Identification of the Cinnamyl Alcohol Dehydrogenase Gene Family in Brassica U-Triangle Species and Its Potential Roles in Response to Abiotic Stress and Regulation of Seed Coat Color in Brassica napus L.
Abstract
1. Introduction
2. Results
2.1. Identification of CAD Family Genes in the Brassica U-Triangle Species
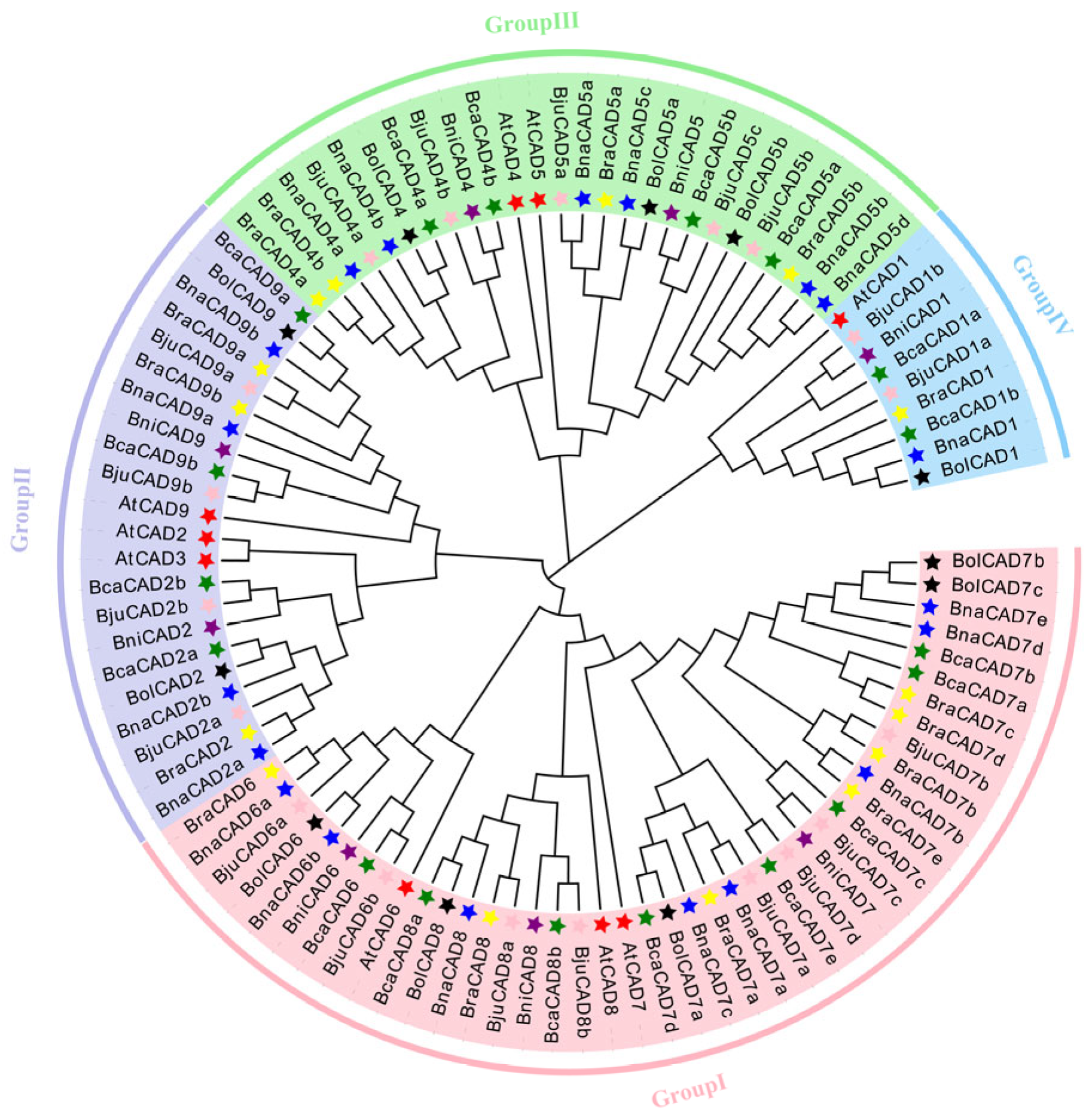
2.2. Phylogenetic Analysis of CAD Proteins in A. thaliana and the Brassica U-Triangle Species
2.3. Multiple Sequence Alignment of CAD Proteins in A. thaliana and the Brassica U-Triangle Species
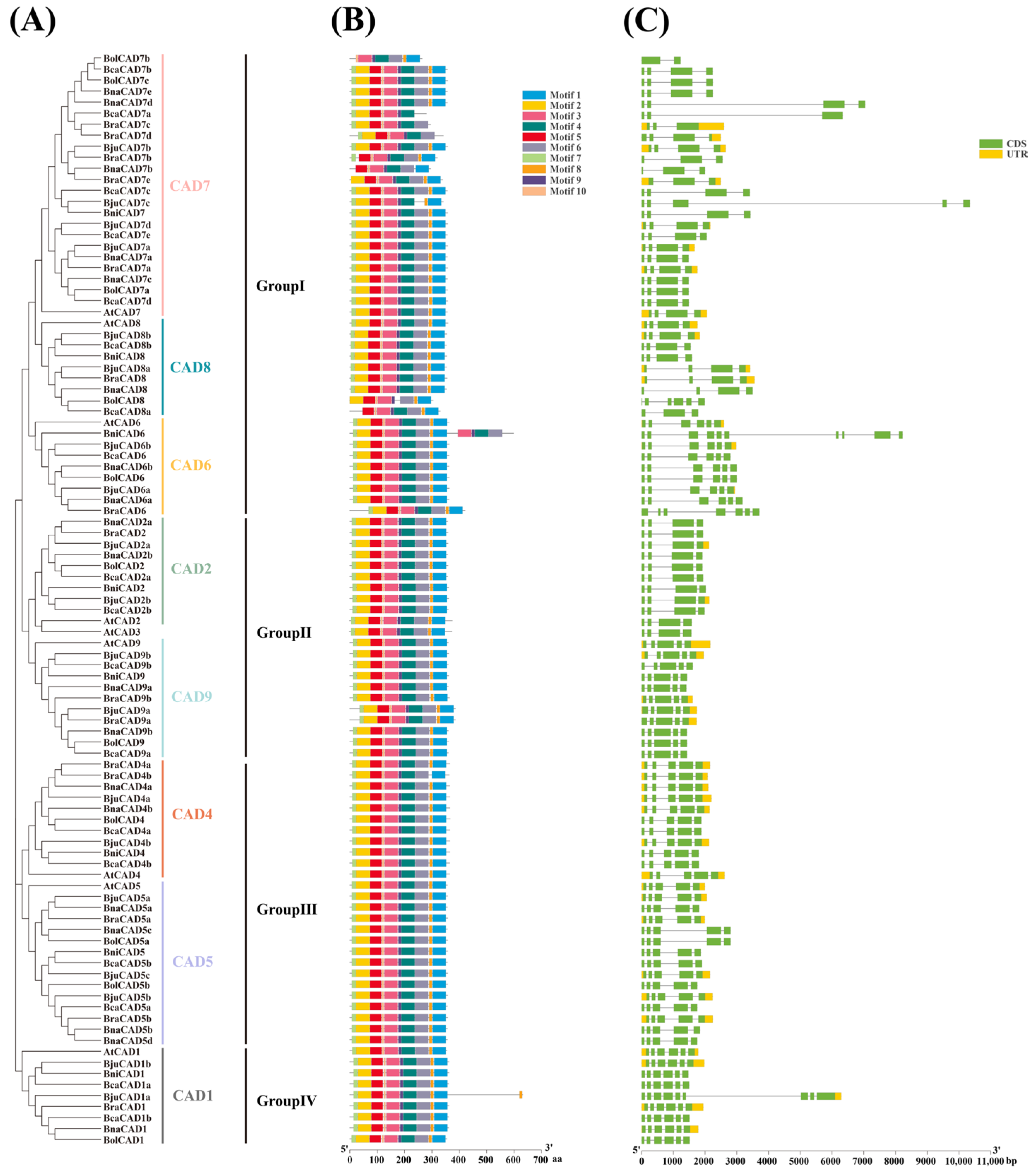
2.4. Conserved Motifs of CAD Proteins and Variations in Gene Structure in A. thaliana and the Brassica U-Triangle Species
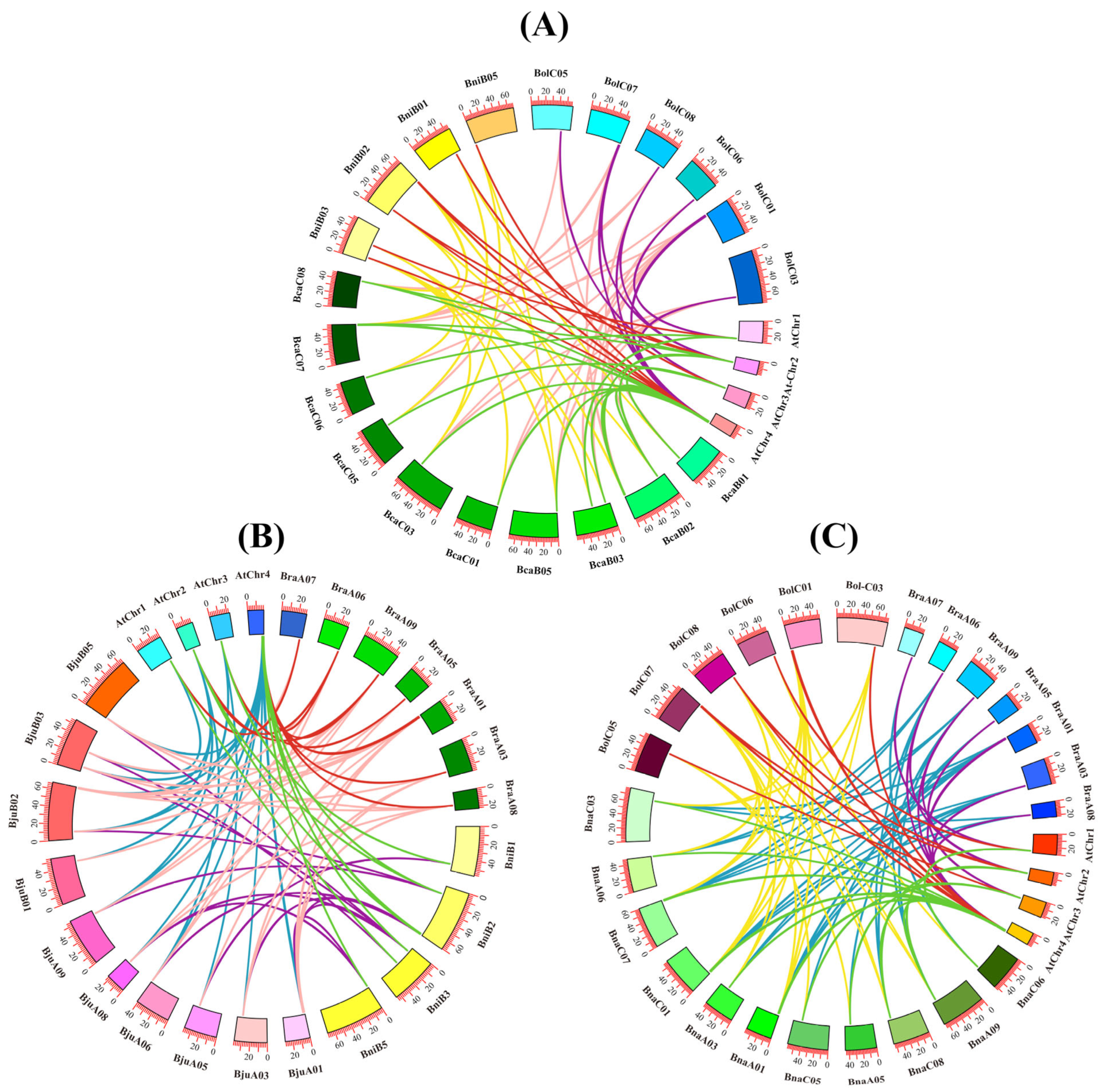
2.5. Chromosomal Distribution of CAD Genes in the Brassica U-Triangle Species
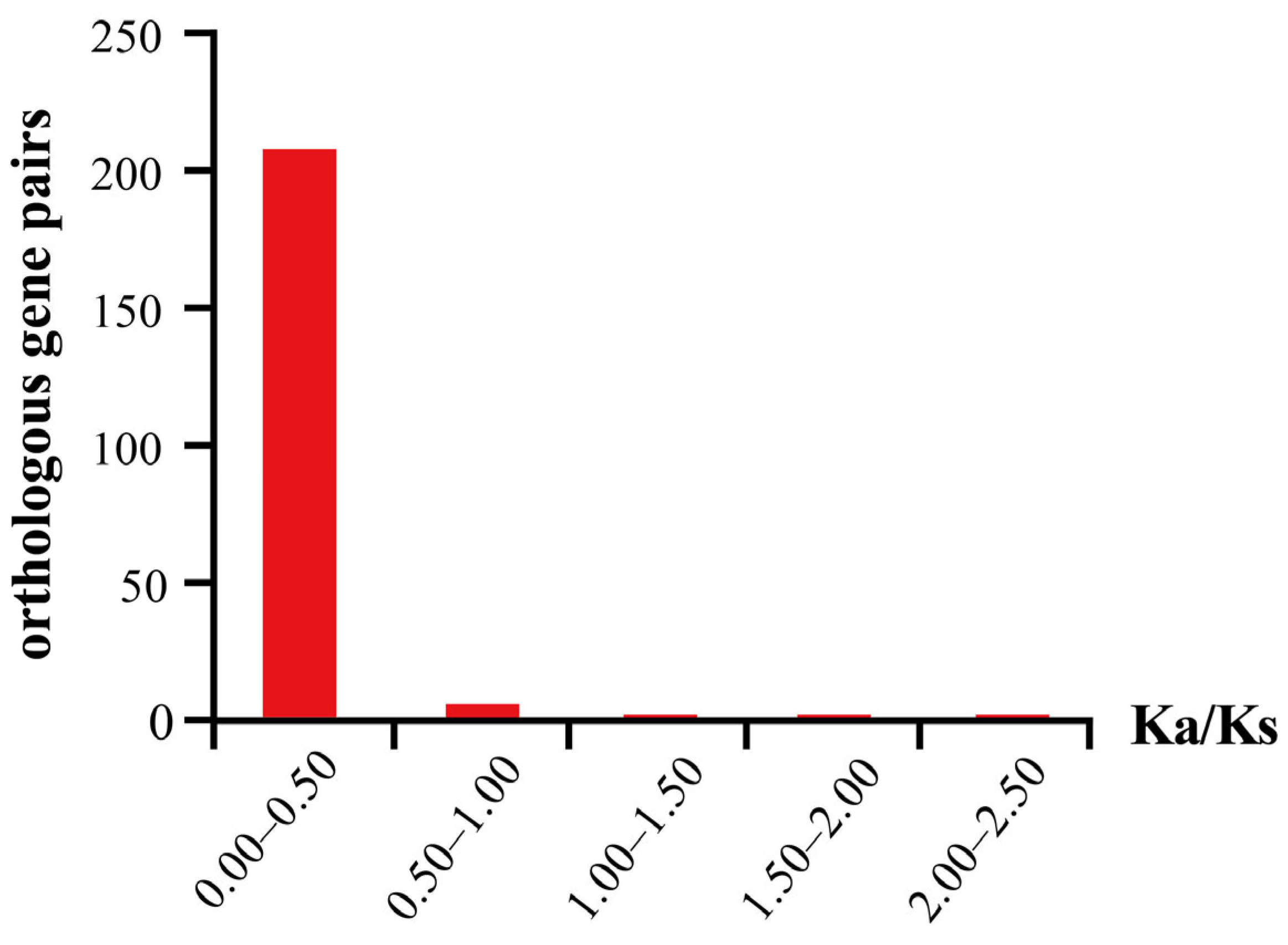
2.6. Collinearity Analysis of CAD Genes in A. thaliana and the Brassica U-Triangle Species
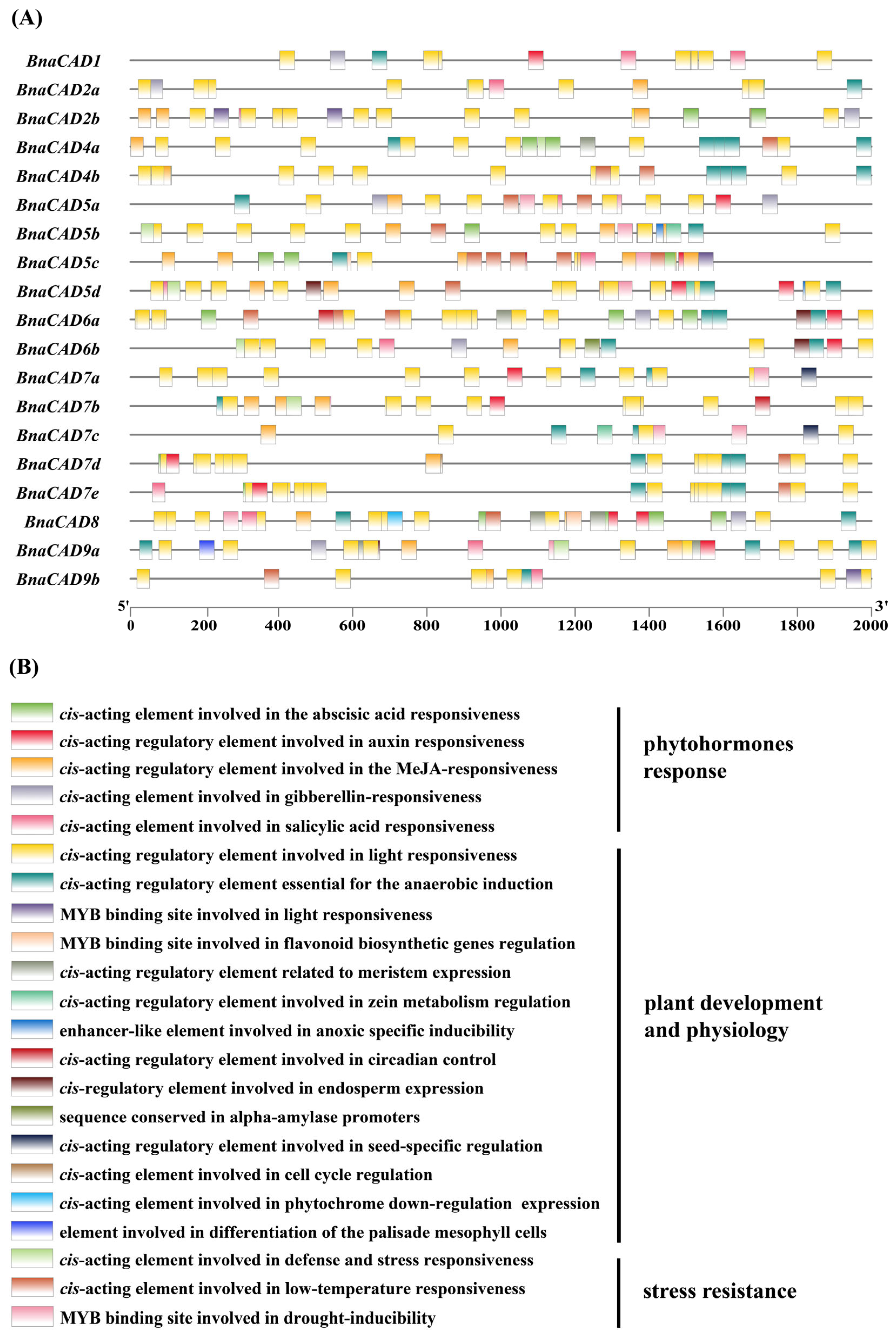
2.7. Cis-Acting Element Analysis of BnaCAD Promoters
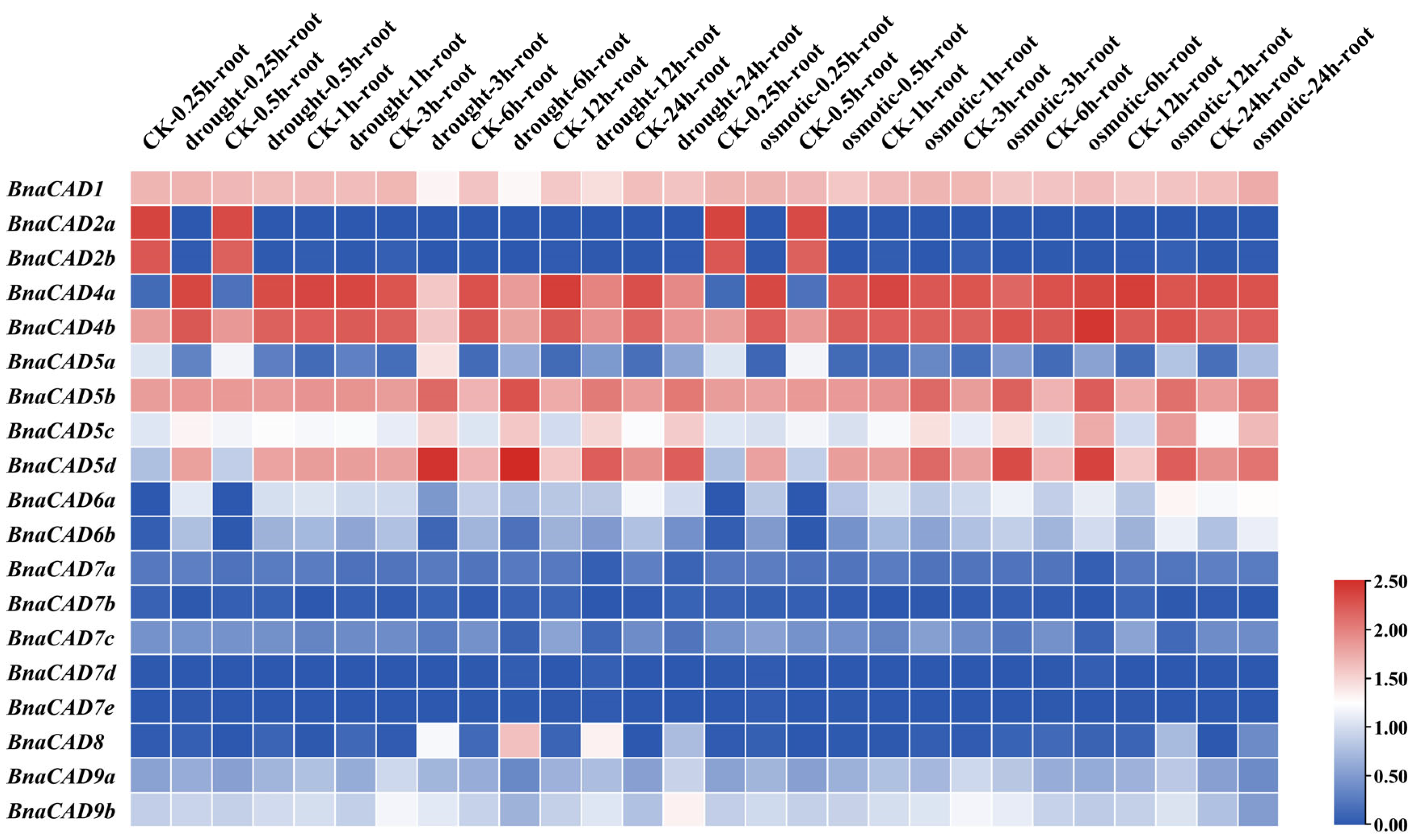
2.8. Expression Profiles of BnaCAD Genes in B. napus Under Drought, Osmotic, and ABA Treatments
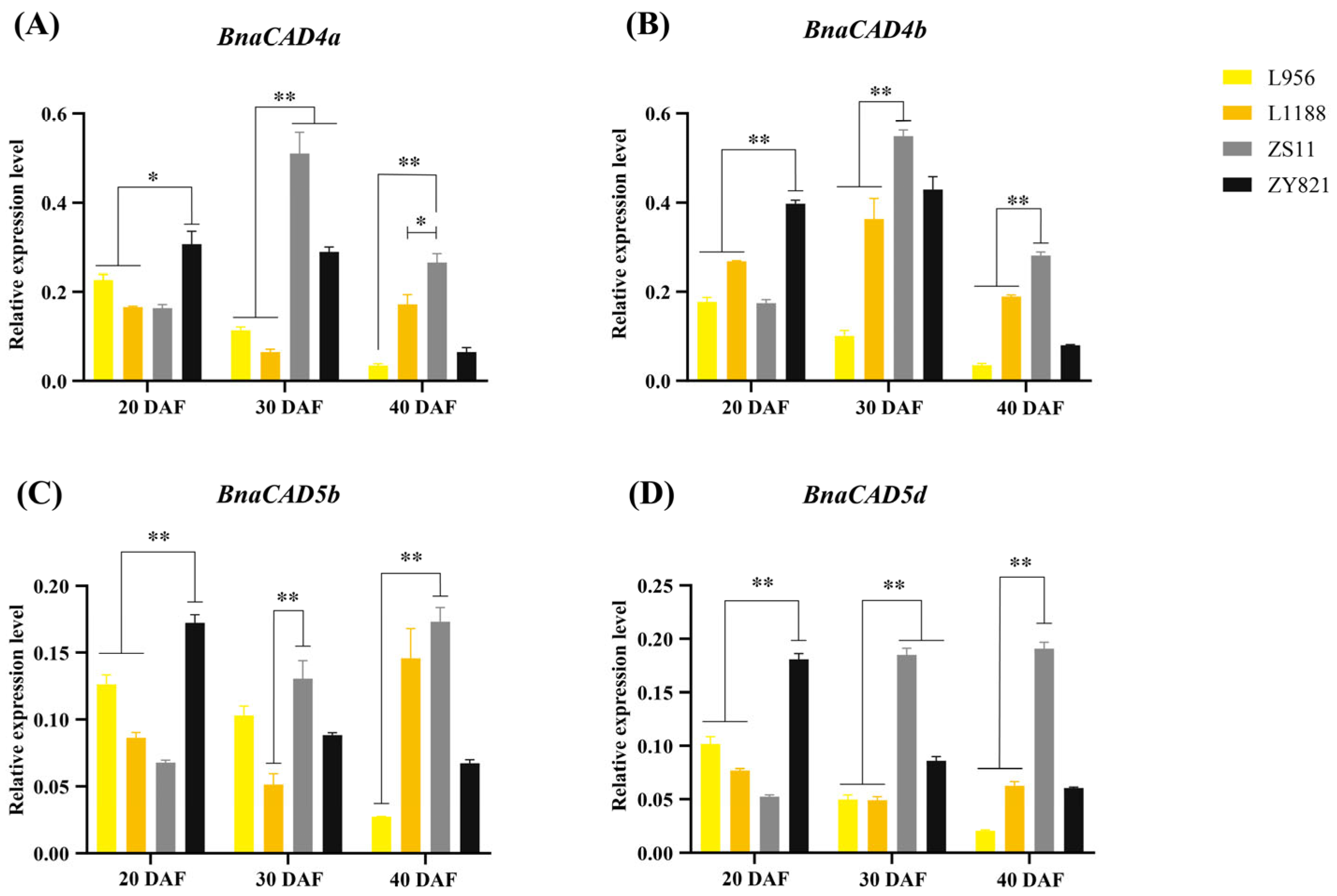
2.9. Expression Pattern of Four BnaCAD Genes in Yellow- and Black-Seeded B. napus
3. Discussion
3.1. Expansion, Loss, and Functional Diversification of the CAD Gene Family in the Brassica U-Triangle Species
3.2. Transcriptional Regulation of BnaCAD Genes in Stress Responses and Seed Coat Color Formation
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification and Annotation of CAD Family Genes
4.3. Comparative Sequence Alignment and Evolutionary Divergence Assessment of CAD Family Genes
4.4. Functional Domain Conservation and Genomic Architecture Profiling in CAD Family Genes
4.5. Chromosomal Distribution and Colinearity Analysis of CAD Genes
4.6. Cis-Element Analysis of BnaCAD Promoters
4.7. Expression Profile Analysis of BnaCAD Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prakash, S.; Bhat, S.R.; Quiros, C.F.; Kirti, P.B.; Chopra, V.L. Brassica and Its Close Allies: Cytogenetics and Evolution. In Plant Breeding Reviews; John Wiley & Sons: Hoboken, NJ, USA, 2009; pp. 21–187. [Google Scholar] [CrossRef]
- Nagaharu, U. Genome Analysis in Brassica with Special Reference to the Experimental Formation of B. napus and Peculiar Mode of Fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early Allopolyploid Evolution in the Post-Neolithic Brassica napus Oilseed Genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The Genome Sequence of Allopolyploid Brassica juncea and Analysis of Differential Homoeolog Gene Expression Influencing Selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Perumal, S.; Koh, C.S.; Jin, L.; Buchwaldt, M.; Higgins, E.E.; Zheng, C.; Sankoff, D.; Robinson, S.J.; Kagale, S.; Navabi, Z.-K.; et al. A High-Contiguity Brassica nigra Genome Localizes Active Centromeres and Defines the Ancestral Brassica Genome. Nat. Plants 2020, 6, 929–941. [Google Scholar] [CrossRef]
- Guo, N.; Wang, S.; Gao, L.; Liu, Y.; Wang, X.; Lai, E.; Duan, M.; Wang, G.; Li, J.; Yang, M.; et al. Genome Sequencing Sheds Light on the Contribution of Structural Variants to Brassica oleracea Diversification. BMC Biol. 2021, 19, 93. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, J.; Chen, H.; Zhang, Z.; Wu, J.; Wang, X. A Near-Complete Genome Assembly of Brassica rapa Provides New Insights into the Evolution of Centromeres. Plant Biotechnol. J. 2023, 21, 1022–1032. [Google Scholar] [CrossRef]
- Song, X.; Wei, Y.; Xiao, D.; Gong, K.; Sun, P.; Ren, Y.; Yuan, J.; Wu, T.; Yang, Q.; Li, X.; et al. Brassica carinata Genome Characterization Clarifies U’s Triangle Model of Evolution and Polyploidy in Brassica. Plant Physiol. 2021, 186, 388–406. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Hu, S.; Zhang, J.; Wang, L.; Ping, X.; Wang, J.; Li, J.; Lu, K.; Tang, Z.; et al. Transcriptome and Proteome Analyses of the Molecular Mechanisms Underlying Changes in Oil Storage under Drought Stress in Brassica napus L. GCB Bioenergy 2021, 13, 1071–1086. [Google Scholar] [CrossRef]
- Qu, C.; Zhao, H.; Fu, F.; Wang, Z.; Zhang, K.; Zhou, Y.; Wang, X.; Wang, R.; Xu, X.; Tang, Z.; et al. Genome-Wide Survey of Flavonoid Biosynthesis Genes and Gene Expression Analysis between Black- and Yellow-Seeded Brassica napus. Front. Plant Sci. 2016, 7, 1755. [Google Scholar] [CrossRef]
- Cheng, Y. Molecular Mechanism of Manipulating Seed Coat Coloration in Oilseed Brassica Species. J. Appl. Genet. 2013, 54, 135–145. [Google Scholar] [CrossRef]
- Ran, X.; Wan, H.; Li, J.; Liang, Y.; Xu, J. An Investigation of the Relationship between Lignin Content and Seed Coat Traits in Yellow-Seeded Brassica napus L. J. Agric. Res. 2015, 3, 1–8. [Google Scholar] [CrossRef]
- Raza, A. Eco-physiological and Biochemical Responses of Rapeseed (Brassica napus L.) to Abiotic Stresses: Consequences and Mitigation Strategies. J. Plant Growth Regul. 2020, 40, 1368–1388. [Google Scholar] [CrossRef]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Díaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H.; et al. Phytohormones Trigger Drought Tolerance in Crop Plants: Outlook and Future Perspectives. Front. Plant Sci. 2022, 12, 799318. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.; Liang, Y.; Li, J. Analysis of the Lignin Contents and Related Enzymes Activities in Seed Coat Between Black-Seeded and Yellow-Seeded Rapes (Brassica napus L.). Agric. Sci. China 2005, 4, 890–897. [Google Scholar]
- Boudet, A.-M.; Hawkins, S.; Rochange, S. The Polymorphism of the Genes/Enzymes Involved in the Last Two Reductive Steps of Monolignol Synthesis: What Is the Functional Significance? C. R. Biol. 2004, 327, 837–845. [Google Scholar] [CrossRef]
- Kim, S.-J.; Kim, M.-R.; Bedgar, D.L.; Moinuddin, S.G.A.; Cardenas, C.L.; Davin, L.B.; Kang, C.; Lewis, N.G. Functional Reclassification of the Putative Cinnamyl Alcohol Dehydrogenase Multigene Family in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 1455–1460. [Google Scholar] [CrossRef]
- Chao, N.; Huang, S.; Kang, X.; Yidilisi, K.; Dai, M.; Liu, L. Systematic Functional Characterization of Cinnamyl Alcohol Dehydrogenase Family Members Revealed Their Functional Divergence in Lignin Biosynthesis and Stress Responses in Mulberry. Plant Physiol. Biochem. 2022, 186, 145–156. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, C.; Liu, W.; Qi, H.; Chen, H.; Cao, S. The Cinnamyl Alcohol Dehydrogenase Gene Family in Melon (Cucumis melo L.): Bioinformatic Analysis and Expression Patterns. PLoS ONE. 2014, 9, e101730. [Google Scholar] [CrossRef]
- Yusuf, C.Y.L.; Nabilah, N.S.; Taufik, N.A.A.M.; Seman, I.A.; Abdullah, M.P. Genome-Wide Analysis of the CAD Gene Family Reveals Two Bona Fide CAD Genes in Oil Palm. 3 Biotech. 2022, 12, 149. [Google Scholar] [CrossRef]
- Barakat, A.; Bagniewska-Zadworna, A.; Choi, A.; Plakkat, U.; DiLoreto, D.S.; Yellanki, P.; Carlsonet, G.E. The Cinnamyl Alcohol Dehydrogenase Gene Family in Populus: Phylogeny, Organization, and Expression. BMC Plant Biol. 2009, 9, 26. [Google Scholar] [CrossRef]
- Tobias, C.M.; Chow, E.K. Structure of the Cinnamyl-Alcohol Dehydrogenase Gene Family in Rice and Promoter Activity of a Member Associated with Lignification. Planta 2005, 220, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-Wide Characterization of the Lignification Toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Goffner, D.; Joffroy, I.; Grima-Pettenati, J.; Halpin, C.; Knight, M.E.; Schuch, W.; Boudet, A.M. Purification and Characterization of Isoforms of Cinnamyl Alcohol Dehydrogenase from Eucalyptus Xylem. Planta 1992, 188, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Aya, K.; Kondo, M.; Okuno, A.; Morinaka, Y.; Matsuoka, M. OsCAD2 Is the Major CAD Gene Responsible for Monolignol Biosynthesis in Rice Culm. Plant Cell Rep. 2012, 31, 91–101. [Google Scholar] [CrossRef]
- Liu, X.; Van Acker, R.; Voorend, W.; Pallidis, A.; Goeminne, G.; Pollier, J.; Morreel, K.; Kim, H.; Muylle, H.; Bosio, M.; et al. Rewired Phenolic Metabolism and Improved Saccharification Efficiency of a Zea Mays Cinnamyl Alcohol Dehydrogenase 2 (Zmcad2) Mutant. Plant J. Cell Mol. Biol. 2021, 105, 1240–1257. [Google Scholar] [CrossRef]
- Kiedrowski, S.; Kawalleck, P.; Hahlbrock, K.; Somssich, I.E.; Dangl, J.L. Rapid Activation of a Novel Plant Defense Gene Is Strictly Dependent on the Arabidopsis RPM1 Disease Resistance Locus. EMBO J. 1992, 11, 4677–4684. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Bae, J.M.; Huh, G.-H. Transcriptional Regulation of the Cinnamyl Alcohol Dehydrogenase Gene from Sweet Potato in Response to Plant Developmental Stage and Environmental Stress. Plant Cell Rep. 2010, 29, 779–791. [Google Scholar] [CrossRef]
- Dwyer, K.G.; Berger, M.T.; Ahmed, R.; Hritzo, M.K.; McCulloch, A.A.; Price, M.J.; Serniak, N.J.; Walsh, L.T.; Nasrallah, J.B.; Nasrallah, M.E. Molecular Characterization and Evolution of Self-Incompatibility Genes in Arabidopsis thaliana: The Case of the Sc Haplotype. Genetics 2013, 193, 985–994. [Google Scholar] [CrossRef]
- Seymour, D.K.; Chae, E.; Arioz, B.I.; Koenig, D.; Weigel, D. Transmission Ratio Distortion is Frequent in Arabidopsis thaliana Controlled Crosses. Heredity 2019, 122, 294–304. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of Duplicate Genes in Exon-Intron Structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and Genome Evolution in Plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.P.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea Genome Reveals the Asymmetrical Evolution of Polyploid Genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Town, C.D.; Cheung, F.; Maiti, R.; Crabtree, J.; Haas, B.J.; Wortman, J.R.; Hine, E.E.; Althoff, R.; Arbogast, T.S.; Tallon, L.J.; et al. Comparative Genomics of Brassica Oleracea and Arabidopsis Thaliana Reveal Gene Loss, Fragmentation, and Dispersal after Polyploidy. Plant Cell 2006, 18, 1348–1359. [Google Scholar] [CrossRef]
- Guo, D.-M.; Ran, J.-H.; Wang, X.-Q. Evolution of the Cinnamyl/Sinapyl Alcohol Dehydrogenase (CAD/SAD) Gene Family: The Emergence of Real Lignin Is Associated with the Origin of Bona Fide CAD. J. Mol. Evol. 2010, 71, 202–218. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.-S.; Wang, J. Correlation Between Ka/Ks and Ks is Related to Substitution Model and Evolutionary Lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- Ibraheem, O.; Botha, C.E.J.; Bradley, G. In Silico Analysis of Cis-Acting Regulatory Elements in 5’ Regulatory Regions of Sucrose Transporter Gene Families in Rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 2010, 34, 268–283. [Google Scholar] [CrossRef]
- Yoshida, T.; Christmann, A.; Yamaguchi-Shinozaki, K.; Grill, E.; Fernie, A.R. Revisiting the Basal Role of ABA—Roles Outside of Stress. Trends Plant Sci. 2019, 24, 625–635. [Google Scholar] [CrossRef]
- Qu, C.; Zhu, M.; Hu, R.; Niu, Y.; Chen, S.; Zhao, H.; Li, C.; Wang, Z.; Yin, N.; Sun, F.; et al. Comparative Genomic Analyses Reveal the Genetic Basis of the Yellow-Seed Trait in Brassica napus. Nat. Commun. 2023, 14, 5194. [Google Scholar] [CrossRef]
- Liu, L.; Stein, A.; Wittkop, B.; Sarvari, P.; Li, J.; Yan, X.; Dreyer, F.; Frauen, M.; Friedt, W.; Snowdon, R.J. A Knockout Mutation in the Lignin Biosynthesis Gene CCR1 Explains a Major QTL for Acid Detergent Lignin Content in Brassica napus Seeds. Theor. Appl. Genet. 2012, 124, 1573–1586. [Google Scholar] [CrossRef]
- Liu, L.; Qu, C.; Wittkop, B.; Yi, B.; Xiao, Y.; He, Y.; Snowdon, R.J.; Li, J. A High-Density SNP Map for Accurate Mapping of Seed Fibre QTL in Brassica napus L. PLoS ONE. 2013, 8, e83052. [Google Scholar] [CrossRef] [PubMed]
- Badani, A.G.; Snowdon, R.J.; Wittkop, B.; Lipsa, F.D.; Baetzel, R.; Horn, R.; De, H.A.; Font, R.; Lühs, W.; Friedt, W. Colocalization of a Partially Dominant Gene for Yellow Seed Colour with a Major QTL Influencing Acid Detergent Fibre (ADF) Content in Different Crosses of Oilseed Rape (Brassica napus). Genome 2006, 49, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chory, J.; Fankhauser, C. Light Signal Transduction in Higher Plants. Annu. Rev. Genet. 2004, 38, 87–117. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, C.; Gaddam, S.R.; Pandey, A.; Trivedi, P.K. COP1 Mediates Light-Dependent Regulation of Flavonol Biosynthesis through HY5 in Arabidopsis. Plant Sci. 2021, 303, 110760. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.-B.; Choi, G.; Park, Y.-I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- Wang, Q.; Gong, X.; Xie, Z.; Qi, K.; Yuan, K.; Jiao, Y.; Pan, Q.; Zhang, S.; Shiratake, K.; Tao, S. Cryptochrome-Mediated Blue-Light Signal Contributes to Lignin Biosynthesis in Stone Cells in Pear Fruit. Plant Sci. 2022, 318, 111211. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A Multi-Omics Database with Various Tools for Brassica napus Research and Breeding. Mol. Plant. 2023, 16, 775–789. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE. 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.H.; Lercher, M.J.; Hu, S.N.; Chen, W.H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, 270–275. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Sun, F.; Chen, Z.; Zhang, Q.; Wan, Y.; Hu, R.; Shen, S.; Chen, S.; Yin, N.; Tang, Y.; Liang, Y.; et al. Genome-Wide Identification of the TIFY Gene Family in Brassiceae and Its Potential Association with Heavy Metal Stress in Rapeseed. Plants 2022, 11, 667. [Google Scholar] [CrossRef]
- Hu, J.; Chen, B.; Zhao, J.; Zhang, F.; Xie, T.; Xu, K.; Gao, G.; Yan, G.; Li, H.; Li, L.; et al. Genomic Selection and Genetic Architecture of Agronomic Traits during Modern Rapeseed Breeding. Nat. Genet. 2022, 54, 694–704. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Weng, Z.; Liu, Y.; Tian, M.; Yang, Y.; Pan, N.; Zhang, M.; Zhao, H.; Du, H.; Yin, N.; et al. Identification of the Cinnamyl Alcohol Dehydrogenase Gene Family in Brassica U-Triangle Species and Its Potential Roles in Response to Abiotic Stress and Regulation of Seed Coat Color in Brassica napus L. Plants 2025, 14, 1184. https://doi.org/10.3390/plants14081184
Liu Y, Weng Z, Liu Y, Tian M, Yang Y, Pan N, Zhang M, Zhao H, Du H, Yin N, et al. Identification of the Cinnamyl Alcohol Dehydrogenase Gene Family in Brassica U-Triangle Species and Its Potential Roles in Response to Abiotic Stress and Regulation of Seed Coat Color in Brassica napus L. Plants. 2025; 14(8):1184. https://doi.org/10.3390/plants14081184
Chicago/Turabian StyleLiu, Yiwei, Ziwuyun Weng, Yuanyuan Liu, Mengjiao Tian, Yaping Yang, Nian Pan, Mengzhen Zhang, Huiyan Zhao, Hai Du, Nengwen Yin, and et al. 2025. "Identification of the Cinnamyl Alcohol Dehydrogenase Gene Family in Brassica U-Triangle Species and Its Potential Roles in Response to Abiotic Stress and Regulation of Seed Coat Color in Brassica napus L." Plants 14, no. 8: 1184. https://doi.org/10.3390/plants14081184
APA StyleLiu, Y., Weng, Z., Liu, Y., Tian, M., Yang, Y., Pan, N., Zhang, M., Zhao, H., Du, H., Yin, N., Qu, C., & Wan, H. (2025). Identification of the Cinnamyl Alcohol Dehydrogenase Gene Family in Brassica U-Triangle Species and Its Potential Roles in Response to Abiotic Stress and Regulation of Seed Coat Color in Brassica napus L. Plants, 14(8), 1184. https://doi.org/10.3390/plants14081184







