A C-Terminally Encoded Peptide, MeCEP6, Promotes Nitrate Uptake in Cassava Roots
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of the Cassava CEP Gene
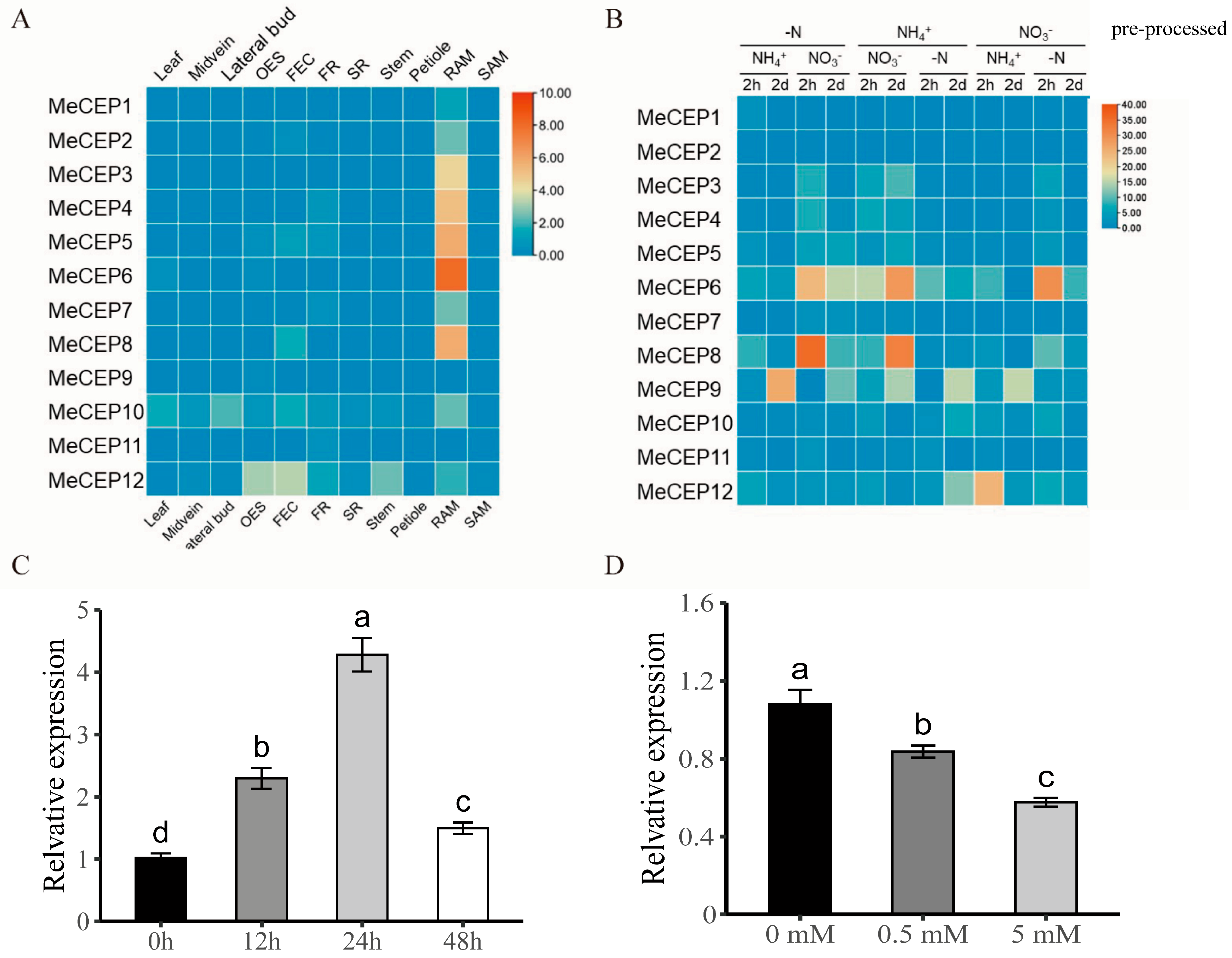
2.2. Expression Patterns of MeCEP Genes in Different Tissues and Nitrogen Treatments
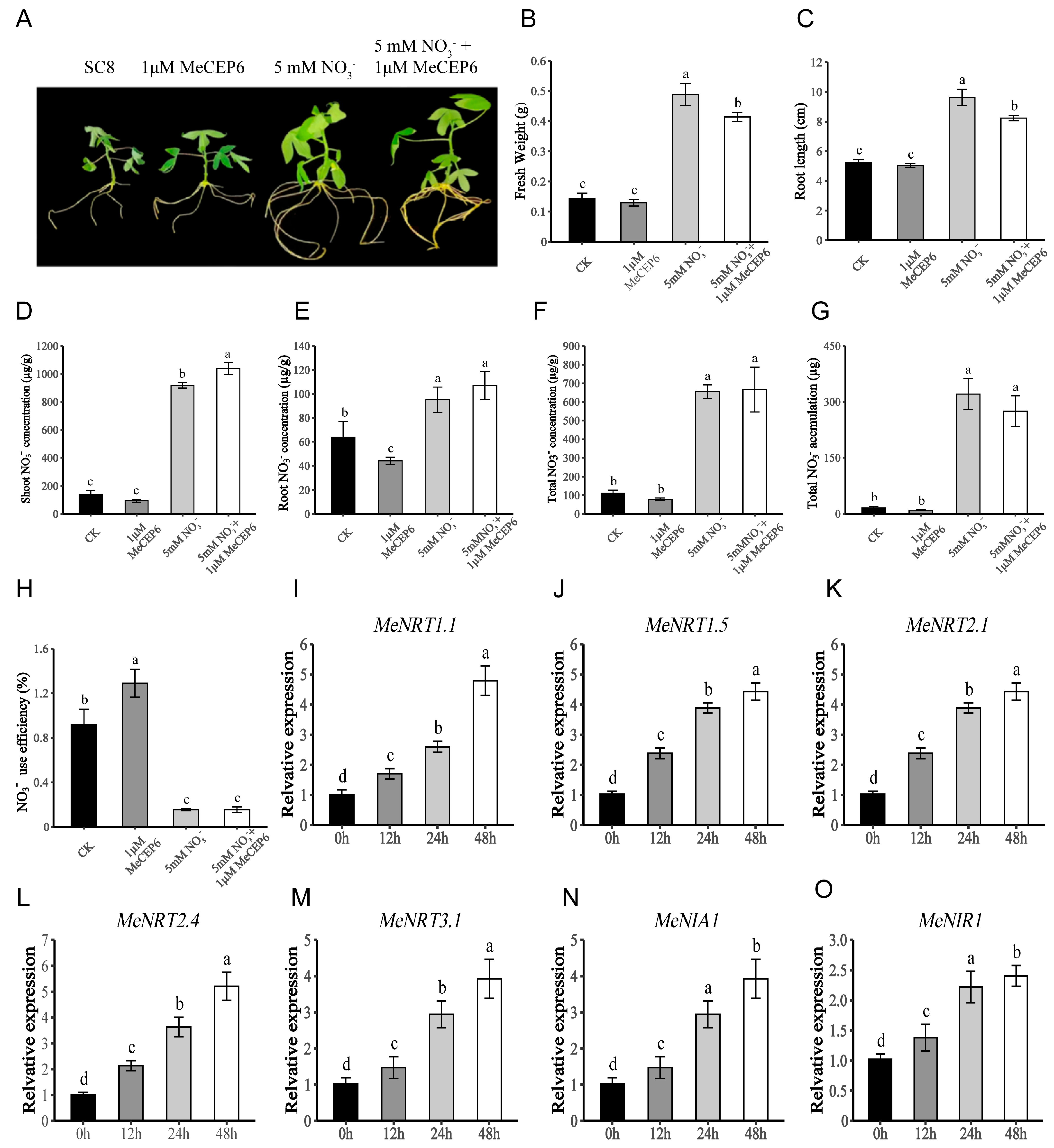
2.3. Exogenous Application of MeCEP6 Promotes Nitrate Uptake by Cassava Roots
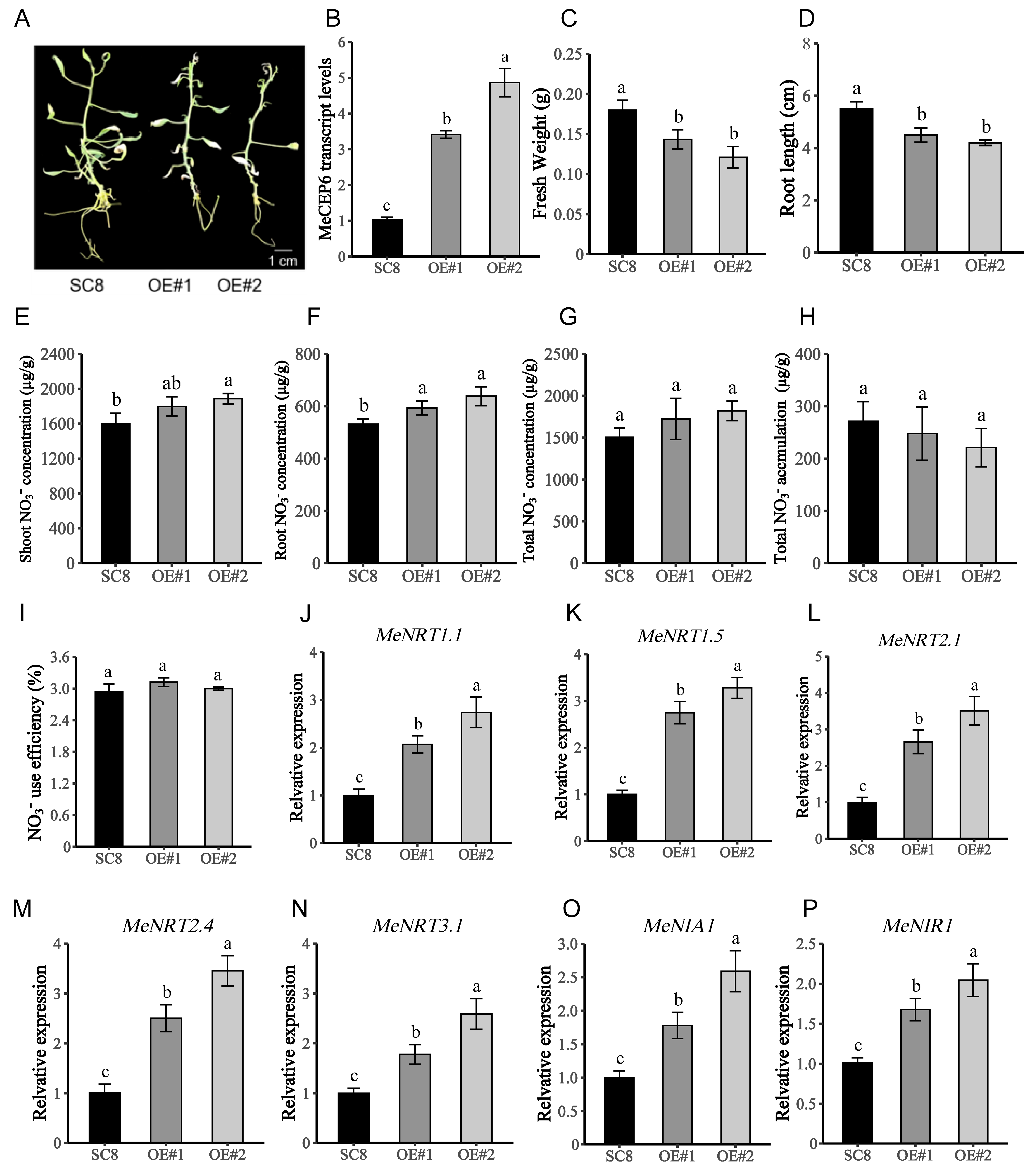
2.4. Overexpression of MeCEP6 Promotes Nitrate Uptake by the Roots of Cassava
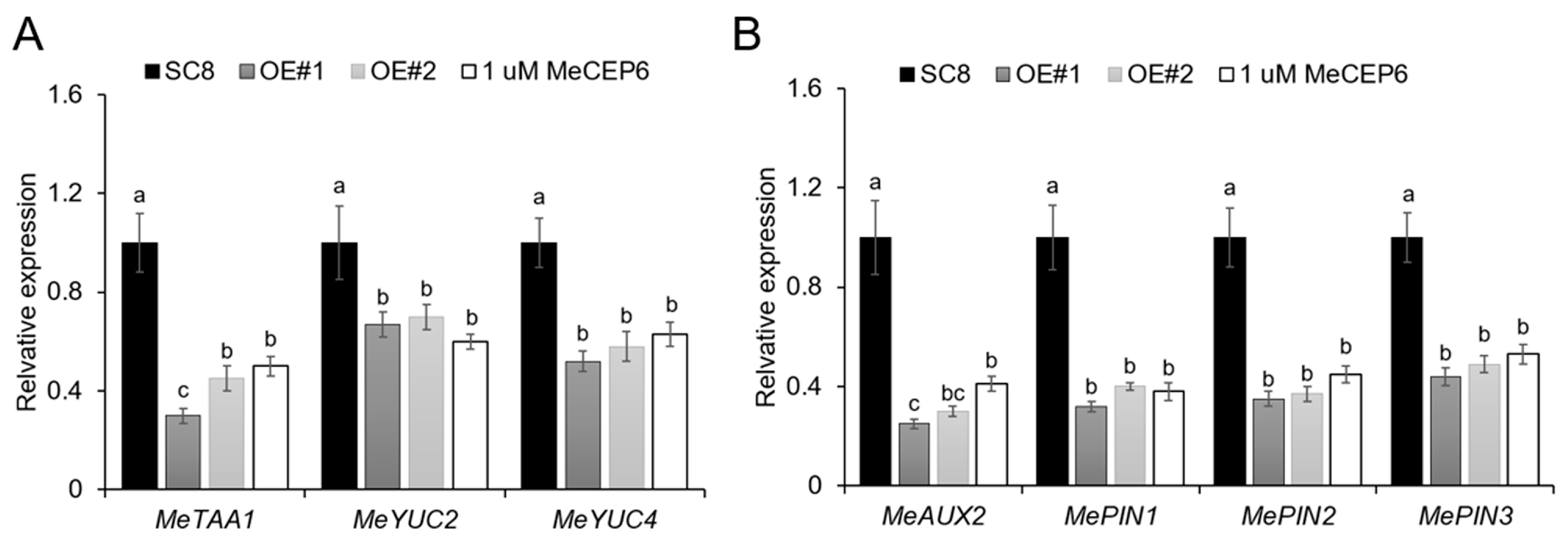
2.5. External Application of MeCEP6 and Its Overexpression Suppress the Gene Expression Associated with Plant Hormones That Regulate Growth
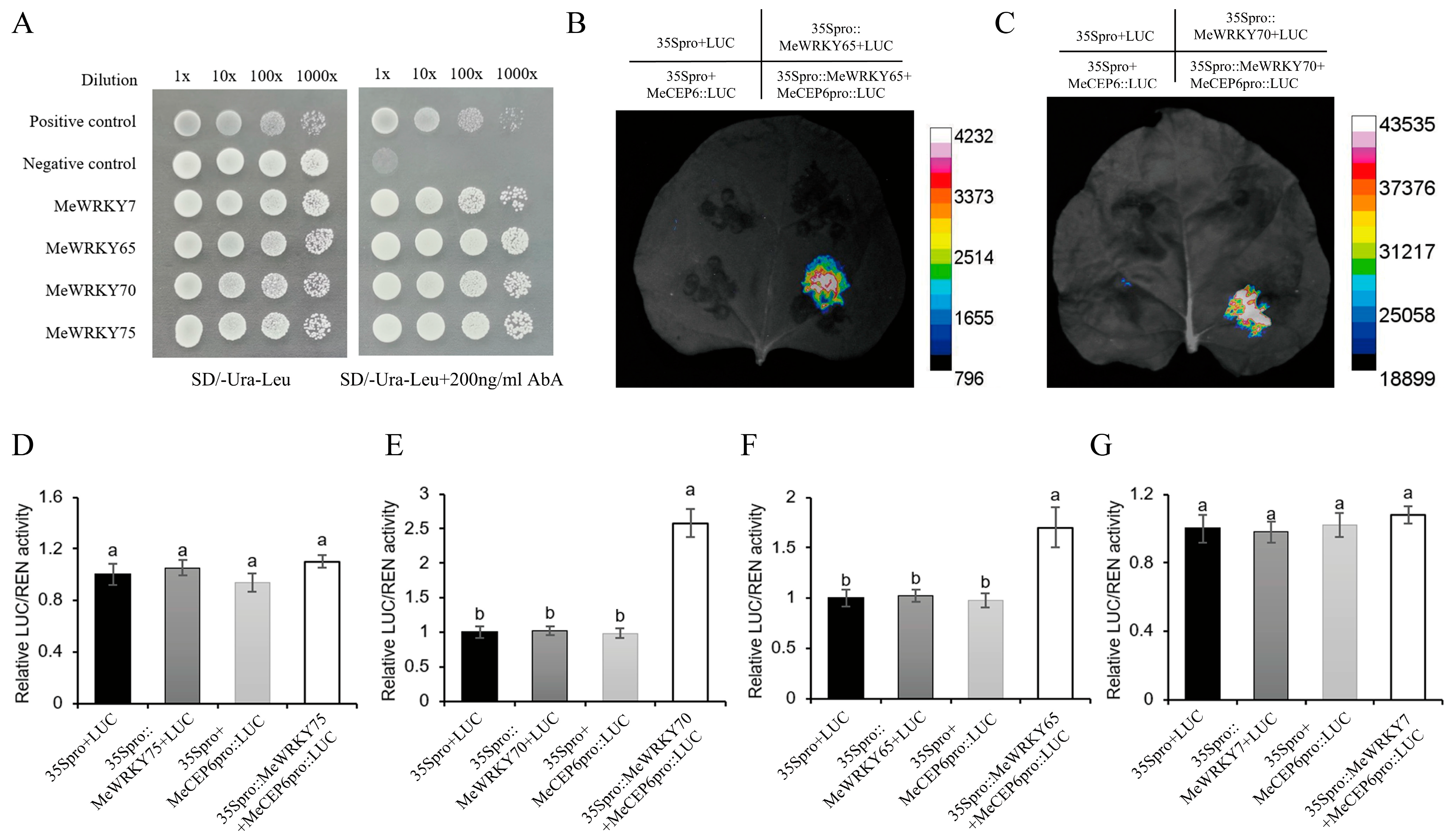
2.6. MeWRKY65 and MeWRKY70 Positively Regulate the Expression of MeCEP6
3. Discussion
3.1. MeCEP6 Promotes Nitrate Uptake by Cassava Roots
3.2. MeCEP6 Inhibits the Growth of Cassava Plants Through Plant Hormones
3.3. The CEP Signaling Pathway Plays a Role in Cassava’s Tolerance to Barrenness
4. Materials and Methods
4.1. Identification of MeCEP Genes in Cassava
4.2. The Creation of Overexpressed Transgenetic MeCEP6 Cassava
4.3. Plant Nitrogen Treatment
4.4. RT-qPCR Analysis
4.5. Phenotypic Characterization
4.6. Yeast One-Hybrid (Y1H) Library Screening
4.7. Luciferase In Vivo Imaging Assay (LCI)
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CEP | C-terminally encoded peptide |
| Pi | Theoretical isoelectric point |
| Kan | Kanamycin |
| Rif | Rifampicin |
| AbA | Aureobasidin A |
| IAA | Auxin (Indole acetic acid) |
Appendix A
| Name | Gene Identifier | aa | Start | Termination | Direction | MW(Da) | pI | GRAVY |
|---|---|---|---|---|---|---|---|---|
| MeCEP1 | Manes.12G100000 | 120 | 25886473 | 25887221 | + | 13,137.72 | 7.17 | −0.592 |
| MeCEP2 | Manes.12G100100 | 322 | 25866490 | 25867763 | + | 34,318.59 | 6.62 | −0.861 |
| MeCEP3 | Manes.12G100200 | 88 | 25811645 | 25812113 | + | 9577.61 | 5.03 | −0.193 |
| MeCEP4 | Manes.12G100300 | 92 | 25805335 | 25805933 | + | 10,220.97 | 9.64 | −0.261 |
| MeCEP5 | Manes.13G126300 | 90 | 33391382 | 33392276 | + | 10,063.72 | 7.73 | −0.227 |
| MeCEP6 | Manes.13G126400 | 93 | 33394734 | 33395016 | + | 10,481.85 | 6.25 | −0.333 |
| MeCEP7 | Manes.13G126500 | 293 | 33397428 | 33398310 | + | 31,134.11 | 6.17 | −0.745 |
| MeCEP8 | Manes.13G126600 | 117 | 33403801 | 33404532 | + | 12,973.67 | 6.70 | −0.517 |
| MeCEP9 | Manes.15G184800 | 89 | 28678844 | 28679114 | + | 9753.32 | 9.77 | −0.237 |
| MeCEP10 | Manes.16G042800 | 96 | 6566461 | 6567257 | + | 10,501.05 | 9.60 | −0.101 |
| MeCEP11 | Manes.17G001300 | 89 | 1491711 | 1492329 | + | 9846.43 | 9.60 | −0.042 |
| MeCEP12 | Manes.17G059300 | 101 | 25812294 | 25813455 | + | 10,718.20 | 9.69 | 0.003 |
| Name | Sequence (5′–3′) | Purpose |
|---|---|---|
| MeCEP1qPCR-F | CGGTAGATGGAAGGCACTTG | qRT-PCR |
| MeCEP1qPCR-R | CCACTAGGAATTGGGATGGC | qRT-PCR |
| MeCEP2qPCR-F | CGTACCGCATTTGCTGATTC | qRT-PCR |
| MeCEP2qPCR-R | CAACTTCAGGGTTCGGACAA | qRT-PCR |
| MeCEP3qPCR-F | CCTCAAGTGTGTGGAATGCT | qRT-PCR |
| MeCEP3qPCR-R | ATCCGTCACTGGGTTCAATG | qRT-PCR |
| MeCEP4qPCR-F | GCAACCATTAGGGCAATGTG | qRT-PCR |
| MeCEP5qPCR-F | GAACGTGTCTGAAGATACCCA | qRT-PCR |
| MeCEP5qPCR-R | TGTTGATGGAGTGACCAACG | qRT-PCR |
| MeCEP6qPCR-F | GCCAACAATGTAGATCGCAC | qRT-PCR |
| MeCEP6qPCR-R | TCCTCTGCTAGGGATTCTCG | qRT-PCR |
| MeCEP7qPCR-F | ATGATCACAGGCCGACAAAG | qRT-PCR |
| MeCEP7qPCR-R | TCAATTGTGGTCCCAGGAGA | qRT-PCR |
| MeCEP8qPCR-F | AGTCCGTAGATGGAAAAAGGC | qRT-PCR |
| MeCEP8qPCR-R | TCACCATGCACGTTATGCTT | qRT-PCR |
| MeCEP9qPCR-F | TCCGCCACCTTTCCCTAATA | qRT-PCR |
| MeCEP9qPCR-R | TTATGACCTACACCAGGGCT | qRT-PCR |
| MeCEP10qPCR-F | CGACTCGTCTCTGTTCCAAG | qRT-PCR |
| MeCEP10qPCR-R | GGAACAGACCGTAGAATCCG | qRT-PCR |
| MeCEP11qPCR-F | AGGCCACTGCATATTCATCC | qRT-PCR |
| MeCEP11qPCR-R | CTATGACCTGGGCTTGTTGG | qRT-PCR |
| MeCEP12qPCR-F | GCAGGAAGCTGTTGATGAGT | qRT-PCR |
| MeCEP12qPCR-R | TTGGAAGGGCACTGGAAATC | qRT-PCR |
| MeNRT1.1qPCR-F | TCAAATCAAGTGTCTCGGGC | qRT-PCR |
| MeNRT1.1qPCR-R | GTGCCAGACAAGAACAGGAT | qRT-PCR |
| MeNRT2.1qPCR-F | TATTTCCGGCATGACTGGTG | qRT-PCR |
| MeNRT2.1qPCR-R | TCTTCGGTGGATTTCACGAC | qRT-PCR |
| MeNRT2.4qPCR-F | GTTCTGGACTGACACAGCTT | qRT-PCR |
| MeNRT2.4qPCR-R | TCTGCTTTTCCTCCTCGTTC | qRT-PCR |
| MeNRT3.1qPCR-F | TCCGGAAAACTCTTGTGGTC | qRT-PCR |
| MeNRT3.1qPCR-R | TTGGATAGGTTGTCCTCGGT | qRT-PCR |
| MeTAA1qPCR-F | CTGGAGGAAGATGGGTGAGA | qRT-PCR |
| MeTAA1qPCR-R | GGTTCGAGAAACCAGCAGAA | qRT-PCR |
| MeYUC1qPCR-F | CCCAAGTACCCAACAAAGCA | qRT-PCR |
| MeYUC1qPCR-R | ATTCAGCTTCTTGAACGGCT | qRT-PCR |
| MeYUC2qPCR-F | TGCATAGCTTCGTTGTGGAA | qRT-PCR |
| MeYUC2qPCR-R | GTCGGGTAGGTAGGGAAACT | qRT-PCR |
| MeAUX1qPCR-F | GTTCGCATGTACACCTCTGT | qRT-PCR |
| MeAUX1qPCR-R | TATGGGTATCACCACAGGCA | qRT-PCR |
| MePIN1qPCR-F | ATGGTGGTGGTTTGGGTAAC | qRT-PCR |
| MePIN1qPCR-R | GCATTATTAGCAGCAACGCC | qRT-PCR |
| MePIN2qPCR-F | CACTCTGGTTATGGGCATCC | qRT-PCR |
| MePIN2qPCR-R | TCAGGAAACTGTTCGCCAAT | qRT-PCR |
| MePIN3qPCR-F | AGGGTGGAGTTGCTGCTAATA | qRT-PCR |
| MePIN3qPCR-R | TGGAGATACAGCCAATCGAAC | qRT-PCR |
| MeCEP6-F | ATGGCCAACAATGTAGATCGC | Carrier construction |
| MeCEP6-R | TTAATGACCAACACCTGGGCT | Carrier construction |
| MeCEP6OE-F | CGCGGATCCATGGCCAACAATGTAGATCGC | Carrier construction |
| MeCEP6OE-R | AACTGCAGATGACCAACACCTGGGCTAT | Carrier construction |
| MeCEP6pro-F | GCCTCATAGGCTCGCTTAAC | Yeast one-hybrid |
| MeCEP6pro-R | ATGACCAACACCTGGGCTAT | Yeast one-hybrid |
| MeWRKY65-F | CATCAGCAGCCACTTAACTCT | Yeast one-hybrid |
| MeWRKY65-R | TATCACCCACACAATTTCCCAT | Yeast one-hybrid |
| MeWRKY70-F | GCGTCGACATGGCAACTCCTTGGCC | Yeast one-hybrid |
| MeWRKY70-R | CGGGATCCTTAGCTTAGGTAGTTGAAATCACTT | Yeast one-hybrid |
| MeWRKY75-F | TGAAGACATGGCTGTGGAAC | Yeast one-hybrid |
| MeWRKY75-R | CTGCTCAGCCCTTAATGAGT | Yeast one-hybrid |
| MeWRKY7-F | TTAGCTTTGGCTTGGTTCTCG | Yeast one-hybrid |
| MeWRKY7-R | AACCCGCGTGATGACAATG | Yeast one-hybrid |
| MeActin-F | TCTTCTCAACTGAGGAGCTGCT | Internal reference primer |
| MeActin-R | CCTTCGTCTGGACCTTGCTG | Internal reference primer |
| Number | Gene Symbol | CDS (bp) | Protein (aa) |
|---|---|---|---|
| 1 | MeWRKY7 | 1077 | 359 |
| 2 | MeWRKY65 | 798 | 266 |
| 3 | MeWRKY70 | 849 | 283 |
| 4 | MeWRKY75 | 582 | 194 |
| 5 | MebHLH96 | 999 | 333 |
| 6 | MeHB6 | 927 | 309 |
| 7 | MeHSFA6B | 1077 | 359 |
| 8 | MeOBP3 | 984 | 328 |
| 9 | MeTGA1 | 1134 | 378 |
| 10 | MeZML2 | 858 | 286 |
| 11 | MeAGL16 | 678 | 226 |
References
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wirén, N.; Takahashi, H. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Pan, J.; Wu, Y.; Han, H.; Zhang, H.; Chen, X.; Zeng, C. Effects of different nitrogen sources on seedling growth and development of four cassava varieties. J. Trop. Biol. 2024, 15, 19–26. [Google Scholar] [CrossRef]
- Ezui, K.; Franke, A.; Mando, A.; Ahiabor, B.; Tetteh, F.; Sogbedji, J.; Janssen, B.; Giller, K. Fertiliser requirements for balanced nutrition of cassava across eight locations in West Africa. Field Crop. Res. 2016, 185, 69–78. [Google Scholar] [CrossRef]
- Arthur, P.; Amandine, C.; Ivan, P.; Ondrej, N.; Gabriel, K.; Benoît, L.; Sandrine, R. Responses to Systemic Nitrogen Signaling in Arabidopsis Roots Involve trans-Zeatin in Shoots. Plant Cell 2018, 30, 1243–1257. [Google Scholar]
- Vidal, E.A.; Álvarez, J.M.; Moyano, T.C.; Gutiérrez, R.A. Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr. Opin. Plant Biol. 2015, 27, 125–132. [Google Scholar] [CrossRef]
- Roy, S.; Griffiths, M.; Torres-Jerez, I.; Sanchez, B.; Antonelli, E.; Jain, D.; Krom, N.; Zhang, S.; York, L.M.; Scheible, W.-R.; et al. Application of Synthetic Peptide CEP1 Increases Nutrient Uptake Rates Along Plant Roots. Front. Plant Sci. 2021, 12, 793145. [Google Scholar] [CrossRef]
- Krouk, G.; Crawford, N.M.; Coruzzi, G.M.; Tsay, Y.-F. Nitrate signaling: Adaptation to fluctuating environments. Curr. Opin. Plant Biol. 2010, 13, 265–272. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.-F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Inge, V.; Tom, B.; Danny, G. Adventitious root induction in Arabidopsis thaliana as a model for in vitro root organogenesis. Methods Mol. Biol. 2013, 959, 159–175. [Google Scholar]
- Kun-Hsiang, L.; Menghong, L.; Ziwei, L.; Zi-Fu, W.; Binqing, C.; Cong, L.; Aping, G.; Mineko, K.; Shuichi, Y.; Gerhard, W.; et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 2022, 377, 1419–1425. [Google Scholar]
- Damion, N.; Gabriel, K.; Daniel, T.; Gloria, M.C. A system biology approach highlights a hormonal enhancer effect on regulation of genes in a nitrate responsive “biomodule”. BMC Syst. Biol. 2009, 3, 59. [Google Scholar]
- Soledad, F.U.; Catalina, I.; Isabel, F.; José, M.; Rodrigo, A.G. Nitrate signaling and early responses in Arabidopsis roots. J. Exp. Bot. 2017, 68, 2541–2551. [Google Scholar]
- Miwa, S.; Mihaela, N.G.; Masaaki, T. A nitrite transporter associated with nitrite uptake by higher plant chloroplasts. Plant Cell Physiol. 2007, 48, 1022–1035. [Google Scholar]
- Zhou, J.-J.; Theodoulou, F.L.; Muldin, I.; Ingemarsson, B.; Miller, A.J. Cloning and Functional Characterization of a Brassica napus Transporter That Is Able to Transport Nitrate and Histidine. J. Biol. Chem. 1998, 273, 12017–12023. [Google Scholar] [CrossRef]
- Kentaro, O.; Mari, O.; Yoshikatsu, M. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. Cell Mol. Biol. 2008, 55, 152–160. [Google Scholar]
- Evan, M.; Stephanie, S.; Ive, D.S. Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 2012, 24, 3198–3217. [Google Scholar]
- Christina, D.; Nijat, I.; Michael, A.D. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J. Exp. Bot. 2013, 64, 5383–5394. [Google Scholar]
- Taleski, M.; Imin, N.; Djordjevic, M.A. CEP peptide hormones: Key players in orchestrating nitrogen-demand signalling, root nodulation, and lateral root development. J. Exp. Bot. 2018, 2606–2617. [Google Scholar] [CrossRef]
- Hussain, S.; Wang, W.; Ahmed, S.; Wang, X.; Adnan Cheng, Y.; Wang, C.; Wang, Y.; Zhang, N.; Tian, H.; Chen, S.; et al. PIP2, An Auxin Induced Plant Peptide Hormone Regulates Root and Hypocotyl Elongation in Arabidopsis. Front Plant Sci. 2021, 12, 646736. [Google Scholar] [CrossRef]
- Ryan, W.; Ana, F.; Ruth, D.G.; Esther, O.; Pierre, H. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18625–18630. [Google Scholar]
- Bergonci, T.; Ribeiro, B.; Ceciliato, P.H.; Guerrero-Abad, J.C.; Silva-Filho, M.C.; Moura, D.S. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 2014, 65, 2219–2230. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Canto, A.M.D.; Ceciliato, P.H.O.; Ribeiro, B.; Morea, F.A.O.; Garcia, A.A.F.; Silva-Filho, M.C.; Moura, D.S. Biological activity of nine recombinant AtRALF peptides: Implications for their perception and function in Arabidopsis. Plant Physiol. Biochem. 2014, 75, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Fugui, Z.; Qinyi, Y.; Hong, C.; Jiangli, D.; Tao, W. Multigene editing reveals that MtCEP1/2/12 redundantly control lateral root and nodule number in Medicago truncatula. J. Exp. Bot. 2021, 72, 3661–3676. [Google Scholar]
- Roberts, I.; Smith, S.; De Rybel, B.; Van Den Broeke, J.; Smet, W.; De Cokere, S.; Mispelaere, M.; De Smet, I.; Beeckman, T. The CEP family in land plants: Evolutionary analyses, expression studies, and role in Arabidopsis shoot development. J. Exp. Bot. 2013, 64, 5371–5381. [Google Scholar] [CrossRef]
- Ruibin, X.; Yufeng, L.; Zhipeng, S.; Tianyu, L.; Wanjun, S.; Ming, Z.; Yirong, Z.; Jiewen, X. A C-Terminal Encoded Peptide, ZmCEP1, is essential for kernel development in maize (Zea mays L.). J. Exp. Bot. 2021, 72, 5390–5406. [Google Scholar]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef]
- Xiaoqian, C.; Mingzhe, L.; Shujuan, Z.; Min, F.; Chao, H.; Fengning, X.; Genying, L.; Yong, W.; Cheng-Bin, X.; Jia-Gang, W.; et al. HBI1-TCP20 interaction positively regulates the CEPs-mediated systemic nitrate acquisition. J. Integr. Plant Biol. 2020, 63, 902–912. [Google Scholar]
- Kawano, K. Thirty Years of Cassava Breeding for Productivity—Biological and Social Factors for Success. Crop. Sci. 2003, 43, 1325–1335. [Google Scholar] [CrossRef]
- Kongsil, P.; Ceballos, H.; Siriwan, W.; Vuttipongchaikij, S.; Kittipadakul, P.; Phumichai, C.; Wannarat, W.; Kositratana, W.; Vichukit, V.; Sarobol, E.; et al. Cassava Breeding and Cultivation Challenges in Thailand: Past, Present, and Future Perspectives. Plants 2024, 13, 1899. [Google Scholar] [CrossRef]
- Shan, Z.; Luo, X.; Wei, M.; Huang, T.; Khan, A.; Zhu, Y. Physiological and proteomic analysis on long-term drought resistance of cassava (Manihot esculenta Crantz). Sci. Rep. 2018, 8, 17982. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Mutka, A.M.; Hummel, A.W.; Berry, J.; Chauhan, R.D.; Vijayaraghavan, A.; Taylor, N.J.; Voytas, D.F.; Chitwood, D.H.; Bart, R.S. Gene expression atlas for the food security crop cassava. New Phytol. 2017, 213, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, R.; Ohkubo, Y.; Izumi, M.; Ota, R.; Yamada, K.; Hayashi, Y.; Yamashita, Y.; Noda, S.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Integration of shoot-derived polypeptide signals by root TGA transcription factors is essential for survival under fluctuating nitrogen environments. Nat. Commun. 2024, 15, 6903. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Ingo, P.; Johanna, P. Efficient production of transgenic cassava using negative and positive selection. Transgenic Res. 2000, 9, 405–415. [Google Scholar]
- Zhipeng, S.; Tianya, W.; Hongjian, L.; Ming, Z.; Yangyang, L.; Ruibin, X.; Guofang, X.; Zhongfu, N.; Mingming, X. Overexpression of Peptide-Encoding OsCEP6.1 Results in Pleiotropic Effects on Growth in Rice (O. sativa). Front. Plant Sci. 2016, 7, 228. [Google Scholar]
- Li, R.; Li, H.; An, J.; You, C.; Wang, X.; Hao, Y. Apple Peptide Hormone and Its Coding Gene MdCEP1 Regulate Root Development in Arabidopsis thaliana. Acta Hortic. Sin. 2017, 44, 1225–1234. [Google Scholar] [CrossRef]
- Lei, Z.; Yue, R.; Qian, X.; Wan, Y.; Zhang, S.; Yang, G.; Huang, J.; Yan, K.; Zheng, C.; Wu, C. SiCEP3, a C-terminally encoded peptide from Setaria italica, promotes ABA import and signaling. J. Exp. Bot. 2021, 72, 6260–6273. [Google Scholar]
- Michael, T.; Kelly, C.; Ondřej, N.; Thomas, S.; Manuel, F.; Michael, A.D. CEP peptide and cytokinin pathways converge on CEPD glutaredoxins to inhibit root growth. Nat. Commun. 2023, 14, 1683. [Google Scholar]
- Fredes, I.; Moreno, S.; Díaz, F.P.; Gutiérrez, R.A. Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol. 2019, 47, 112–118. [Google Scholar] [CrossRef]
- Abualia, R.; Riegler, S.; Benkova, E. Nitrate, Auxin and Cytokinin—A Trio to Tango. Cells 2023, 12, 14. [Google Scholar] [CrossRef]
- Vega, A.; O’Brien, J.A.; Gutiérrez, R.A. Nitrate and hormonal signaling crosstalk for plant growth and development. Curr. Opin. Plant Biol. 2019, 52, 155–163. [Google Scholar] [CrossRef]
- Zhang, Y.; Duan, X.; Wang, Z.; Lv, Y.; Qi, W.; Li, L.; Le Luo Xuan, W. CEPs suppress auxin signaling but promote cytokinin signaling to inhibit root growth in Arabidopsis. Biochem. Biophys. Res. Commun. 2024, 711, 149934. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Chen, H.; Jiang, J.F.; Zhao, Y.; Xu, M.L.; Xu, Y.Y.; Tan, K.H.; Xu, Z.H.; Chong, K. Overexpression of OsRAA1 Causes Pleiotropic Phenotypes in Transgenic Rice Plants, including Altered Leaf, Flower, and Root Development and Root Response to Gravity. Plant Physiol. 2004, 135, 1502–1513. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Z.; Guo, X.; Duan, C.; Ma, W.J.; Zhang, Y.G.; Xu, P.; Gao, Z.Q.; Wang, Z.F.; Yan, H.; Zhang, Y.F.; et al. Comparative analysis of microarray and qRT-PCR data for validating gene expression profiles. Plant Methods 2015, 11, 45. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, F.; Wang, X.; Liu, B.; Lin, H.; Ai, L.; Mai, W.; Liu, X.; Zhang, H.; Zhao, J.; Khan, L.; et al. A C-Terminally Encoded Peptide, MeCEP6, Promotes Nitrate Uptake in Cassava Roots. Plants 2025, 14, 1264. https://doi.org/10.3390/plants14081264
Lu F, Wang X, Liu B, Lin H, Ai L, Mai W, Liu X, Zhang H, Zhao J, Khan L, et al. A C-Terminally Encoded Peptide, MeCEP6, Promotes Nitrate Uptake in Cassava Roots. Plants. 2025; 14(8):1264. https://doi.org/10.3390/plants14081264
Chicago/Turabian StyleLu, Fabao, Xiuning Wang, Bo Liu, Hongxin Lin, Li Ai, Weitao Mai, Xiaochen Liu, Huaifang Zhang, Jinling Zhao, Luqman Khan, and et al. 2025. "A C-Terminally Encoded Peptide, MeCEP6, Promotes Nitrate Uptake in Cassava Roots" Plants 14, no. 8: 1264. https://doi.org/10.3390/plants14081264
APA StyleLu, F., Wang, X., Liu, B., Lin, H., Ai, L., Mai, W., Liu, X., Zhang, H., Zhao, J., Khan, L., Wang, W., Zeng, C., & Chen, X. (2025). A C-Terminally Encoded Peptide, MeCEP6, Promotes Nitrate Uptake in Cassava Roots. Plants, 14(8), 1264. https://doi.org/10.3390/plants14081264






