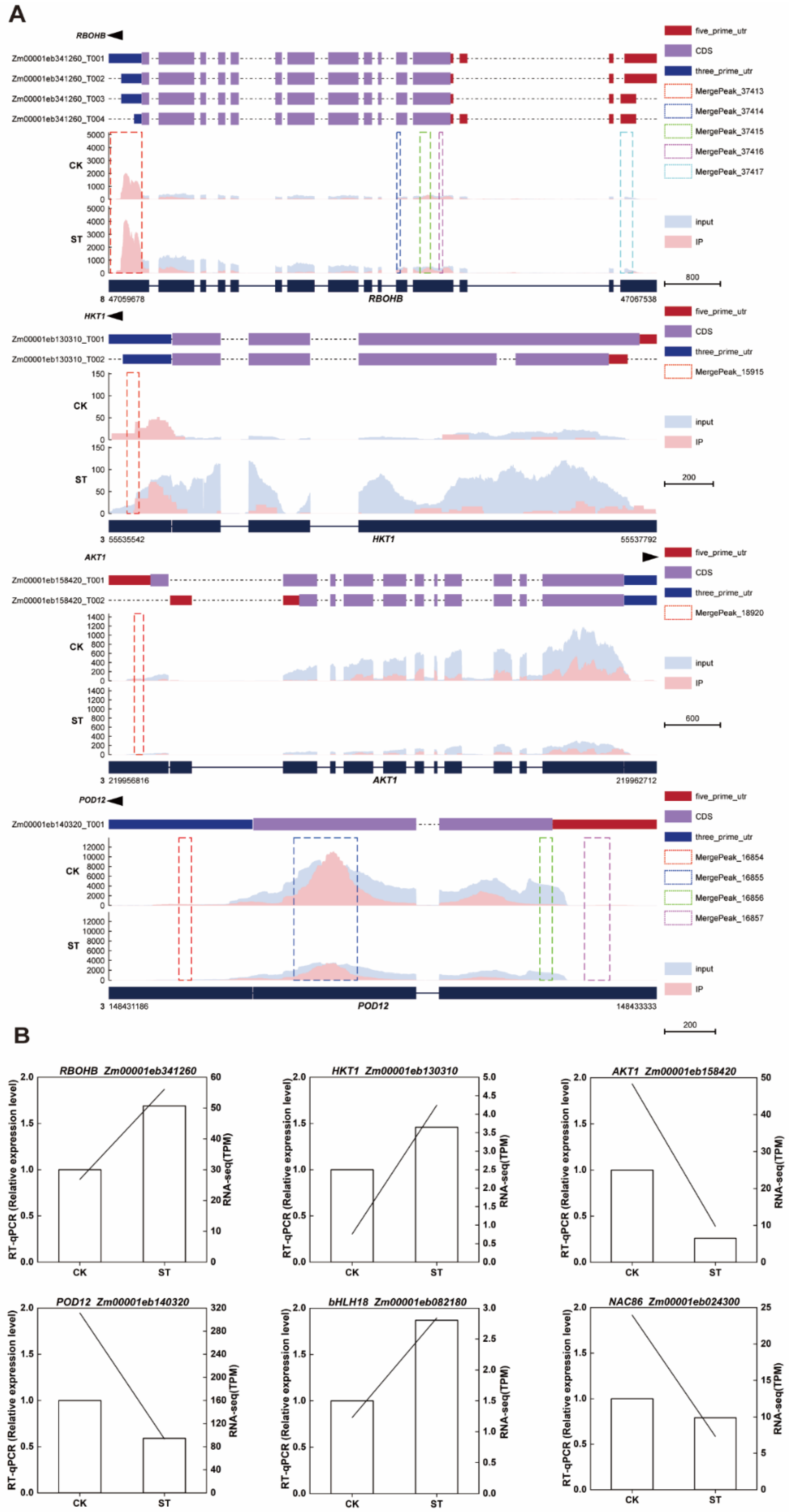
2.7. m6A Modification Characteristics and Expression Validation of Key Salt-Tolerance Related Genes
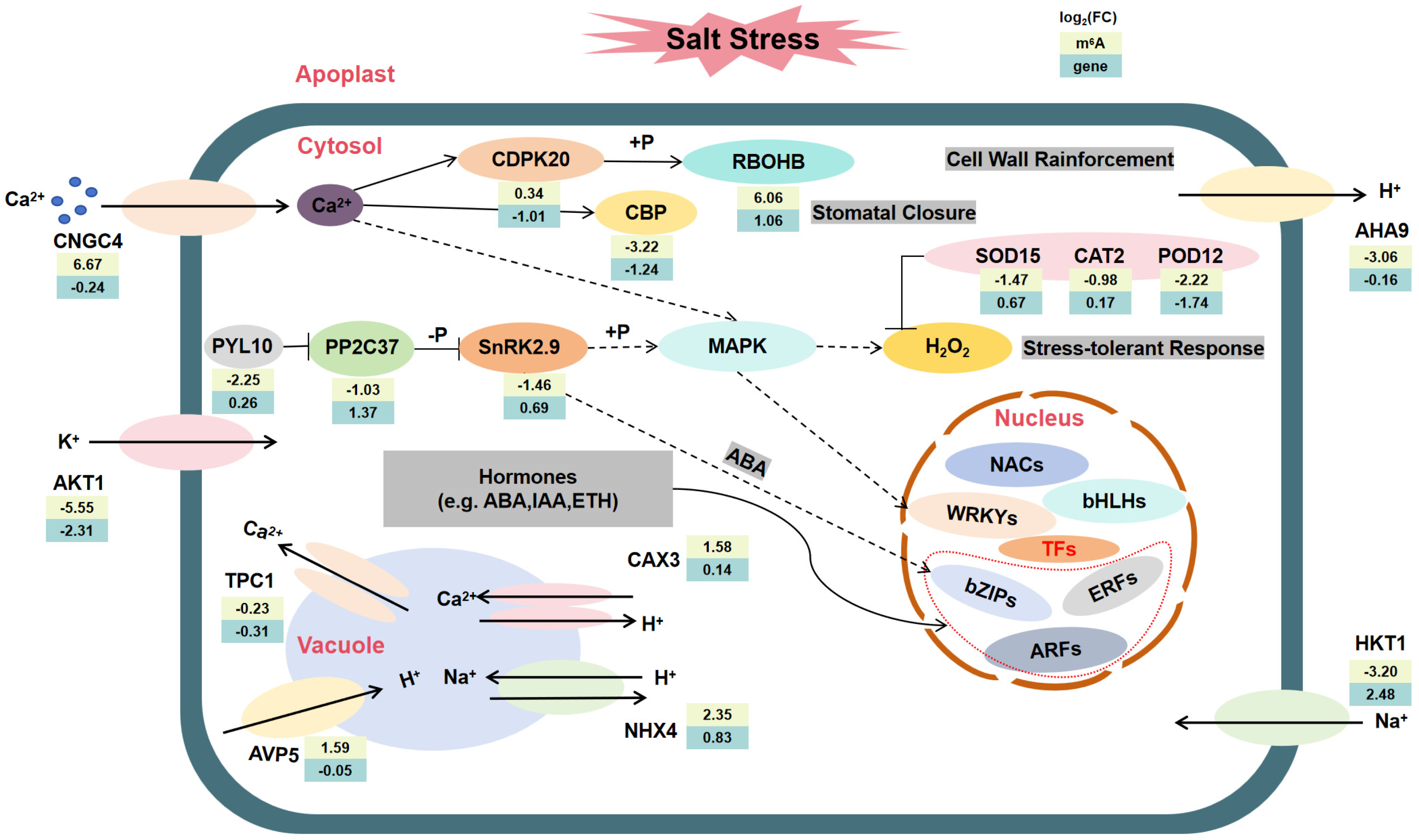
3. Discussion
4. Materials and Methods
4.1. Experimental Materials and Design
4.2. Measurement of Growth and Physiological Parameters
4.3. Ion Concentration Determination
4.4. RNA Extraction, Library Construction, and Sequencing
4.5. Bioinformatic Analysis of MeRIP-Seq Data
4.6. Bioinformatics Analysis of RNA-Seq Data
4.7. GO and KEGG Enrichment Analysis
4.8. qRT-PCR Validation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H.M. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustainable Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef]
- Machado, R.; Serralheiro, R. Soil salinity: Effect on vegetable crop growth. management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- He, B.Z.; Ding, J.L.; Huang, W.J.; Ma, X. Spatiotemporal variation and future predictions of soil salinization in the werigan-kuqa river delta oasis of China. Sustainability 2023, 15, 13996. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Alharbi, K.; Al-Osaimi, A.A.; Alghamdi, B.A. Sodium Chloride (NaCl)-Induced Physiological Alteration and Oxidative Stress Generation in Pisum sativum (L.): A Toxicity Assessment. ACS Omega 2022, 7, 20819–20832. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.X.; Fan, Q.L.; Tian, L.X.; Jiang, H.Y.; Wang, C.; Fu, X.Q.; Li, X.Z.; Li, M.; Zhang, S.Y.; Zhang, Y.B.; et al. Evaluation of cucumber seed germination vigor under salt stress environment based on improved YOLOv8. Front. Plant Sci. 2024, 15, 16. [Google Scholar] [CrossRef]
- Dadach, M.; Ahmed, M.Z.; Bhatt, A.; Radicetti, E.; Mancinelli, R. Effects of Chloride and Sulfate Salts on Seed Germination and Seedling Growth of Ballota hirsuta Benth. and Myrtus communis L. Plants 2023, 12, 3906. [Google Scholar] [CrossRef]
- Dong, Z.; Huang, J.; Qi, T.; Meng, A.; Fu, Q.; Fu, Y.; Xu, F. Exogenous γ-Aminobutyric Acid Can Improve Seed Germination and Seedling Growth of Two Cotton Cultivars under Salt Stress. Plants 2023, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Galon, Y.; Finkler, A.; Fromm, H. Calcium-regulated transcription in plants. Mol. Plant 2010, 3, 653–669. [Google Scholar] [CrossRef]
- Raddatz, N.; Morales de los Ríos, L.; Lindahl, M.; Quintero, F.J.; Pardo, J.M. Coordinated Transport of Nitrate, Potassium, and Sodium. Front. Plant Sci. 2020, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 2016, 67, 1663–1676. [Google Scholar] [CrossRef]
- Chen, Z.P.; Xie, Y.J.; Gu, Q.; Zhao, G.; Zhang, Y.H.; Cui, W.T.; Xu, S.; Wang, R.; Shen, W.B.A. The AtrbohF-dependent regulation of ROS signaling is required for melatonin-induced salinity tolerance in Arabidopsis. Free Radical Biol. Med. 2017, 108, 465–477. [Google Scholar] [CrossRef]
- Liang, P.; Ning, J.; Wang, W.L.; Zhu, P.; Gui, L.Y.; Xie, W.; Zhang, Y.J. Catalase promotes whitefly adaptation to high temperature by eliminating reactive oxygen species. Insect Sci. 2023, 30, 1293–1308. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A.; Correa, S.; Sevilla, F. Identification of Superoxide Dismutase (SOD) Isozymes in Plant Tissues. Methods Mol. Biol. 2024, 2798, 205–212. [Google Scholar] [CrossRef]
- Thapar, R.; Bacolla, A.; Oyeniran, C.; Brickner, J.R.; Chinnam, N.B.; Mosammaparast, N.; Tainer, J.A. RNA modifications: Reversal mechanisms and cancer. Biochemistry 2018, 58, 312–329. [Google Scholar] [CrossRef]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef]
- Liang, Z.; Ye, H.; Ma, J.; Wei, Z.; Wang, Y.; Zhang, Y.; Huang, D.; Song, B.; Meng, J.; Rigden, D.J.; et al. m6A-Atlas v2.0: Updated resources for unraveling the N6-methyladenosine (m6A) epitranscriptome among multiple species. Nucleic Acids Res. 2024, 52, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.E.; Zhang, S.; Xu, T.; Zhang, Y.; Teo, Z.W.N.; Yan, A.; Shen, L.; Yu, H. Shaping the landscape of N6-methyladenosine RNA methylation in Arabidopsis. Plant Physiol. 2023, 191, 2045–2063. [Google Scholar] [CrossRef]
- Cai, J.; Hu, J.; Xu, T.; Kang, H. FIONA1-mediated mRNA m6A methylation regulates the response of Arabidopsis to salt stress. Plant Cell Environ. 2024, 47, 900–912. [Google Scholar] [CrossRef]
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. N. Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m6A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef]
- Wei, L.-H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.-C.; Jia, G. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef]
- Rennie, S.; Arribas-Hernández, L.; Köster, T.; Porcelli, C.; Lewinski, M.; Staiger, D.; Andersson, R.; Brodersen, P. Principles of mRNA targeting via the Arabidopsis m6A-binding protein ECT2. Elife 2021, 10, e72375. [Google Scholar] [CrossRef]
- Luo, J.-H.; Guo, T.; Wang, M.; Liu, J.-H.; Zheng, L.-M.; He, Y. RNA m6A modification facilitates DNA methylation during maize kernel development. Plant Physiol. 2024, 194, 2165–2182. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, Y.; Sun, W.; Ding, P.; Bu, Y.; Qi, Y.; Shi, T.; Jia, C.; Lei, B.; Ma, C. The N6-methyladenosine reader ECT1 regulates seed germination via gibberellic acid- and phytochrome B-mediated signaling. Plant Physiol. 2025, 198, 180. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.K.; Duan, X.Y.; Zhou, L.L.; Gao, G.T.; Liu, J.Y.; Wang, Y.Y.; Shao, X.F.; Qin, G.Z. The FvABF3-FvALKBH10B-FvSEP3 cascade regulates fruit ripening in strawberry. Nat. Commun. 2024, 15, 10912. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wong, C.E.; Shen, L.; Yu, H. N6-methyladenosine modification underlies messenger RNA metabolism and plant development. Curr. Opin. Plant Biol. 2021, 63, 102047. [Google Scholar] [CrossRef]
- Zheng, H.; Dang, Y.; Gao, Y.; Li, S.; Wu, F.; Zhang, F.; Wang, X.; Du, X.; Wang, L.; Song, J.; et al. An mRNA methylase and demethylase regulate sorghum salt tolerance by mediating N6-methyladenosine modification. Plant Physiol. 2024, 196, 3048–3070. [Google Scholar] [CrossRef]
- Li, B.; Zhang, M.; Sun, W.; Yue, D.; Ma, Y.; Zhang, B.; Duan, L.; Wang, M.; Lindsey, K.; Nie, X.; et al. N6-methyladenosine RNA modification regulates cotton drought response in a Ca2+ and ABA-dependent manner. Plant Biotechnol. J. 2023, 21, 1270–1285. [Google Scholar] [CrossRef]
- Yang, D.D.; Xu, H.C.; Liu, Y.; Li, M.Z.; Ali, M.; Xu, X.Y.; Lu, G. RNA N6-methyladenosine responds to low-temperature stress in tomato anthers. Front. Plant Sci. 2021, 12, 687826. [Google Scholar] [CrossRef]
- Su, T.; Fu, L.; Kuang, L.; Chen, D.; Zhang, G.; Shen, Q.; Wu, D. Transcriptome-wide m6A methylation profile reveals regulatory networks in roots of barley under cadmium stress. J. Hazard. Mater. 2022, 423, 127140. [Google Scholar] [CrossRef]
- Xia, Z.; Zhang, S.; Guo, H.; Gao, X.; Hao, K.; Dong, X.; Guo, J.; Li, J.; Wang, Z.; An, M.; et al. N6-methyladenosine RNA modification regulates maize resistance to maize chlorotic mottle virus infection. J. Agric. Food Chem. 2024, 72, 21935–21945. [Google Scholar] [CrossRef]
- Chen, D.Y.; Fu, L.B.; Su, T.T.; Xiong, J.Y.; Chen, Y.K.; Shen, Q.F.; Kuang, L.H.; Wu, D.Z. N6-methyladenosine methylation analysis reveals transcriptome-wide expression response to salt stress in rice roots. Environ. Exp. Bot. 2022, 201, 104945. [Google Scholar] [CrossRef]
- Li, J.; Pang, Q.; Yan, X. Unique features of the m6A methylome and its response to salt stress in the roots of sugar beet (Beta vulgaris). Int. J. Mol. Sci. 2023, 24, 11659. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.B.; Zhou, X.Y.; Song, H.F.; Zhang, M.; Jiang, C.F. Advances in deciphering salt tolerance mechanism in maize. Crop J. 2023, 11, 1001–1010. [Google Scholar] [CrossRef]
- Hu, D.D.; Dong, S.T.; Zhang, J.W.; Zhao, B.; Ren, B.Z.; Liu, P. Endogenous hormones improve the salt tolerance of maize by inducing root architecture and ion balance optimizations. J. Agron. Crop Sci. 2022, 208, 662–674. [Google Scholar] [CrossRef]
- Zhou, H.P.; Shi, H.F.; Yang, Y.Q.; Feng, X.X.; Chen, X.; Xiao, F.; Lin, H.H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genomics 2024, 51, 16–34. [Google Scholar] [CrossRef]
- Hu, J.Z.; Manduzio, S.; Kang, H.S. Epitranscriptomic RNA methylation in plant development and abiotic stress responses. Front. Plant Sci. 2019, 10, 500. [Google Scholar] [CrossRef]
- Miao, Z.Y.; Zhang, T.; Qi, Y.H.; Song, J.; Han, Z.X.; Ma, C. Evolution of the RNA N6-methyladenosine methylome mediated by genomic duplication. Plant Physiol. 2020, 182, 345–360. [Google Scholar] [CrossRef]
- Zhang, G.; Lv, Z.; Diao, S.; Liu, H.; Duan, A.; He, C.; Zhang, J. Unique features of the m6A methylome and its response to drought stress in sea buckthorn (Hippophae rhamnoides Linn.). RNA Biology 2021, 18, 794–803. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, K.J.; Tian, Y.T.; Jia, K.H.; El-Kassaby, Y.A.; Wu, Y.; Liu, J.; Si, H.Y.; Sun, Y.H.; Li, Y. N6-methyladenosine mRNA methylation positively regulated the response of poplar to salt stress. Plant Cell Environ. 2024, 47, 1797–1812. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Jia, Y.; Jin, X.; Wu, H.; Yang, S.; Zhao, L.; Zhang, H.; Gu, L. The RNA m6A methyltransferase PheMTA1 and PheMTA2 of moso bamboo regulate root development and resistance to salt stress in plant. Plant Cell Environ. 2025, 48, 5184–5197. [Google Scholar] [CrossRef]
- Li, Y.B.; Yin, M.; Wang, J.; Zhao, X.Q.; Xu, J.L.; Wang, W.S.; Fu, B.Y. Epitranscriptome profiles reveal participation of the RNA methyltransferase gene OsMTA1 in rice seed germination and salt stress response. BMC Plant Biol. 2025, 25, 115. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, S.; Acosta-Motos, J.R.; Sánchez-Blanco, M.J. Morphological performance and seasonal pattern of water relations and gas exchange in Pistacia lentiscus plants subjected to salinity and water deficit. Front. Plant Sci. 2023, 14, 1237332. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.B.; Shen, Q.F.; Kuang, L.H.; Wu, D.Z.; Zhang, G.P. Transcriptomic and alternative splicing analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Environ. Exp. Bot. 2019, 166, 103810. [Google Scholar] [CrossRef]
- Yang, W.Q.; Zhang, W.; Wang, X.X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef]
- Pérez-Salamó, I.; Papdi, C.; Rigó, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horváth, B.; Domoki, M.; Darula, Z.; et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Yu, K.J.; Zhang, S.W.; Li, Y.; Xu, C.W.; Qian, H.P.; Guo, Y.Y.; Zhang, X.; Li, R.L.; Dixon, R.A.; et al. Poplar glutathione S-transferase PtrGSTF8 contributes to reactive oxygen species scavenging and salt tolerance. Plant Physiol. Biochem. 2024, 212, 108766. [Google Scholar] [CrossRef]
- Wang, X.; Qi, X.; Zhuang, Z.; Bian, J.; Li, J.; Chen, J.; Li, Z.; Peng, Y. Interactions between brassinosteroids and strigolactones in alleviating salt stress in maize. Int. J. Mol. Sci. 2024, 25, 10505. [Google Scholar] [CrossRef] [PubMed]
- Haida, Z.; Hakiman, M. A comprehensive review on the determination of enzymatic assay and nonenzymatic antioxidant activities. Food Sci. Nutr. 2019, 7, 1555–1563. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Stark, R.; Brown, G. DiffBind: Differential binding analysis of ChIP-Seq peak data. R Package Version 2011, 100, 2–21. [Google Scholar]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
| Gene_Id | Symbol | Type | m6A | Gene_Expression | ||||
|---|---|---|---|---|---|---|---|---|
| Diff. log2 (FC) | p-Value | Up/Down | log2 (FC) | p-Value | Up/Down | |||
| Zm00001eb346760 | ALKBH10B | eraser | −3.07 | 0.088 | Down | −0.17 | 0.477 | NC |
| Zm00001eb287600 | ALKBH10B | eraser | −0.92 | 0.170 | NC | −0.07 | 0.164 | NC |
| Zm00001eb010150 | ALKBH10B | eraser | −0.44 | 0.493 | NC | 0.45 | 0.000 ** | NC |
| Zm00001eb380830 | ALKBH2 | eraser | −0.36 | 0.552 | NC | 0.92 | 0.045 * | NC |
| Zm00001eb047810 | ALKBH6 | eraser | −0.29 | 0.552 | NC | −0.14 | 0.486 | NC |
| Zm00001eb071740 | ALKBH8 | eraser | −0.36 | 0.475 | NC | 0.15 | 0.062 | NC |
| Zm00001eb377750 | ALKBH9B | eraser | −0.70 | 0.115 | NC | 0.45 | 0.000 ** | NC |
| Zm00001eb403260 | ECT2 | reader | 0.16 | 0.733 | NC | 0.30 | 0.001 ** | NC |
| Zm00001eb360390 | ECT2 | reader | 0.11 | 0.847 | NC | 0.71 | 0.000 ** | NC |
| Zm00001eb117310 | ECT2 | reader | −1.81 | 0.217 | Down | −0.74 | 0.843 | NC |
| Zm00001eb057150 | ECT2 | reader | 0.29 | 0.355 | NC | 0.01 | 0.154 | NC |
| Zm00001eb414030 | ECT3 | reader | −1.23 | 0.090 | Down | −0.21 | 0.566 | NC |
| Zm00001eb156740 | ECT3 | reader | −1.19 | 0.077 | Down | 1.03 | 0.000 ** | Up |
| Zm00001eb300970 | ECT4 | reader | 0.54 | 0.235 | NC | 0.58 | 0.000 ** | NC |
| Zm00001eb171010 | ECT4 | reader | 1.24 | 0.086 | Up | −0.14 | 0.297 | NC |
| Zm00001eb117080 | ECT4 | reader | 0.53 | 0.235 | NC | 0.60 | 0.000 ** | NC |
| Zm00001eb039740 | ECT4 | reader | 1.30 | 0.037 * | Up | −0.03 | 0.152 | NC |
| Zm00001eb231610 | FIP37 | writer | 1.69 | 0.002 ** | Up | −0.07 | 0.266 | NC |
| Zm00001eb209200 | FIP37 | writer | −0.39 | 0.320 | NC | 0.15 | 0.038 * | NC |
| Zm00001eb221140 | HAKAI | writer | 0.33 | 0.449 | NC | 0.50 | 0.000 ** | NC |
| Zm00001eb044800 | HAKAI | writer | −0.27 | 0.494 | NC | 0.15 | 0.008 ** | NC |
| Zm00001eb003840 | MTB | writer | −0.13 | 0.790 | NC | 0.30 | 0.011 * | NC |
| Zm00001eb339360 | MTB1 | writer | −2.02 | 0.087 | Down | −0.01 | 0.134 | NC |
| Zm00001eb119000 | MTB1 | writer | 0.46 | 0.253 | NC | 0.08 | 0.049 * | NC |
| Zm00001eb155410 | MTB2 | writer | −1.42 | 0.011 * | Down | −0.69 | 0.087 | NC |
| Zm00001eb220020 | VIR | writer | −0.34 | 0.472 | NC | −0.21 | 0.863 | NC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Ta, W.; Zhuang, Z.; Bian, J.; Ren, Z.; Hao, X.; Zhang, L.; Peng, Y. N6-Methyladenosine (m6A) Methylation-Mediated Transcriptional Regulation in Maize Root Response to Salt Stress. Plants 2026, 15, 36. https://doi.org/10.3390/plants15010036
Ta W, Zhuang Z, Bian J, Ren Z, Hao X, Zhang L, Peng Y. N6-Methyladenosine (m6A) Methylation-Mediated Transcriptional Regulation in Maize Root Response to Salt Stress. Plants. 2026; 15(1):36. https://doi.org/10.3390/plants15010036
Chicago/Turabian StyleTa, Wanling, Zelong Zhuang, Jianwen Bian, Zhenping Ren, Xiaojia Hao, Lei Zhang, and Yunling Peng. 2026. "N6-Methyladenosine (m6A) Methylation-Mediated Transcriptional Regulation in Maize Root Response to Salt Stress" Plants 15, no. 1: 36. https://doi.org/10.3390/plants15010036
APA StyleTa, W., Zhuang, Z., Bian, J., Ren, Z., Hao, X., Zhang, L., & Peng, Y. (2026). N6-Methyladenosine (m6A) Methylation-Mediated Transcriptional Regulation in Maize Root Response to Salt Stress. Plants, 15(1), 36. https://doi.org/10.3390/plants15010036





