Selection of Optimal Diagnostic Positions for Early Nutrient Deficiency in Cucumber Leaves Based on Spatial Distribution of Raman Spectra
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions and Experimental Design
2.2. Measurement of Leaf NPK Content
2.3. Raman Spectroscopy Data Collection
2.4. Spectral Similarity Calculation Method
2.5. Diagnostic Model Development and Evaluation Methods
3. Results
3.1. Identification of Nutrient Deficiency in Cucumber
3.2. Evaluation and Analysis of Spectral Similarity at the Same Position
3.3. Analysis of Spectral Similarity Outliers at Different Positions
3.4. Analysis of Spatial Distribution Characteristics of Spectral Similarity on Cucumber Leaves
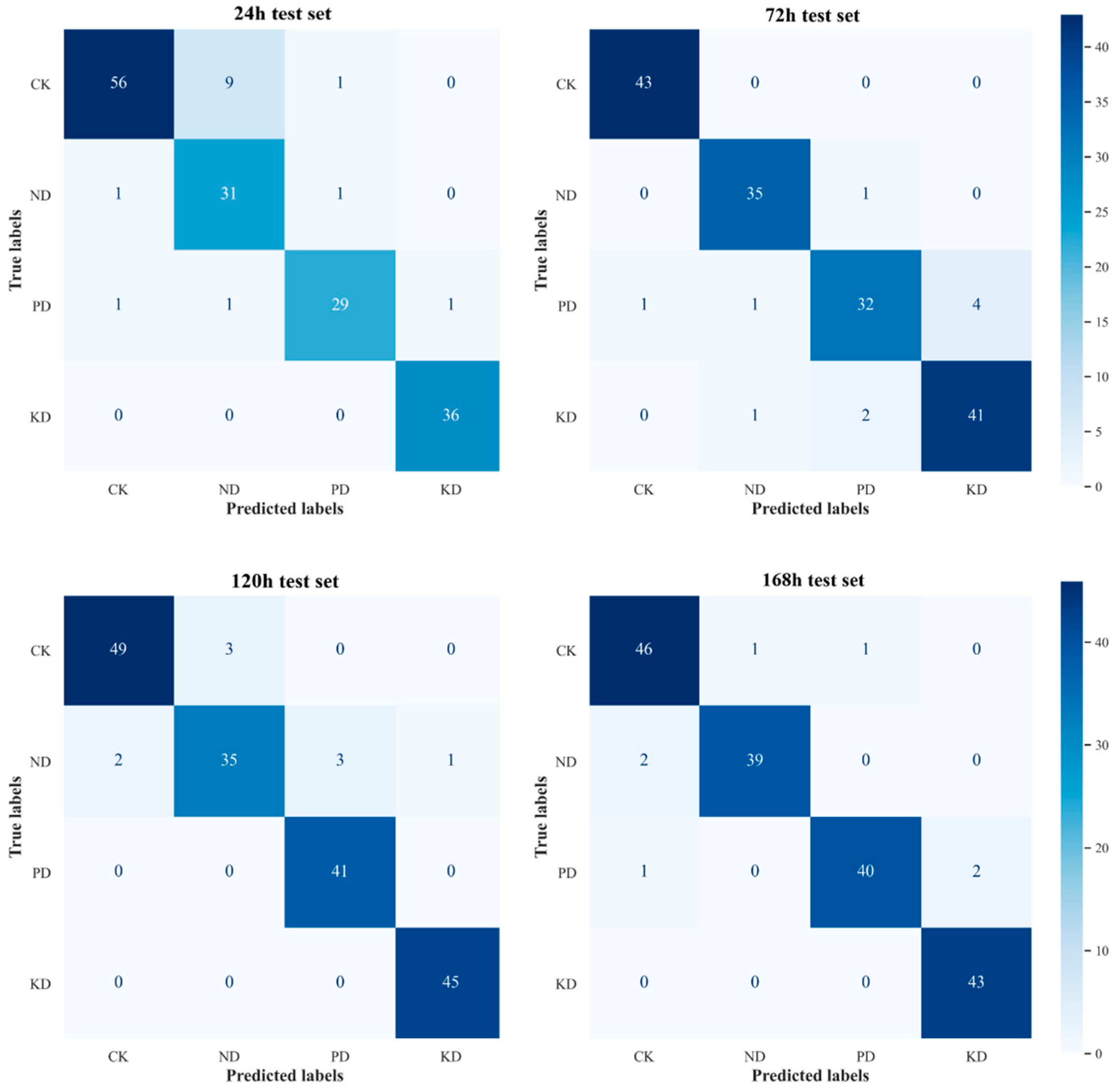
3.5. Establishment of an Early Diagnostic Model for Nutrient Deficiency in Cucumber
3.6. Selection and Validation Analysis of Early Diagnostic Positions for Nutrient Deficiencies in Cucumber
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gebbers, R.; Adamchuk, V.I.J. Precision agriculture and food security. Science 2010, 327, 828–831. [Google Scholar] [PubMed]
- Lemaire, G.; Tang, L.; Bélanger, G.; Zhu, Y.; Jeuffroy, M.-H. Forward new paradigms for crop mineral nutrition and fertilization towards sustainable agriculture. Eur. J. Agron. 2021, 125, 126248. [Google Scholar]
- Kant, S.; Bi, Y.-M.; Rothstein, S.J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1499–1509. [Google Scholar] [PubMed]
- Ohyama, T. Nitrogen as a major essential element of plants. Nitrogen Assim. Plants 2010, 37, 1–17. [Google Scholar]
- Ellsworth, D.S.; Crous, K.Y.; Lambers, H.; Cooke, J. Phosphorus recycling in photorespiration maintains high photosynthetic capacity in woody species. Plant Cell Environ. 2014, 38, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Sardans, J.; Peñuelas, J. Potassium Control of Plant Functions: Ecological and Agricultural Implications. Plants 2021, 10, 419. [Google Scholar] [CrossRef]
- Sáez-Plaza, P.; Navas, M.J.; Wybraniec, S.; Michałowski, T.; Asuero, A.G.J. An overview of the Kjeldahl method of nitrogen determination. Part II. Sample preparation, working scale, instrumental finish, and quality control. Crit. Rev. Anal. Chem. 2013, 43, 224–272. [Google Scholar]
- Peverill, K.I. Soil Analysis: An Interpretation Manual; CSIRO Publishing: Clayton, Australia, 1999. [Google Scholar]
- Altangerel, N.; Ariunbold, G.O.; Gorman, C.; Alkahtani, M.H.; Borrego, E.J.; Bohlmeyer, D.; Hemmer, P.; Kolomiets, M.V.; Yuan, J.S.; Scully, M.O. In vivo diagnostics of early abiotic plant stress response via Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2017, 114, 3393–3396. [Google Scholar]
- Heraud, P.; Wood, B.R.; Beardall, J.; McNaughton, D. Effects of pre-processing of Raman spectra on in vivo classification of nutrient status of microalgal cells. J. Chemom. A J. Chemom. Soc. 2006, 20, 193–197. [Google Scholar]
- Sanchez, L.; Pant, S.; Xing, Z.; Mandadi, K.; Kurouski, D. Rapid and noninvasive diagnostics of Huanglongbing and nutrient deficits on citrus trees with a handheld Raman spectrometer. Anal. Bioanal. Chem. 2019, 411, 3125–3133. [Google Scholar] [CrossRef]
- Higgins, S.; Biswas, S.; Goff, N.K.; Septiningsih, E.M.; Kurouski, D.J. Raman spectroscopy enables non-invasive and confirmatory diagnostics of aluminum and iron toxicities in rice. Front. Plant Sci. 2022, 13, 754735. [Google Scholar]
- Butler, H.J.; McAinsh, M.R.; Adams, S.; Martin, F.L. Application of vibrational spectroscopy techniques to non-destructively monitor plant health and development. Anal. Methods 2015, 7, 4059–4070. [Google Scholar]
- de Bang, T.C.; Husted, S.; Laursen, K.H.; Persson, D.P.; Schjoerring, J.K.J. The molecular–physiological functions of mineral macronutrients and their consequences for deficiency symptoms in plants. New Phytol. 2021, 229, 2446–2469. [Google Scholar]
- de Mello Prado, R.; Rozane, D.E. Leaf analysis as diagnostic tool for balanced fertilization in tropical fruits. In Fruit Crops; Srivastava, A.K., Hu, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 131–143. [Google Scholar]
- Huang, C.H.; Singh, G.P.; Park, S.H.; Chua, N.-H.; Ram, R.J.; Park, B.S. Early Diagnosis and Management of Nitrogen Deficiency in Plants Utilizing Raman Spectroscopy. Front. Plant Sci. 2020, 11, 663. [Google Scholar] [CrossRef]
- Sanchez, L.; Ermolenkov, A.; Biswas, S.; Septiningsih, E.M.; Kurouski, D. Raman Spectroscopy Enables Non-invasive and Confirmatory Diagnostics of Salinity Stresses, Nitrogen, Phosphorus, and Potassium Deficiencies in Rice. Front. Plant Sci. 2020, 11, 573321. [Google Scholar]
- Zhao, X.; Cai, L. Early detection of zinc deficit with confocal Raman spectroscopy. J. Raman Spectrosc. 2018, 49, 1706–1712. [Google Scholar]
- Zhang, X.; Liu, F.; He, Y.; Gong, X. Detecting macronutrients content and distribution in oilseed rape leaves based on hyperspectral imaging. Biosyst. Eng. 2013, 115, 56–65. [Google Scholar]
- Hu, J.; Li, C.; Wen, Y.; Gao, X.; Shi, F.; Han, L. Spatial distribution of SPAD value and determination of the suitable leaf for N diagnosis in cucumber. IOP Conf. Ser. Earth Environ. Sci. 2018, 108, 022001. [Google Scholar] [CrossRef]
- Yuan, Z.; Cao, Q.; Zhang, K.; Ata-Ul-Karim, S.T.; Tian, Y.; Zhu, Y.; Cao, W.; Liu, X.J. Optimal leaf positions for SPAD meter measurement in rice. Front. Plant Sci. 2016, 7, 719. [Google Scholar]
- Mostafapour, S.; Dörfer, T.; Heinke, R.; Rösch, P.; Popp, J.; Bocklitz, T.J. Investigating the effect of different pre-treatment methods on Raman spectra recorded with different excitation wavelengths. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123100. [Google Scholar]
- Zhao, X.; Xu, M.; Zhang, W.; Liu, G.; Tong, L.J. Identification of zinc pollution in rice plants based on two characteristic variables. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 261, 120043. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Tang, Y.; Qian, Z.; Wang, J.; Guo, H.J. Deeply-recursive convolutional neural network for Raman spectra identification. RSC Adv. 2022, 12, 5053–5061. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Min, C.; Kim, H.; Cho, S.; Oh, J.; Ha, S.-H.; Yi, J.-h. Structural Health Monitoring with Sensor Data and Cosine Similarity for Multi-Damages. Sensors 2019, 19, 3047. [Google Scholar] [CrossRef]
- Kwak, B.I.; Han, M.L.; Kim, H.K. Cosine similarity based anomaly detection methodology for the CAN bus. Expert Syst. Appl. 2021, 166, 114066. [Google Scholar] [CrossRef]
- Irimatsugawa, T.; Shimizu, Y.; Okubo, S.; Inaba, H. Cosine similarity for quantitatively evaluating the degree of change in an optical frequency comb spectra. Opt. Express 2021, 29, 35613–35622. [Google Scholar] [CrossRef]
- Smith, I.; Ortmann, J.; Abbas-Aghababazadeh, F.; Smirnov, P.; Haibe-Kains, B. On the distribution of cosine similarity with application to biology. arXiv 2023, arXiv:2310.13994. [Google Scholar]
- Dong, W.; Sheng, K.; Huang, B.; Xiong, K.; Liu, K.; Cheng, X. Stretchable Self-Powered TENG Sensor Array for Human-Robot Interaction Based on Conductive Ionic Gels and LSTM Neural Network. IEEE Sens. J. 2024, 24, 37962–37969. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1938, 347, 39. [Google Scholar]
- Antoszewski, G.; Guenther, J.F.; Roberts, J.K.; Adler, M.; Dalle Molle, M.; Kaczmar, N.S.; Miller, W.B.; Mattson, N.S.; Grab, H. Non-Invasive Detection of Nitrogen Deficiency in Cannabis sativa Using Hand-Held Raman Spectroscopy. Agronomy 2024, 14, 2390. [Google Scholar] [CrossRef]
- Yu, M.; Bai, X.; Bao, J.; Wang, Z.; Tang, Z.; Zheng, Q.; Zhi, J. The Prediction Model of Total Nitrogen Content in Leaves of Korla Fragrant Pear Was Established Based on Near Infrared Spectroscopy. Agronomy 2024, 14, 1284. [Google Scholar] [CrossRef]
- Peng, X.; Chen, D.; Zhou, Z.; Zhang, Z.; Xu, C.; Zha, Q.; Wang, F.; Hu, X. Prediction of the Nitrogen, Phosphorus and Potassium Contents in Grape Leaves at Different Growth Stages Based on UAV Multispectral Remote Sensing. Remote Sens. 2022, 14, 2659. [Google Scholar] [CrossRef]
- Kelly, C.; Price, T.D. Comparative methods based on species mean values. Math. Biosci. 2004, 187, 135–154. [Google Scholar] [CrossRef]
- Lieber, C.A.; Mahadevan-Jansen, A. Automated Method for Subtraction of Fluorescence from Biological Raman Spectra. Appl. Spectrosc. 2016, 57, 1363–1367. [Google Scholar] [CrossRef]
- Wong, K.H.; Razmovski-Naumovski, V.; Li, K.M.; Li, G.Q.; Chan, K.J. Differentiation of Pueraria lobata and Pueraria thomsonii using partial least square discriminant analysis (PLS-DA). J. Pharm. Biomed. Anal. 2013, 84, 5–13. [Google Scholar] [PubMed]
- Huang, X.; Li, H.; Ruan, Y.; Li, Z.; Yang, H.; Xie, G.; Yang, Y.; Du, Q.; Ji, K.; Yang, M.J. An integrated approach utilizing raman spectroscopy and chemometrics for authentication and detection of adulteration of agarwood essential oils. Front. Chem. 2022, 10, 1036082. [Google Scholar]
- Guo, P.; Li, T.; Gao, H.; Chen, X.; Cui, Y.; Huang, Y. Evaluating calibration and spectral variable selection methods for predicting three soil nutrients using Vis-NIR spectroscopy. Remote Sens. 2021, 13, 4000. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, X.; Liu, J.; Li, H.; Jiang, J.; Li, Y.; Diao, S. Prediction of cyanidin 3-rutinoside content in Michelia crassipes based on near-infrared spectroscopic techniques. Front. Plant Sci. 2024, 15, 1346192. [Google Scholar]
- Smith, A.P.; Fontenot, E.B.; Zahraeifard, S.; DiTusa, S.F. Molecular components that drive phosphorus-remobilisation during leaf senescence. Annu. Plant Rev. Phosphorus Metab. Plants 2015, 48, 159–186. [Google Scholar]
- Ingestad, T.; Ågren, G.I. Plant nutrition and growth: Basic principles. Plant Soil 1995, 168, 15–20. [Google Scholar]
- Payne, W.Z.; Kurouski, D.J. Raman spectroscopy enables phenotyping and assessment of nutrition values of plants: A review. Plant Methods 2021, 17, 78. [Google Scholar]
- Shi, J.-Y.; Zou, X.-B.; Zhao, J.-W.; Wang, K.-L.; Chen, Z.-W.; Huang, X.-W.; Zhang, D.-T.; Holmes, M. Nondestructive diagnostics of nitrogen deficiency by cucumber leaf chlorophyll distribution map based on near infrared hyperspectral imaging. Sci. Hortic. 2012, 138, 190–197. [Google Scholar]
- Synytsya, A.; Čopíková, J.; Matějka, P.; Machovič, V.J. Fourier transform Raman and infrared spectroscopy of pectins. Carbohydr. Polym. 2003, 54, 97–106. [Google Scholar]
- Edwards, H.; Farwell, D.; Webster, D.J. FT Raman microscopy of untreated natural plant fibres. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 1997, 53, 2383–2392. [Google Scholar]
- Schulz, H.; Baranska, M.; Baranski, R.J. Potential of NIR-FT-Raman spectroscopy in natural carotenoid analysis. Biopolym. Orig. Res. Biomol. 2005, 77, 212–221. [Google Scholar]
- Agarwal, U.P.J. Raman imaging to investigate ultrastructure and composition of plant cell walls: Distribution of lignin and cellulose in black spruce wood (Picea mariana). Planta 2006, 224, 1141–1153. [Google Scholar]
- Lei, K.; Kai, W.; Xiaodong, L.; Bo, Z. High Pressure Structural Investigation of Benzoic Acid: Raman Spectroscopy and X-ray Diffraction. J. Phys. Chem. C 2016, 120, 14758–14766. [Google Scholar]
- Devitt, G.; Howard, K.; Mudher, A.; Mahajan, S.J. Raman spectroscopy: An emerging tool in neurodegenerative disease research and diagnosis. ACS Chem. Neurosci. 2018, 9, 404–420. [Google Scholar]
- Yu, M.M.; Schulze, H.G.; Jetter, R.; Blades, M.W.; Turner, R.F.J. Raman microspectroscopic analysis of triterpenoids found in plant cuticles. Appl. Spectrosc. 2007, 61, 32–37. [Google Scholar]




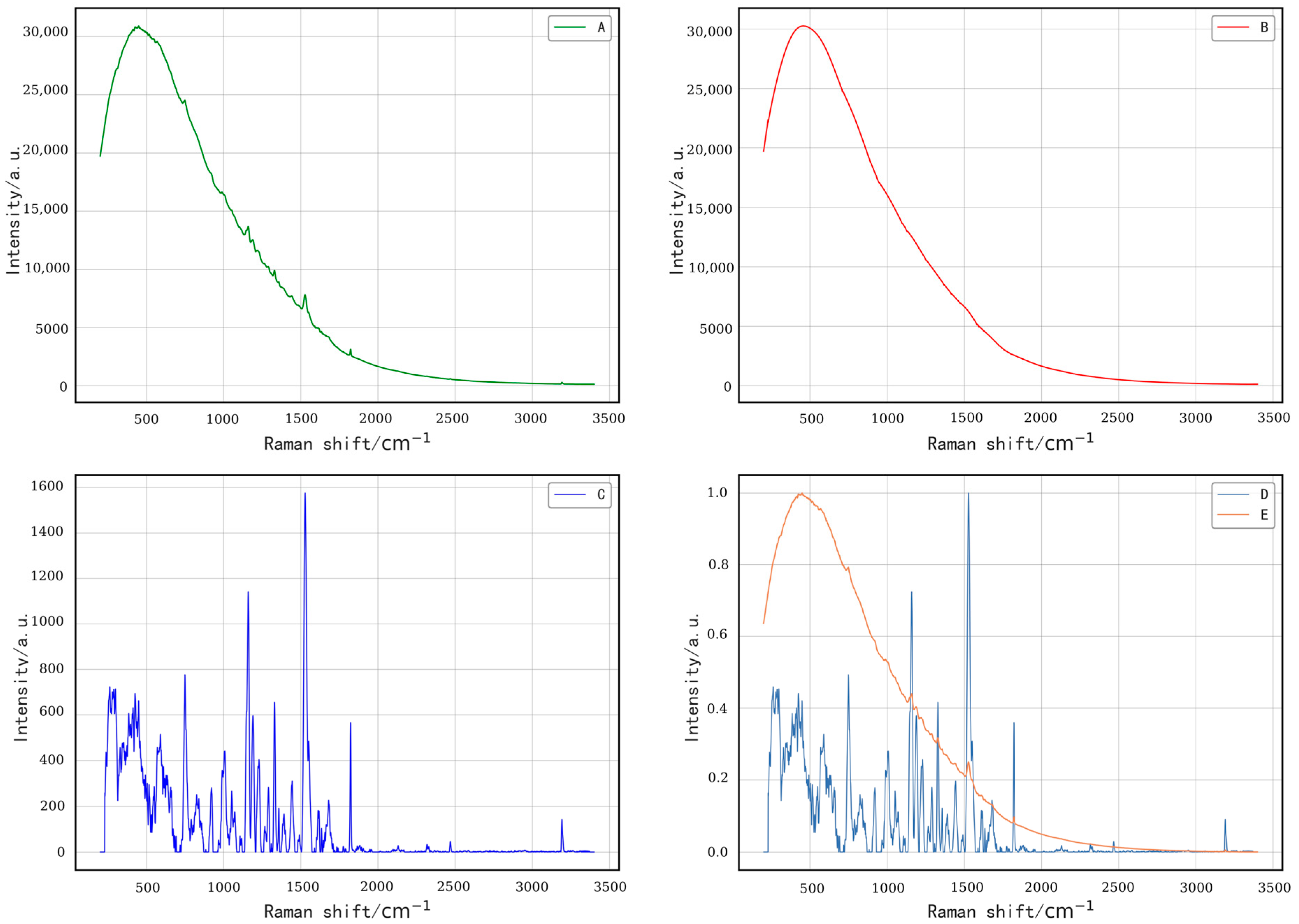

| Positions | 10 Times | 20 Times | 30 Times |
|---|---|---|---|
| P1 | 0.99211 | 0.99139 | 0.99135 |
| P2 | 0.99191 | 0.99153 | 0.99134 |
| P3 | 0.99273 | 0.99215 | 0.99203 |
| Mean value | 0.99225 | 0.99169 | 0.99157 |
| Duration of Stress | Treatment Groups | Total | |||
|---|---|---|---|---|---|
| CK | ND | PD | KD | ||
| 24 h | 37 | 9 | 4 | 1 | 51 |
| 72 h | 2 | 3 | 0 | 0 | 5 |
| 120 h | 1 | 2 | 1 | 1 | 5 |
| 168 h | 1 | 0 | 0 | 1 | 2 |
| Total | 41 | 14 | 5 | 3 | 63 |
| Duration of Stress | Treatment Groups | Total | |||
|---|---|---|---|---|---|
| CK | ND | PD | KD | ||
| 24 h | 107 | 35 | 23 | 6 | 171 |
| 72 h | 22 | 28 | 19 | 7 | 76 |
| 120 h | 21 | 23 | 11 | 6 | 61 |
| 168 h | 15 | 11 | 4 | 4 | 34 |
| Total | 165 | 97 | 57 | 23 | 342 |
| Group Names | Sample Size | LVs | MDs | Cross-Validation | Test Set Evaluation | ||||
|---|---|---|---|---|---|---|---|---|---|
| Macro-P (%) | Macro-R (%) | Macro-F1 (%) | Macro-P (%) | Macro-R (%) | Macro-F1 (%) | ||||
| 24 h | 335 | 11 | 15 | 90.99 | 89.31 | 89.56 | 90.75 | 92.35 | 91.20 |
| 24 h-cleaned | 254 | 7 | 8 | 92.32 | 91.52 | 90.57 | 92.70 | 94.76 | 93.30 |
| 72 h | 323 | 7 | 10 | 91.71 | 90.55 | 90.70 | 93.72 | 93.65 | 93.64 |
| 72 h-cleaned | 243 | 7 | 5 | 92.68 | 91.92 | 91.67 | 95.78 | 95.87 | 95.80 |
| 120 h | 358 | 8 | 9 | 91.21 | 90.92 | 90.91 | 94.80 | 94.90 | 94.78 |
| 120 h-cleaned | 270 | 6 | 5 | 91.80 | 90.97 | 91.00 | 96.34 | 96.30 | 96.31 |
| 168 h | 351 | 6 | 7 | 92.21 | 92.55 | 91.45 | 96.12 | 95.99 | 96.03 |
| 168 h-cleaned | 265 | 5 | 3 | 93.64 | 94.24 | 92.95 | 97.84 | 97.93 | 97.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Z.; Wang, Y.; Tan, F.; Gao, J.; Jiao, F.; Su, C.; Zheng, X. Selection of Optimal Diagnostic Positions for Early Nutrient Deficiency in Cucumber Leaves Based on Spatial Distribution of Raman Spectra. Plants 2025, 14, 1199. https://doi.org/10.3390/plants14081199
Hou Z, Wang Y, Tan F, Gao J, Jiao F, Su C, Zheng X. Selection of Optimal Diagnostic Positions for Early Nutrient Deficiency in Cucumber Leaves Based on Spatial Distribution of Raman Spectra. Plants. 2025; 14(8):1199. https://doi.org/10.3390/plants14081199
Chicago/Turabian StyleHou, Zhaolong, Yaxuan Wang, Feng Tan, Jiaxin Gao, Feng Jiao, Chunjie Su, and Xin Zheng. 2025. "Selection of Optimal Diagnostic Positions for Early Nutrient Deficiency in Cucumber Leaves Based on Spatial Distribution of Raman Spectra" Plants 14, no. 8: 1199. https://doi.org/10.3390/plants14081199
APA StyleHou, Z., Wang, Y., Tan, F., Gao, J., Jiao, F., Su, C., & Zheng, X. (2025). Selection of Optimal Diagnostic Positions for Early Nutrient Deficiency in Cucumber Leaves Based on Spatial Distribution of Raman Spectra. Plants, 14(8), 1199. https://doi.org/10.3390/plants14081199







