Abstract
ABA Insensitive 5 (ABI5) is a basic leucine zipper (bZIP) transcription factor (TF) that plays a critical role in seed dormancy and germination, particularly under stress conditions. This study identified ABI5 as an important candidate gene regulating seed germination under drought stress during early germination in rapeseed (Brassica napus L.) seeds through Genome-Wide Association Study (GWAS). Using Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9 (CRISPR/CAS9) technology, ABI5 mutant plants were generated, showing higher germination rates and more developed root systems at 72 h. Transcriptomic analysis of wild-type (WT) and mutant seeds under water, 2μM of abscisic acid (ABA), and 10% PEG treatments after 0, 24, 48, and 72 h revealed complex changes in gene regulatory networks due to ABI5 mutation. Differential expression analysis showed that the number of downregulated differentially expressed genes (DEGs) in the mutant was significantly higher than upregulated DEGs at multiple time points and treatments, indicating a negative regulatory role for ABI5 in gene expression. Weighted Gene Co-Expression Network Analysis (WGCNA) revealed that genes related to ABA content, such as those in the glutathione metabolism pathway, were similarly downregulated in the ABI5 mutants. Key genes, including BnA03g0120550.1 (GST), BnA09g0366300.1 (GST), BnA10g0413960.1 (gshA), and BnC02g0518750.1 (GST), were identified as potential candidates in ABI5-regulated drought responses. Additionally, TFs involved in regulating the glutathione metabolism pathway were identified, providing insights into the collaboration of ABI5 with other TF. This comprehensive transcriptomic analysis of ABI5 mutant plants highlights how ABI5 affects gene expression in multiple pathways, impacting seed germination and drought resistance, offering a foundation for improving drought tolerance in rapeseed.
1. Introduction
Drought stress is one of the most common abiotic stresses affecting global agricultural production, particularly in regions with limited water resources []. Over the course of evolution, plants have developed various mechanisms to cope with drought, including morphological and biochemical adaptations such as changes in leaf morphology, thickening of the waxy cuticle, and osmoregulation [,,]. Additionally, plant hormones, including jasmonic acid, ethylene, and abscisic acid (ABA), play crucial regulatory roles in drought responses [,].
The relative levels of plant hormones regulate seed dormancy and germination. Gibberellins (GA), brassinosteroids, ethylene, and cytokinins have been shown to break seed dormancy and promote germination [,,]. In contrast, ABA is the only known hormone capable of inducing and maintaining seed dormancy [], and it plays a key role in regulating drought tolerance, seed dormancy, and germination [,]. Other plant hormones, such as GA, ethylene, cytokinins, and brassinosteroids, as well as their antagonistic interactions with ABA, positively regulate the seed germination process [,,]. During seed germination, ABA acts as an inhibitor, promoting seed dormancy, which is vital for preventing premature germination under unfavorable conditions []. The balance between ABA and GA during seed germination determines the outcome of this process []. High concentrations of ABA inhibit germination, while GA promotes it. Under drought stress, ABA levels increase, activating ABA-responsive genes and enhancing plant tolerance to drought. ABA plays an essential role in both seed germination and resistance to environmental stresses, making it a key regulator of seed germination and early seedling growth [,].
ABI5 is a bZIP TF that plays a pivotal role in the ABA signaling pathway []. It is activated by SnRK2 kinases (SnRK2.2, SnRK2.3, and SnRK2.6), which phosphorylate the transactivation domain of ABI5 in vegetative tissues under stress conditions []. Phosphorylation alters the conformation of ABI5, enhancing its ability to interact with other proteins []. Once activated, ABI5 binds to the promoter regions of ABA-responsive genes, such as RD29A, RD29B, and EM6, and upregulates their expression []. Notably, EM1, EM6, and LEA D-34 (encoding late embryogenesis abundant (LEA) proteins) were among the first identified direct targets of ABI5 []. Yeast one-hybrid assays further confirmed that ABI5 directly binds to the ABRE motifs in the EM6 promoter [,]. Several ABI5 target genes, such as PGIP1 and PGIP2, are involved in seed germination by inhibiting polygalacturonase activity, thereby delaying seed coat rupture and suppressing germination []. In this process, ABI5 is negatively regulated by PED3, a peroxisomal ABC transporter involved in fatty acid β-oxidation.
Plant developmental processes such as dormancy and germination, as well as interactions with the environment, are regulated by phytohormones and other signaling molecules. Previous studies have demonstrated that plant hormones and other signaling compounds play crucial roles in regulating both developmental pathways and responses to abiotic stresses []. Glutathione (GSH), one of the major antioxidant molecules, is essential for plant survival under stress conditions by detoxifying excess reactive oxygen species (ROS), maintaining cellular redox homeostasis, and modulating protein function []. Recently, GSH has also emerged as an important signaling molecule that modulates ABA signaling and related developmental events, as well as stress responses. In Arabidopsis guard cells, GSH was shown to negatively regulate ABA signaling components, including excluding ROS production and Ca2+ oscillations, thereby suppressing ABA-induced stomatal closure []. Increased levels of GSH-related proteins/compounds, such as glutaredoxins (GRXs) and S-nitrosoglutathione (GSNO), have been shown to suppress ABA signaling during seed germination []. Moreover, glutathione transferases (GSTs) involved in the GSH metabolic pathway have been reported to affect the expression of ABI5 [], suggesting a complex interaction between GSH and ABA signaling, particularly during the critical phase of seed germination. Given that ABI5 is a key gene in ABA-mediated seed dormancy and germination, it is, therefore, essential to investigate the potential regulatory relationship between ABI5 and glutathione metabolism.
Rapeseed (Brassica napus L.) is an important oilseed crop, widely cultivated for edible oil and meal production. The drought tolerance of rapeseed is crucial for its agricultural performance. Although ABA and ABI5 play important roles in drought tolerance, the specific functions of BnABI5 (the homolog of ABI5) in seed germination and early seedling growth in rapeseed have not been fully elucidated. In this study, Genome-Wide Association Studies (GWAS) of drought phenotypes during seed germination and early seedling growth identified BnABI5 as being associated with drought stress in rapeseed. To explore the role of BnABI5 in drought tolerance during seed germination and early seedling growth, we generated BnABI5 mutants using Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9 (CRISPR/Cas9) technology and compared their germination and early growth performance under normal and drought stress conditions with WT plants. To investigate the impact of the BnABI5 mutation on seed germination and drought tolerance in rapeseed, we analyzed 60 RNA-seq samples collected from two genotypes, three treatments, and four time points. Differential expression analysis and Weighted Gene Co-Expression Network Analysis (WGCNA) were performed to uncover the regulatory networks involving BnABI5 under drought conditions. These findings provide a foundation for the development of drought-tolerant germplasm in rapeseed breeding.
2. Materials and Methods
2.1. Plant Materials and Germination Rate Calculation Under Drought Treatment
In this study, 196 rapeseed accessions were obtained from the Crop Research Institute at Zhejiang University, China. All plant materials were planted in September 2022 at the Wuchang experimental base of the Oilseed Crop Research Institute, Chinese Academy of Agricultural Sciences (114.14° E, 30.32° N). Within each row, individual plants were spaced 0.15 m apart, with row spacing maintained at 0.25 m. Seeds were sown at the end of September and harvested in early May of the following year. Mature seeds were collected, sun-dried, and stored at −20 °C until further use. For each accession, 50 viable seeds were placed in petri dishes for germination, with each dish lined with two layers of filter paper (Shanghai Experiment Reagent Co., Shanghai, China). As a control, 7 mL of deionized water was used to moisten the filter paper, or a polyethylene glycol (PEG) 6000 (Solarbio, Beijing, China) solution was used to adjust the osmotic potential to −0.8 MPa to simulate drought stress conditions. The petri dishes were incubated in a growth chamber set (Jinggong Industrial Co., Shanghai, China) at a constant temperature of 25 °C, with light intensity of 100 μmol m−2 s−1, a 12-h light/12-h dark photoperiod, and 70% relative humidity. A completely randomized design was employed to ensure three replicates for each accession under both control and drought conditions. After seven days, the survival rate of each accession was recorded.
2.2. GWAS Analysis
We obtained a total of 2,404,340 SNPs from the rapeseed SNP database (BnaSNPDB: https://bnapus-zju.com/bnasnpdb/, accessed on 14 June 2023), filtering out SNPs with a minor allele frequency (MAF) less than 0.05 and a missing genotype rate greater than 0.5 to ensure high-quality data. GWAS analysis of drought survival rate phenotypes for each accession was performed using the Efficient Mixed-Model Association eXpedited (EMMAX) method [] available on the BnaVGD platform (https://bnapus-zju.com/gwas/; accessed on 9 November 2023). This model accounts for population structure and relatedness, improving the accuracy of the association tests. Manhattan and quantile-quantile (Q-Q) plots were generated based on the GWAS results on the BnaVGD platform, with a significance threshold of −log10 (p-value) = 5.75 to minimize false-positive associations while capturing biologically meaningful signals.
2.3. CRISPR/Cas9-Mediated Editing of ABI5 in the B. napus
We performed gene editing of ABI5 in B. napus following the method described by Li et al. [], with the specific procedures as follows: We designed two sgRNAs with minimal off-target effects for the conserved sequences of three copies of BnABI5 genes (BnA04g0179250.1, BnA05g0194800.1, and BnC04g0676540.1) using CRISPR-P 2.0: sgRNA 1 (TTTGGTCTGAGATACATAGAGG) and sgRNA 2 (GTATGGAGTTGATATGGGAGGG). The sgRNA assembly and vector construction followed protocols from previous studies [,].
Genome editing vectors were introduced into Agrobacterium tumefaciens strain GV3101 competent cells (Sangon Biotech, Shanghai, China) following the manufacturer’s protocol. The vectors containing the two sgRNAs were then introduced into the rapeseed variety “Zhongshuang 6” (ZS6) [] using Agrobacterium-mediated transformation, with kanamycin as the selection marker. In the T0 generation, putative transgenic plants were first selected on kanamycin-containing medium, and PCR amplification using NPTII (a kanamycin resistance marker) primers (forward: 5′-GATGGATTGCACGCAGGT-3′; reverse: 5′-TCGTCAAGAAGGCGATAGA-3′) was performed to confirm the presence of the transgene and eliminate false positives. Verified T0 plants were transferred to a growth chamber (16 h light/8 h dark, 22 °C) for further growth and seed collection. T1 and T2 generation plants were grown under controlled conditions (22 °C, 16 h light/8 h dark) or at the Hanchuan Experimental Station (OCRI-CAAS). Genomic DNA was extracted from the leaves of these plants using the CTAB method []. For mutation detection, genomic regions spanning the CRISPR target sites were first amplified using gene-specific primers DT1-BsF (5′-ATATATGGTCTCGATTGtttggtctgagatacatagGTT-3′) and DT1-BsR (5′-ATTATTGGTCTCGAAACTCCCATATCAACTCCATACCAA-3′). The PCR products were then screened via PAGE gel electrophoresis to determine the presence of heterozygous or biallelic mutations based on banding patterns. Samples with distinct mutation bands were subsequently selected for Sanger sequencing. To ensure accurate mutation characterization, a nested PCR was performed using secondary primers DT1-F0 (5′-TGtttggtctgagatacatagGTTTTAGAGCTAGAAATAGC-3′) and DT1-R0 (5′-AACTCCCATATCAACTCCATACCAATCTCTTAGTCGACTCTAC-3′) to produce shorter amplicons (150–350 bp) for sequencing.
2.4. Stress Treatment Experiment for WT and BnABI5 CRISPR-Edited Lines
First, surface sterilization of WT (ZS6) and BnABI5 CRISPR-edited line seeds was performed by soaking them in a 10% sodium hypochlorite (Ganzhou Jinfu Fine Chemical Technology Co., Ltd., Ganzhou, China) solution for 5 min. The seeds were then evenly placed on moist filter paper (Sunnyo Environmental Technology, Shanghai, China), with 50 seeds per germination box. Three treatment groups were set up: the control group (water treatment), the ABA treatment group (2 µM ABA solution), and the PEG treatment group (10% PEG solution). For each treatment group, the corresponding solution was added to maintain moisture on the filter paper. The treated seeds, along with the filter paper, were placed in 10 cm × 10 cm Petri dishes (Bickman Biotechnology, Changsha, China), with three replicates per group to ensure reliable results. Finally, the Petri dishes were placed in a plant growth chamber with a light/dark cycle of 16 h light/8 h dark and a light intensity of 300 µmol·m−2·s−1.
2.5. Phytohormone Quantification
The endogenous levels of indole-3-acetic acid (IAA), abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), gibberellins (GA1, GA3, GA4, GA7), and 1-aminocyclopropane-1-carboxylic acid (ACC) were determined using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Fresh plant tissues from WT and BnABI5 CRISPR-edited lines at 0, 24, 48, and 72 h under ABA, PEG treatments, and controls were lyophilized, homogenized, and extracted with methanol/water/formic acid (15:4:1, v/v/v) containing deuterated internal standards (e.g., D2-IAA, D6-ABA). After centrifugation and purification via C18 solid-phase extraction, the supernatant was evaporated and reconstituted in methanol (Sigma-Aldrich, St. Louis, MO, USA). Chromatographic separation was performed on a C18 column with a gradient of methanol and 0.1% formic acid. Analytes were detected in multiple reaction monitoring (MRM) mode with electrospray ionization (ESI), using optimized precursor-to-product ion transitions for each compound (TSQ Fortis platform, Thermo Fisher Scientific, Waltham, MA, USA). Quantification was achieved by comparing peak areas to standard curves normalized against corresponding internal standards. Three biological replicates were analyzed per treatment.
2.6. Library Preparation and Transcriptome Sequencing
Total RNA was extracted from 60 samples (WT and BnABI5 CRISPR-edited lines at 0, 24, 48, and 72 h under two treatments) using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China). RNA quality was assessed using a NanoDrop® 2000 (Thermo Fisher Scientific, Waltham, MA, USA), and treated with DNase I (RNase-free) according to the manufacturer’s instructions. RNA purity and integrity were evaluated by agarose gel electrophoresis. RNA concentration was measured using the Qubit RNA BR Assay Kit (Q10210, Thermo Fisher Scientific, Waltham, MA, USA), and RNA integrity was checked using an Agilent Bioanalyzer 2100 system (Santa Clara, CA, USA). For RNA-seq library preparation, 5 μg of RNA from each sample was used. RNA libraries were constructed by Qingke Biotechnology Co. (Wuhan, China), using the Illumina TruSeq Stranded Library Prep Kit, and sequencing was performed on the Illumina HiSeq 2500 platform (San Diego, CA, USA). In brief, poly(A) mRNA was isolated from total RNA using Oligo-(dT) magnetic beads. RNA fragments were generated in fragmentation buffer, and the first-strand cDNA was synthesized using six-base random primers. Then, double-stranded cDNA (ds-cDNA) was synthesized using a buffer, dNTPs, RNase H, DNA polymerase I, and RNase H. Library fragments were purified using AMPure XP beads (Beckman Coulter, Brea, CA, USA) to obtain cDNA fragments of approximately 250–300 bp in length. To ensure library quality, PCR products were purified with AMPure XP beads and assessed on an Agilent Bioanalyzer 2100 system. The index-labeled samples were clustered using the cBot clustering system of the TruSeq PE Cluster Kit v4-cBot-HS (Illumina, San Diego, CA, USA). Finally, the libraries were sequenced on the Illumina HiSeq-PE150 platform (Illumina, San Diego, CA, USA).
2.7. Transcriptome Analysis
Raw RNA sequencing data were processed using fastp v0.23.4 [] with default parameters to remove low-quality reads and adapters, generating high-quality clean reads. Clean reads were then aligned to the B. napus reference genome (Brana_ZS_V2.0, http://brassicadb.cn:82/download_genome/Brassica_Genome_data/Brana_ZS_V2.0/; accessed on 12 February 2024) using STAR v2.7.10b [] with default settings. Gene read counts were obtained using featureCounts v2.0.3 [], and gene expression levels were quantified in terms of Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) using HTSeq 2.0 [] with default parameters. Differentially expressed genes (DEGs) between samples were detected using DESeq v1.42.0 [] in R v4.3.3, with the filtering criteria set to Padj < 0.05 and |log2(FoldChange)| > 2. Cluster analysis of the 0, 24, 48, and 72-h time-point samples was performed using Mfuzz v2.60.0 []. Gene expression heatmaps were generated using the R package pheatmap v1.0.12 [] (scale = “row”). GO and KEGG annotations of the protein sequences from Brana_ZS_V2.0 were performed using eggnog-mapper v2 [] and kobas [], respectively. Enrichment analysis was conducted with the R package clusterProfiler 4.0 [], and results were visualized using ggplot2 v3.5.1 [].
2.8. WGCNA
Gene co-expression network analysis was performed using the R package WGCNA v1.73 []. From the expression matrix of 50,488 genes, the top 25% of genes with the highest variance in expression across samples were selected as the input dataset. Hierarchical clustering of all samples was conducted using the hclust function with the average linkage method (method = “average”). The soft-thresholding power (β) was determined to be 16 using the pickSoftThreshold function. Co-expression modules were identified using the blockwiseModules function with TOMtype set to “unsigned”, minModuleSize set to 30, and mergeCutHeight set to 0.25. Module eigengenes (MEs) were calculated using the moduleEigengenes function, and the Pearson correlation coefficients (PCCs) between MEs and phenotypic traits such as ABA levels were computed with the cor.test function in R. Modules with a correlation coefficient greater than 0.65 and a p-value less than 0.05 were considered significantly associated with the phenotype.
2.9. Co-Expression Analysis of TFs and Genes
Protein sequences of TFs from all plant species were downloaded from the PlantTFDB 4.0 database []. These TF sequences were compared against the protein sequences of Brana_ZS_V2.0 using DIAMOND [] with the blastp algorithm to identify TFs in the B. napus genome. The PCC between differentially expressed TFs and genes was calculated in R. Gene-TF pairs with |PCC| > 0.8 and Q-value < 0.05 were retained. Co-expression networks of these gene-TF pairs were visualized using Cytoscape v3.9.1 [].
3. Results
3.1. BnABI5 as a Potential Regulator of Seed Germination Under Drought Stress in B. napus
In response to drought stress, B. napus seeds often regulate their germination through dormancy mechanisms, a strategy that helps the plant conserve energy and survive under adverse conditions. However, how B. napus seeds regulate their germination mechanisms under drought stress remains unclear. To identify potential factors influencing seed germination under drought stress in B. napus, we measured the germination rate of 196 B. napus accessions under simulated drought stress (Supplementary Table S1) and performed a GWAS using 2,404,340 SNPs obtained from the BnaSNPDB. The results identified two significant loci (C4:6775140 and C4:6775750) on chromosome C04, which are located inside the gene BnaC04g09030D (Darmor-bzh genome, B. napus v4.1, https://yanglab.hzau.edu.cn/BnIR/germplasm_info?id=Darmor.v4.1/, accessed on 9 November 2023) (Figure 1A). This gene is homologous to the Arabidopsis gene AT2G36270 (ABI5), which is reported to play a crucial role in regulating plant development and responses to abiotic stresses []. Using homology searches, we identified three homologous genes in the ZS6 reference genome: BnC04g0676540.1, BnA04g0179250.1, and BnA05g0194800.1. Multiple sequence alignments revealed that these genes, like AtABI5, contain the typical bZIP domain (Figure 1B). These findings suggest that the ABI5 homologs may play important roles in regulating seed dormancy and germination under drought stress in B. napus.
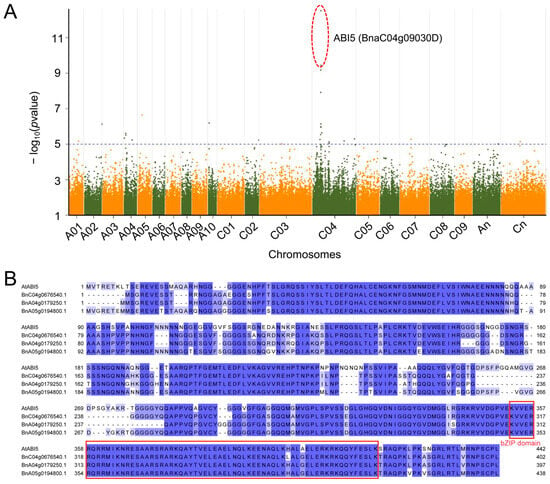
Figure 1.
(A) The Manhattan plot shows the GWAS results for the drought survival rate phenotype. The dashed line represents the significance threshold, with a value of −log10 (p-value) = 5.75. The x-axis indicates the chromosomal positions, and the y-axis shows −log10 (p-value). The circled points indicate those SNPs located within BnaC04g09030D. (B) Multiple sequences alignment of putative amino encoded by AtABI5 and the three ABI5 homologous genes in the ZS6 reference genome. Identical amino acid residues across all four sequences are colored blue, with variants shown in light blue. The bZIP domain is indicated by a red box.
3.2. CRISPR Editing of BnABI5 Promotes Seed Germination Under Drought Stress Treatment
To further validate the function of ABI5 homologs in B. napus, we designed sgRNAs targeting the three homologs of ABI5 in the ZS6 reference genome and utilized the CRISPR-Cas9 gene-editing system for genome editing (Supplementary Figure S1). The flanking sequences of the target sites were amplified and sequenced, confirming a 1 bp deletion at the sgRNA target site of BnABI5 (Figure 2A, Supplementary Figure S2). To evaluate the impact of drought stress treatment on seed germination in the edited lines, we treated WT and BnABI5 CRISPR-edited lines with water, ABA, and PEG. After 24 h of treatment, the BnABI5 CRISPR-edited lines displayed stronger growth compared to WT (Figure 2B). All seeds of both WT and the edited lines germinated after 48 and 72 h under different treatments. However, the BnABI5 CRISPR-edited lines exhibited faster growth and longer shoots than WT under all treatments, with the most pronounced differences observed under water and PEG treatments (Figure 2C,D). Furthermore, we examined the germination rates of rapeseed seeds at multiple time points following treatment. The results indicated that the BnABI5 CRISPR-edited lines displayed an accelerated germination rate during the early stages under ABA and PEG treatments (Figure 2E).
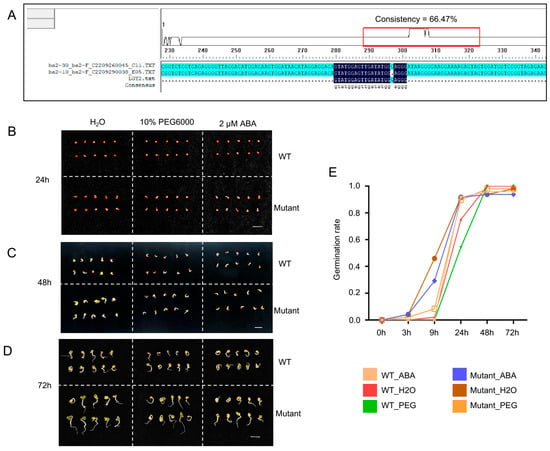
Figure 2.
Gene editing of ABI5 in rapeseed. (A) Sanger sequencing results of CRISPR-CAS9-edited BnABI5. In the alignment, blue highlights identical PCR product sequences, while the dark background indicates regions matching the sgRNA. (B–D) Phenotypes of WT and mutant (BnABI5 CRISPR-edited lines) rapeseed seeds subjected to water, PEG, and ABA treatments at 24 h (B), 48 h (C), and 72 h (D). (E) Germination rates of rapeseed seeds under different treatments after different times. Mutant represents BnABI5 CRISPR-edited lines.
3.3. Comparative Transcriptome Analysis of WT and BnABI5 CRISPR-Edited Lines Under Different Treatments
ABI5, as a TF, exerts systemic effects on the entire plant by regulating multiple downstream genes. To investigate the impact of BnABI5 gene editing on plant responses to drought, this study performed transcriptomic sequencing on two genotypes of rapeseed under different treatments (H2O, ABA, PEG) at three time points (24 h, 48 h, and 72 h). Additionally, we compared the differential expression of genes in BnABI5 CRISPR-edited lines relative to the WT under different treatments at these time points. With the exception of the 48 h PEG treatment, the number of downregulated DEGs in BnABI5 CRISPR-edited lines was significantly higher than the upregulated DEGs at most time points (Figure 3A). This suggests that gene editing of BnABI5 may disrupt some of the pathways in which BnABI5 is involved. The enrichment analysis of these DEGs revealed that the downregulated genes in BnABI5 CRISPR-edited lines were primarily enriched in GO terms associated with drought stress, such as S-glycoside metabolic process, defense response by cell wall thickening, and defense response by callose deposition in the cell wall (Figure 3B). Interestingly, in all treatments at different time points, the downregulated DEGs in BnABI5 CRISPR-edited lines were consistently enriched in the glutathione metabolism pathway (Figure 3C). This indicates that BnABI5 plays a crucial role in regulating cellular stress responses. Furthermore, after 24 h of PEG treatment, the upregulated genes in BnABI5 CRISPR-edited lines were mainly enriched in pathways related to photosynthesis, glycolysis/gluconeogenesis, and carbon fixation in photosynthetic organisms (Supplementary Figure S3). This may be due to the faster germination of seeds in BnABI5 CRISPR-edited lines compared to WT, which are likely in a phase of rapid growth and development.
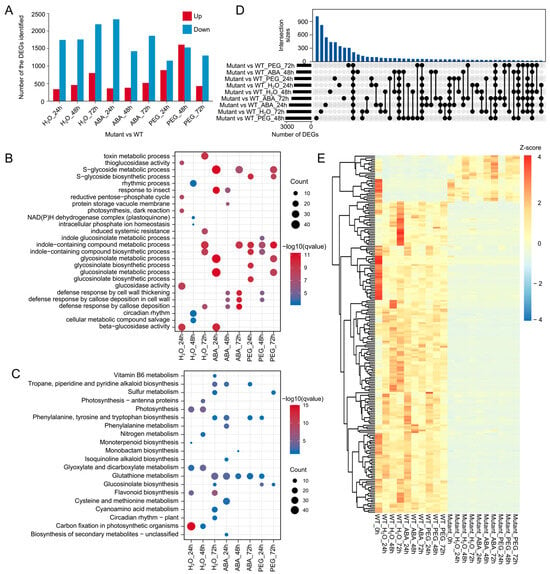
Figure 3.
Differential expression analysis of BnABI5 CRISPR-edited lines compared to WT under different treatments. (A) The number of upregulated and downregulated genes in BnABI5 CRISPR-edited lines (mutant) under various treatments after different time points. (B,C) GO (B) and KEGG (C) enrichment analysis of downregulated genes in different treatment groups. (D) UpSet plot showing the number of unique and overlapping DEGs across 9 comparison groups. (E) Heatmap of gene expression for the 211 DEGs that are differentially expressed across all treatments. The color scale represents the Z-score of the gene expression level.
Additionally, we analyzed the overlap of DEGs between BnABI5 CRISPR-edited lines and WT under different conditions. As the treatment time increased, the DEGs exhibited distinct overlap patterns, with varying numbers of overlapping DEGs between the BnABI5 CRISPR-edited lines and WT under different treatment conditions. The PEG treatment at 72 h induced the most gene expression changes (Figure 3D), suggesting that BnABI5 plays a significant role in regulating gene expression responses under prolonged drought stress. A total of 211 genes showed differential expression across all treatments. The cluster heatmap divided these genes into two groups: 184 genes that were highly expressed only in the WT, and 27 genes that were highly expressed in BnABI5 CRISPR-edited lines (Figure 3E). These results provide further evidence that BnABI5 gene editing alters the expression of key genes related to stress tolerance pathways, influencing the plant’s ability to respond to drought stress.
3.4. Complex Gene Expression Patterns in BnABI5 CRISPR-Edited Lines and WT Plants
To further analyze the temporal and treatment-specific response patterns of DEGs under stress treatments, we performed clustering analysis using mfuzz on 1179 DEGs that were differentially expressed in at least four out of six treatment groups. Based on the time-course dataset, these genes were assigned to 8 clusters, each containing between 33 and 286 genes (Figure 4A). The expression patterns of these clusters revealed that genes in cluster1, cluster4, cluster6, and cluster8 were predominantly highly expressed in WT plants, whereas genes in cluster2 and cluster7 were more highly expressed in BnABI5 CRISPR-edited lines (Figure 4B).
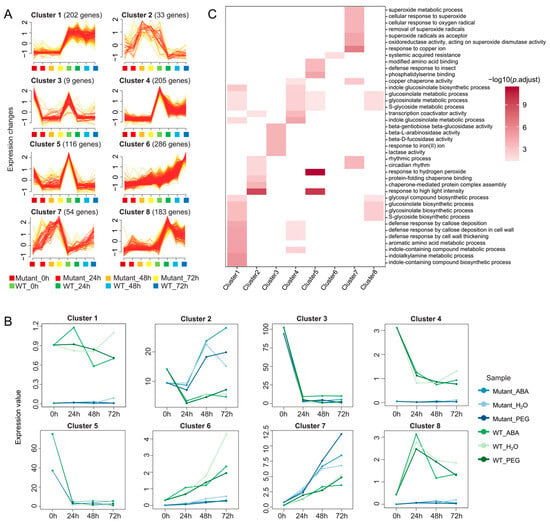
Figure 4.
Mfuzz Clustering Analysis of DEGs. (A) Clustering of 1179 DEGs based on their expression patterns across four time points (0 h, 24 h, 48 h, and 72 h). The x-axis represents treatment duration, while the y-axis indicates expression changes. (B) Expression patterns of genes in the eight clusters under different treatments. The x-axis denotes treatment duration, while the y-axis shows the median expression level of all genes in each cluster. (C) Heatmap of the GO enrichment results of genes from eight distinct clusters.
To further explore the functions of genes within these clusters, we conducted enrichment analysis for the genes in each cluster. The results showed that genes in cluster1 and cluster4 were mainly involved in biological processes related to indole compound metabolism (such as indole-containing compound biosynthetic process, indolalkylamine metabolic process, and indole-containing compound metabolic process) and defense responses associated with callose deposition and cell wall thickening (such as defense response by cell wall thickening, defense response by callose deposition in cell wall). These processes are likely linked to plant defense mechanisms. In particular, the metabolism of indole compounds and the deposition of callose are important pathways through which BnABI5 regulates plant stress responses. Additionally, genes in cluster8 were enriched in processes related to S-glycoside and glucosinolate metabolism, further suggesting that BnABI5 may influence plant drought responses through the regulation of these metabolic pathways (Figure 4C). Furthermore, all DEGs in cluster7 showed higher expression levels in BnABI5 CRISPR-edited lines compared to WT, and their expression increased over the course of seed germination. These genes were enriched in biological processes related to copper ion response (response to copper ion) and superoxide (superoxide), which may be related to the ability of plants to enhance stress tolerance under drought by regulating these pathways. The analysis of gene expression patterns and functions in these clusters provides strong evidence for understanding the effects of BnABI5 gene editing on seed germination and the dynamic changes in gene expression during seed germination.
3.5. Gene Expression Regulatory Networks Involving Multiple Hormones
The above analysis indicates that the mutation of ABI5 induces dysregulation of several genes, particularly under stress conditions, revealing a complex regulatory network between ABI5 and other genes. This network likely involves interactions with various plant hormones, such as ABA, jasmonic acid (JA), and 1-aminocyclopropane-1-carboxylic acid (ACC), which are known to play critical roles in stress responses. To further explore this complex gene-hormone interaction, we conducted a WGCNA to identify gene modules with similar expression patterns across all samples (Figure 5A). These genes were grouped into 12 modules, with the largest module, MEturquoise, containing 3927 genes, and the smallest module, MEgreenyellow, containing only 44 genes (Figure 5B).
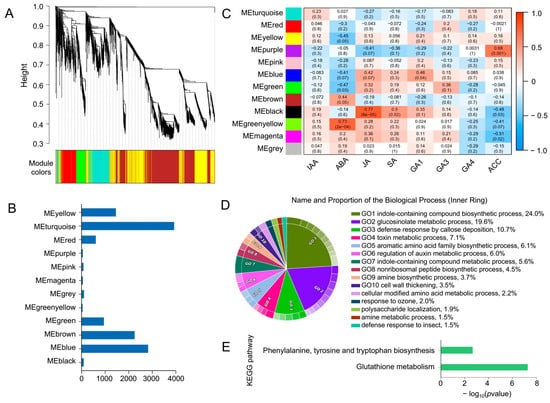
Figure 5.
WGCNA of transcriptome and 8 hormone phenotypes. (A) Dendrogram displaying 12 co-expression modules identified by WGCNA across all samples. (B) Heatmap showing the correlation between the 12 modules and the levels of 9 plant hormones. Each row corresponds to a module represented by different colors, and each column represents one hormone. Red indicates a positive correlation between the module and the hormone, while blue indicates a negative correlation. (C) Bar chart illustrating the number of genes in each module. (D,E) GO (D) and KEGG (E) enrichment analysis results for the 44 genes in the MEgreenyellow module, which are significantly correlated with ABA.
Since plant responses to stress are often initiated by rapid changes in hormone levels, we calculated the correlation between eight plant hormones: indole-3-acetic acid (IAA), JA, ABA, ACC, gibberellin 1 (GA1), gibberellin 2 (GA2), gibberellin 4 (GA4), and salicylic acid (SA), and the identified expression modules (Supplementary Table S2). Analysis of the relationship between the modules and hormonal traits revealed significant correlations, with MEgreenyellow being significantly associated with ABA, MEblack with JA, and MEpurple with ACC (p < 0.05) (Figure 5C). Thus, the MEgreenyellow module emerges as a key module containing genes related to ABA synthesis and ABA-regulated genes involved in seed growth and development in B. napus. We further performed functional enrichment analysis on the genes in the MEgreenyellow module. The results indicated that 44 genes were enriched in biological processes associated with stress responses, including indole-containing compound biosynthetic process (24%), glucosinolate metabolic process (19.6%), and defense response by callose deposition (10.7%) (Figure 5D). KEGG pathway enrichment showed significant enrichment in glutathione metabolism and phenylalanine, tyrosine, and tryptophan biosynthesis pathways. These findings suggest that genes in the MEgreenyellow module may influence ABA synthesis and drought resistance in B. napus by regulating the metabolism and biosynthesis of glutathione, phenylalanine, tyrosine, and tryptophan.
3.6. Regulation of Glutathione Metabolism Pathway Genes by TFs
Given the important role of GSH in the ABA signaling pathway, this study further investigates the co-expression relationship between genes enriched in the glutathione metabolism pathway and TFs identified through WGCNA. We first identified TFs in the B. napus genome, identifying a total of 7702 TFs. The results show that 9 genes in the glutathione metabolism pathway are co-expressed with 99 TFs, which belong to 32 different TF families. The MYB family contains the highest number of TFs (11), followed by C2H2 (9), Trihelix (7), NAC (7), bZIP (7), and WRKY (6) (Figure 6A, Supplementary Table S3).
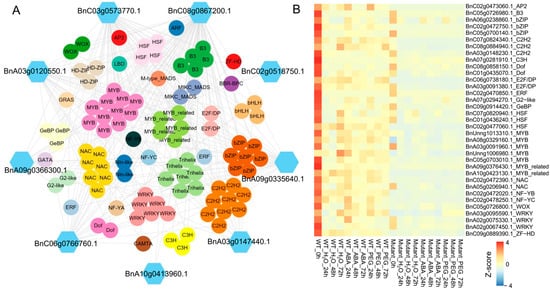
Figure 6.
Co-expression of glutathione metabolism pathway genes and TFs. (A) Co-expression regulatory network between 9 genes in the glutathione metabolism pathway and TFs. (B) Heatmap of the expression of differentially expressed TFs in the co-expression network. The color scale represents the Z-score of the gene expression level.
Further analysis revealed that 34 TFs exhibit differential expression between BnABI5 CRISPR-edited lines and WT, including 5 MYB, 3 WRKY, 3 HSF, 3 C2H2, and 3 bZIP TFs. Most of these differentially expressed TFs show the highest expression levels in the water treatment at 0 h, with expression levels decreasing in the samples after different treatments. Furthermore, the expression levels of these differentially expressed TFs in WT are generally higher than in BnABI5 CRISPR-edited lines. This suggests that editing the BnABI5 gene may affect the expression of these TFs, thereby regulating the gene expression in the glutathione metabolism pathway and influencing the ABA signaling and drought response mechanisms in plants. These findings provide new insights into the molecular mechanisms by which BnABI5 regulates the glutathione metabolism pathway and may offer theoretical support for improving drought resistance in B. napus.
3.7. The Role of Glutathione-Mediated ABA Signaling in Drought Resistance
Drought-induced increases in GSH levels enhance the expression of ZEP (zeaxanthin epoxidase) and NCED (9-cis-epoxycarotenoid dioxygenase), promoting the conversion of β-carotene to abscisic aldehyde. Additionally, GST (glutathione S-transferase) mediates detoxification reactions and promotes the expression of ABI3 and ABI5, thereby activating ABA-responsive genes and enhancing plant drought resistance [] (Figure 7A). In the GSH-mediated drought regulatory pathway, 70 genes were differentially expressed in BnABI5 CRISPR-edited lines compared to WT, including 39 GST, 13 PP2C (protein phosphatase 2C), 11 PYR/PYL, 3 ABI5, 3 ABI3, 1 ZEP, and 1 NCED. Notably, BnC09g0896470.1 (ZEP) and BnA05g0215860.1 (NCED) showed increased expression levels with prolonged ABA and PEG treatments (Figure 7B), and ZEP expression was higher in BnABI5 CRISPR-edited lines than in WT. Two ABI3 genes (BnC03g0586980.1 and BnA03g0136860.1) and three ABI5 genes (BnA05g0194800.1, BnA04g0179250.1, and BnC04g0676540.1) exhibited high expression only at 0 h under water treatment, with reduced expression after treatment. Among the 39 GST genes, genes such as BnC08g0878140.1, BnA08g0329980.1, and BnA07g0302050.1 had higher expression levels in BnABI5 CRISPR-edited lines than in WT under drought (PEG) treatment. For PP2C, BnA07g0297030.1 showed elevated expression only in BnABI5 CRISPR-edited lines after 72 h of PEG treatment, while the other 12 PP2C genes were generally expressed at low levels. Three PYR/PYL/RCAR genes (BnA07g0285090.1, BnC03g0563340.1, and BnC06g0754380.1) exhibited increased expression after stress treatments, with overall higher expression in BnABI5 CRISPR-edited lines compared to WT. This suggests that GSH enhances ABA synthesis, thereby promoting the expression of PYR/PYL/RCAR while suppressing PP2C.
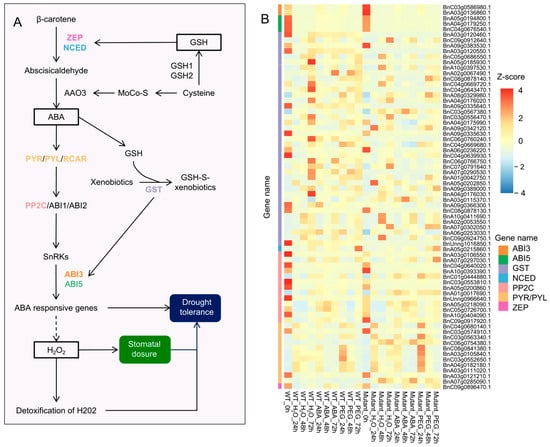
Figure 7.
Glutathione-mediated ABA signaling regulates stomatal closure and drought resistance. (A) A pathway diagram illustrating the regulatory role of glutathione-mediated ABA signaling in stomatal closure and drought resistance. (B) Heatmap showing the expression profiles of 70 DEGs involved in the pathway. The color scale represents the Z-score of the gene expression level.
4. Discussion
This study identified BnABI5 as a key candidate gene for seed germination under drought stress through GWAS. ABI5 is a well-known functional gene that plays a crucial role in both seed germination and drought stress responses in plants []. By generating stable genetic lines of BnABI5 CRISPR-edited plants, we observed an increased seed germination rate at 24 h under water, PEG, and ABA treatments. Although at 72 h the germination rates of the mutant plants were similar to those of the WT plants, the BnABI5 CRISPR-edited lines exhibited stronger growth, suggesting that BnABI5 knockout facilitates seed germination under drought conditions.
As a TF, the phenotypic changes caused by the mutation of BnABI5 are mediated through its involvement in a complex gene regulatory network. To investigate which genes are affected by the BnABI5-mediated regulatory network, this study designed time-course RNA-seq datasets under two genotypes and various treatment conditions to provide insights into the role of BnABI5 in seed germination and drought stress. The analysis revealed that, under most treatment conditions and time-points, the number of downregulated DEGs in the mutant plants was significantly higher than the number of upregulated DEGs. Further analysis of the expression trends of DEGs between the BnABI5 CRISPR-edited lines and WT plants highlighted the functions of these genes, especially those showing lower expression levels in the WT (DEGs in clusters 1, 4, 6, and 8). Notably, the genes in these clusters (except cluster 6) were enriched in the glucosinolate biosynthesis pathway, in which enzymes are involved in regulating ABA levels and the synthesis of other secondary metabolites [,]. After successful ABI5 gene editing, the plants exhibited stronger root development at 72 h, possibly due to the disruption of ABA homeostasis resulting from the downregulation of certain enzymes in the glucosinolate biosynthesis pathway, which accelerated the seed germination process.
The results of this study demonstrate that BnABI5 editing influences phenotypes through a complex regulatory network, affecting numerous genes across various pathways. For instance, it was observed that, at most stress treatment time points, downregulated genes in the mutant plants were enriched in the glutathione metabolism pathway. Moreover, genes in the MEgreenyellow module, identified through WGCNA as significantly associated with ABA levels, were also enriched in the glutathione metabolism pathway. GSH is an important signaling molecule that regulates ABA signaling and its role in seed dormancy and drought resistance []. Increased levels of phenylalanine, tyrosine, and tryptophan are linked to enhanced drought tolerance mechanisms in plants, including stomatal regulation, plant hormone synthesis, and oxidative stress responses [,]. Furthermore, ABA and GSH are closely related in mediating plant responses to drought stress [,,]. GSH plays a role in regulating ABA-induced downstream signaling of H2O2 in guard cells, preventing stomatal closure. Additionally, GSH is involved in detoxifying H2O2 through the AsA-GSH pathway [,]. Through differential expression analysis and WGCNA, this study identified key genes in the glutathione metabolism pathway, including BnC02g0518750.1 (GST), BnA03g0120550.1 (GST), BnA10g0413960.1 (gshA), and BnA03g0147440.1 (gshA). These genes provide potential candidates for further investigation into how ABI5 regulates downstream genes to achieve fine-tuned responses to drought stress. These results offer promising molecular markers for marker-assisted selection (MAS). For example, favorable alleles of BnABI5 or its downstream targets could be used to develop drought-tolerant rapeseed cultivars through conventional breeding or gene editing approaches. Furthermore, the regulatory network revealed in this study provides a framework for selecting gene combinations that balance germination vigor and drought resilience, which could enhance seedling establishment in water-limited environments.
Several studies have shown that ABI5 can interact with other TFs to regulate target genes. For example, in apple, ABI5 interacts with MdbHLH3, enhancing the expression of target genes in the anthocyanin biosynthesis pathway and thereby promoting anthocyanin production []. The WD40-repeat protein XIW1 in Arabidopsis has been shown to regulate ABI5 stability and contribute to the modulation of ABA signaling []. To explore how ABI5 affects target genes through TF interactions, this study conducted a detailed analysis of the co-expression relationships between genes in the glutathione metabolism pathway and TFs. The results revealed that TFs co-expressed with genes in this pathway generally exhibited lower expression levels in BnABI5 CRISPR-edited lines compared to WT plants (under most drought stress treatment conditions). Previous studies have shown that several TFs in the glutathione metabolism pathway negatively regulate ABI5. For instance, in Arabidopsis, G6PD5 inhibits ABI5, thereby affecting ABA-mediated seed germination and root growth []. CycC1;1 negatively regulates the ABA signaling pathway during seed germination by inhibiting ABI5 []. Our results suggest that the expression of these TFs may be influenced by ABI5 or its signaling pathway. The regulatory relationship between these TFs and ABI5 warrants further experimental investigation. In summary, this study systematically elucidated how the candidate gene ABI5 influences multiple genes and pathways through its regulatory network, thereby affecting germination and drought resistance phenotypes in rapeseed. This was achieved through an integrative approach, starting from germination rate assessments under drought treatment, candidate gene identification, mutant construction, and transcriptomic analysis. These findings lay a foundation and provide clues for identifying additional drought-resistance candidate genes. These findings lay a theoretical and molecular foundation for integrating ABI5-related genes into breeding programs aimed at improving drought tolerance in B. napus. Future studies could focus on validating these markers in diverse genetic backgrounds and testing their effectiveness under field conditions to accelerate their application in practical breeding strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants14091276/s1, Supplementary Figure S1: A schematic diagram of BnABI5 gene editing vector construction. (A) Blue rectangular boxes indicate exons of BnA05g0194800.1, BnC04g0676540.1, and BnA04g0179250.1 copies; black horizontal lines represent introns; thickened red vertical lines show sgRNA1 (S1) targeting Exon2; thickened yellow lines indicate sgRNA2 (S2) targeting Exon3. (B) A schematic diagram of the BnABI5 gene editing vector. sgRNA1 is initiated and terminated by U6-26p and U6-26t, respectively, sgRNA2 is initiated and terminated by U6-29p and U6-26t, respectively. Supplementary Figure S2: Identification and characterization of CRISPR-Cas9-induced BnABI5 mutants by Sanger sequencing in the T1 generation. Mutations at two target sites (WT-1: TTTGGTCTGAGATACATAGAGG; WT-2: GTATGGAGTTGATATGGGAGGG) were detected. Deletions were detected at three copies of the BnABI5 gene. Supplementary Figure S3: GO (left) and KEGG (right) enrichment analysis of upregulated genes in different treatment groups. Supplementary Table S1: Survival rate of 196 germplasms under drought stress. Supplementary Table S2: Determination of hormone content in samples under different treatment conditions and time. Supplementary Table S3: Co-expression relationship between 9 GSHs and 99 TFs.
Author Contributions
Conceptualization, G.L., H.F. and Y.L.; methodology, D.L., Q.H. and M.C.; software, H.L.; validation, D.L. and Q.H.; formal analysis, G.L. and D.L.; investigation, D.L.; resources, G.L., Y.L. and H.F.; data curation, D.L. and G.L.; writing—original draft preparation, G.L.; writing—review and editing, G.L., H.F. and Y.L.; funding acquisition, G.L and Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was jointly funded by the National Natural Science Foundation of China (32072131), Hubei Provincial Agricultural Science and Technology Innovation Center Innovation Team (2024-620-000-001-031), and the Talent Introduction Project of Guangdong University of Petrochemical Technology (2021rc009).
Data Availability Statement
The data are presented in the manuscript and the Supporting Materials. The raw read data are submitted to the Short Read Archive (SRA) and BioProject accession number PRJNA1227215.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Shabala, L.; Brodribb, T.J.; Zhou, M.; Shabala, S. Understanding physiological and morphological traits contributing to drought tolerance in barley. J. Agron. Crop Sci. 2019, 205, 129–140. [Google Scholar] [CrossRef]
- Bi, H.; Kovalchuk, N.; Langridge, P.; Tricker, P.J.; Lopato, S.; Borisjuk, N. The impact of drought on wheat leaf cuticle properties. BMC Plant Biol. 2017, 17, 85. [Google Scholar] [CrossRef]
- Furlan, A.L.; Bianucci, E.; Giordano, W.; Castro, S.; Becker, D.F. Proline metabolic dynamics and implications in drought tolerance of peanut plants. Plant Physiol. Biochem. 2020, 151, 566–578. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic Interaction between Abscisic Acid and Jasmonate-Ethylene Signaling Pathways Modulates Defense Gene Expression and Disease Resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef]
- Fujii, H. Abscisic acid implication in plant growth and stress responses. In Phytohormones: A Window to Metabolism, Signaling and Biotechnological Applications; Springer: New York, NY, USA, 2014; ISBN 978-1-4939-0490-7. [Google Scholar]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Ye, T.; Zhao, S.; Liu, Z.; Feng, Y.-Q.; Wu, Y. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 2011, 68, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.-Q.; Luan, S.; Li, J.; He, Z.-H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef]
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA Transport and Plant Water Stress Responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of Basal ABA in Plant Growth and Development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef]
- Hermann, K.; Meinhard, J.; Dobrev, P.; Linkies, A.; Pesek, B.; Heß, B.; Macháčková, I.; Fischer, U.; Leubner-Metzger, G. 1-Aminocyclopropane-1-carboxylic acid and abscisic acid during the germination of sugar beet (Beta vulgaris L.): A comparative study of fruits and seeds. J. Exp. Bot. 2007, 58, 3047–3060. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D.L. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2022, 35, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Vishal, B.; Kumar, P.P. Regulation of Seed Germination and Abiotic Stresses by Gibberellins and Abscisic Acid. Front. Plant Sci. 2018, 9, 838. [Google Scholar] [CrossRef]
- Aleem, M.; Razzaq, M.K.; Aleem, M.; Yan, W.; Sharif, I.; Siddiqui, M.H.; Aleem, S.; Iftikhar, M.S.; Karikari, B.; Ali, Z.; et al. Genome-wide association study provides new insight into the underlying mechanism of drought tolerance during seed germination stage in soybean. Sci. Rep. 2024, 14, 20765. [Google Scholar] [CrossRef] [PubMed]
- Thabet, S.G.; Safhi, F.A.; Börner, A.; Alqudah, A.M. Genetic associations of transgenerational stress memory in wheat under drought stress. Environ. Exp. Bot. 2024, 226, 105920. [Google Scholar] [CrossRef]
- Collin, A.; Daszkowska-Golec, A.; Szarejko, I. Updates on the Role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA Signaling in Different Developmental Stages in Plants. Cells 2021, 10, 1996. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Xie, Q. Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 2015, 20, 569–575. [Google Scholar] [CrossRef]
- Nakamura, S.; Lynch, T.J.; Finkelstein, R.R. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001, 26, 627–635. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis Abscisic Acid Response Gene ABI5 Encodes a Basic Leucine Zipper Transcription Factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Carles, C.; Bies-Etheve, N.; Aspart, L.; Léon-Kloosterziel, K.M.; Koornneef, M.; Echeverria, M.; Delseny, M. Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 2002, 30, 373–383. [Google Scholar] [CrossRef]
- Kanai, M.; Nishimura, M.; Hayashi, M. A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 2010, 62, 936–947. [Google Scholar] [CrossRef]
- Edreva, A. Generation and scavenging of reactive oxygen species in chloroplasts: A submolecular approach. Agric. Ecosyst. Environ. 2005, 106, 119–133. [Google Scholar] [CrossRef]
- Okuma, E.; Jahan, M.S.; Munemasa, S.; Hossain, M.A.; Muroyama, D.; Islam, M.M.; Ogawa, K.; Watanabe-Sugimoto, M.; Nakamura, Y.; Shimoishi, Y.; et al. Negative regulation of abscisic acid-induced stomatal closure by glutathione in Arabidopsis. J. Plant Physiol. 2011, 168, 2048–2055. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Tang, J.; Gao, S.; Cheng, X.; Du, L.; Chu, C. Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant J. 2019, 100, 1036–1051. [Google Scholar] [CrossRef]
- Paiva, A.L.S.; Passaia, G.; Jardim-Messeder, D.; Nogueira, F.C.S.; Domont, G.B.; Margis-Pinheiro, M. The mitochondrial isoform glutathione peroxidase 3 (OsGPX3) is involved in ABA responses in rice plants. J. Proteom. 2021, 232, 104029. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Li, C.; Sang, S.; Sun, M.; Yang, J.; Shi, Y.; Hu, X.; Li, Y.; Hao, M.; Chu, W.; Zhang, H.; et al. Direct modification of multiple gene homoeologs in Brassica oleracea and Brassica napus using doubled haploid inducer-mediated genome-editing system. Plant Biotechnol. J. 2021, 19, 1889–1891. [Google Scholar] [CrossRef]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hao, M.; Wang, W.; Wang, H.; Chen, F.; Chu, W.; Zhang, B.; Mei, D.; Cheng, H.; Hu, Q. An Efficient CRISPR/Cas9 Platform for Rapidly Generating Simultaneous Mutagenesis of Multiple Gene Homoeologs in Allotetraploid Oilseed Rape. Front. Plant Sci. 2018, 9, 442. [Google Scholar] [CrossRef]
- Turaki, A.A.; Ahmad, B.; Magaji, U.F.; Abdulrazak, U.K.; Yusuf, B.A.; Hamza, A.B. Optimised cetyltrimethylammonium bromide (CTAB) DNA extraction method of plant leaf with high polysaccharide and polyphenolic compounds for downstream reliable molecular analyses. Afr. J. Biotechnol. 2017, 16, 1354–1365. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Putri, G.H.; Anders, S.; Pyl, P.T.; Pimanda, J.E.; Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 2021, 38, 2943–2945. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Gu, Z. Complex heatmap visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- Koramutla, M.K.; Negi, M.; Ayele, B.T. Roles of Glutathione in Mediating Abscisic Acid Signaling and Its Regulation of Seed Dormancy and Drought Tolerance. Genes 2021, 12, 1620. [Google Scholar] [CrossRef]
- Li, X.; Zhong, M.; Qu, L.; Yang, J.; Liu, X.; Zhao, Q.; Liu, X.; Zhao, X. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021, 310, 110983. [Google Scholar] [CrossRef]
- Du, W.; Yang, J.; Li, Q.; Jiang, W.; Pang, Y. Medicago truncatula β-glucosidase 17 contributes to drought and salt tolerance through antioxidant flavonoid accumulation. Plant. Cell Environ. 2024, 47, 3076–3089. [Google Scholar] [CrossRef]
- Xiong, D.-M.; Liu, Z.; Chen, H.; Xue, J.-T.; Yang, Y.; Chen, C.; Ye, L.-M. Profiling the dynamics of abscisic acid and ABA-glucose ester after using the glucosyltransferase UGT71C5 to mediate abscisic acid homeostasis in Arabidopsis thaliana by HPLC–ESI-MS/MS. J. Pharm. Anal. 2014, 4, 190–196. [Google Scholar] [CrossRef]
- Wei, L.; Wang, L.; Yang, Y.; Wang, P.; Guo, T.; Kang, G. Abscisic acid enhances tolerance of wheat seedlings to drought and regulates transcript levels of genes encoding ascorbate-glutathione biosynthesis. Front. Plant Sci. 2015, 6, 458. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Zhou, Q.; Xing, H.; Jiang, H.; Wang, S. Early Abscisic Acid Accumulation Regulates Ascorbate and Glutathione Metabolism in Soybean Leaves Under Progressive Water Stress. J. Plant Growth Regul. 2016, 35, 865–876. [Google Scholar] [CrossRef]
- Cheng, M.-C.; Ko, K.; Chang, W.-L.; Kuo, W.-C.; Chen, G.-H.; Lin, T.-P. Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 2015, 83, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Redox Modulation Matters: Emerging Functions for Glutaredoxins in Plant Development and Stress Responses. Plants 2014, 3, 559–582. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y. The Roles of Glutaredoxin GRXS17 in Improving Chilling Tolerance in Tomato and Drought Tolerance in Rice via Different Mechanisms. Ph.D. Thesis, Kansas State University, Manhattan, KS, USA, 2015. [Google Scholar]
- An, J.-P.; Zhang, X.-W.; Liu, Y.-J.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 2021, 72, 1460–1472. [Google Scholar] [CrossRef]
- Xu, X.; Wan, W.; Jiang, G.; Xi, Y.; Huang, H.; Cai, J.; Chang, Y.; Duan, C.-G.; Mangrauthia, S.K.; Peng, X.; et al. Nucleocytoplasmic Trafficking of the Arabidopsis WD40 Repeat Protein XIW1 Regulates ABI5 Stability and Abscisic Acid Responses. Mol. Plant 2019, 12, 1598–1611. [Google Scholar] [CrossRef]
- Yang, L.; Wang, S.; Sun, L.; Ruan, M.; Li, S.; He, R.; Zhang, W.; Liang, C.; Wang, X.; Bi, Y. Involvement of G6PD5 in ABA response during seed germination and root growth in Arabidopsis. BMC Plant Biol. 2019, 19, 44. [Google Scholar] [CrossRef]
- Guo, J.-X.; Song, R.-F.; Lu, K.-K.; Zhang, Y.; Chen, H.-H.; Zuo, J.-X.; Li, T.-T.; Li, X.-F.; Liu, W.-C. CycC1;1 negatively modulates ABA signaling by interacting with and inhibiting ABI5 during seed germination. Plant Physiol. 2022, 190, 2812–2827. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).