Decoding G-Quadruplexes Sequence in Vitis vinifera: Regulatory Region Enrichment, Drought Stress Adaptation, and Sugar–Acid Metabolism Modulation
Abstract
1. Introduction
2. Results
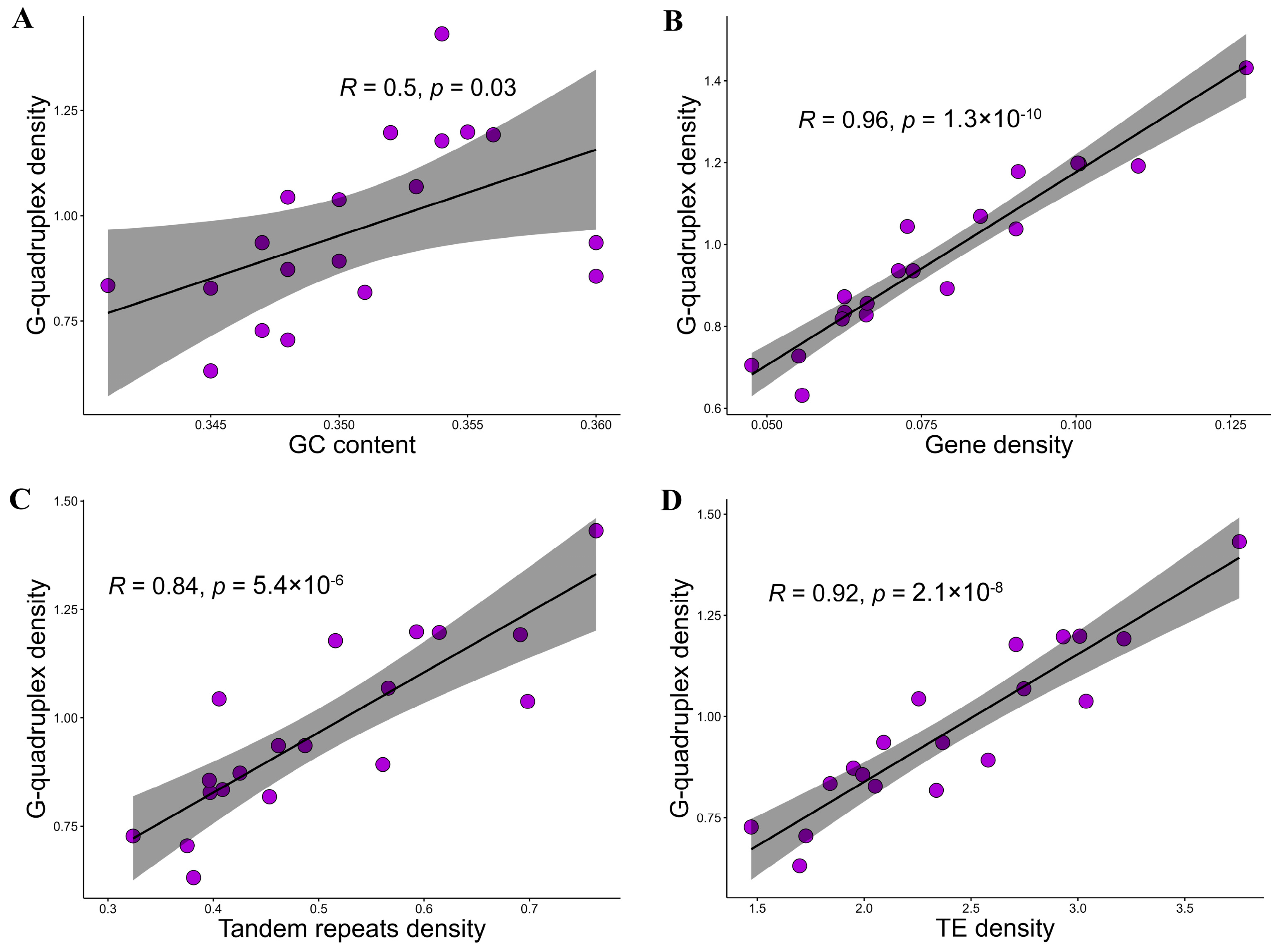
2.1. General Landscape of G-Quadruplexes in Vitis vinifera Genome
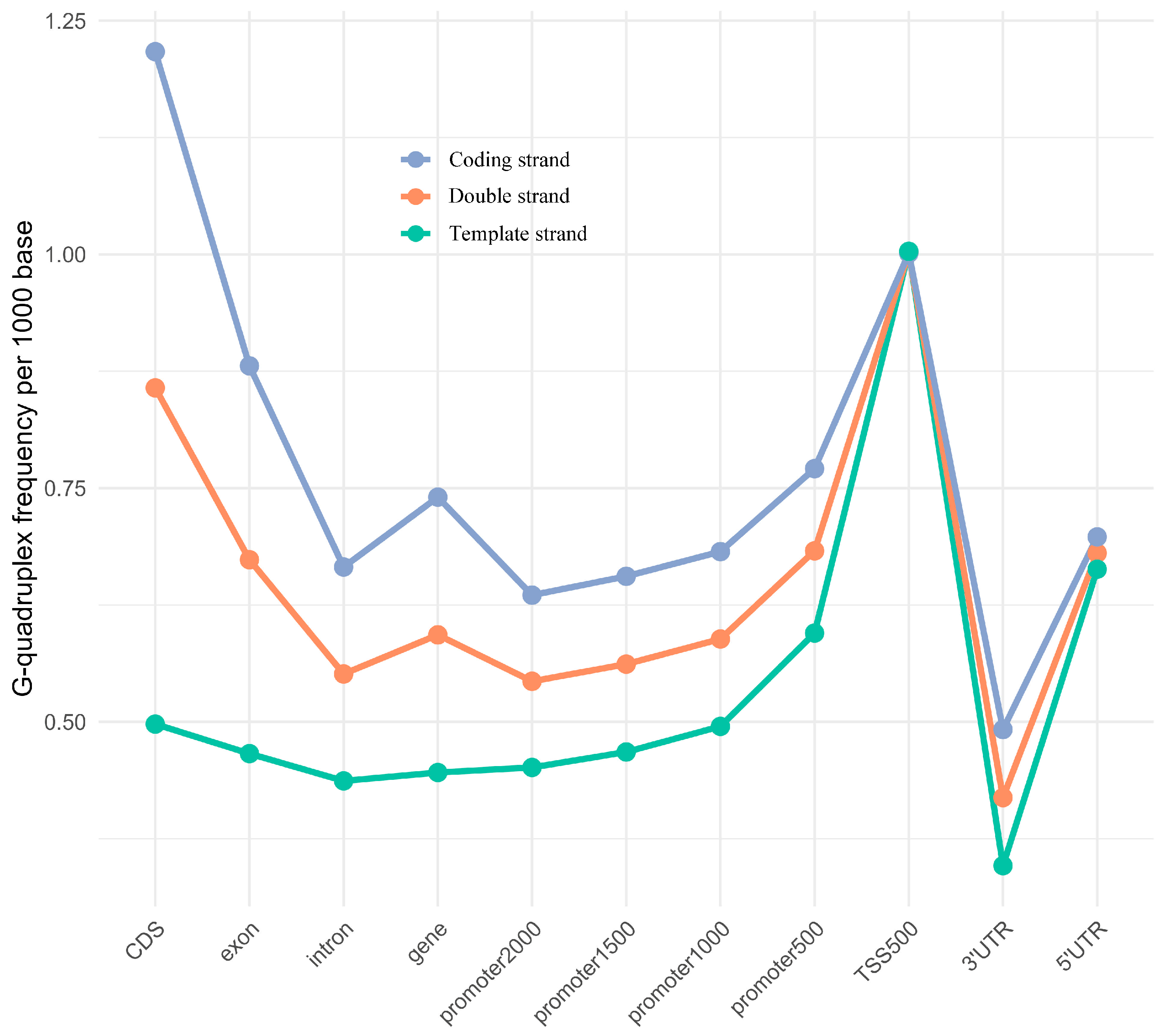
2.2. Distribution Patterns of G-Quadruplexes in the Genomic Feature Regions
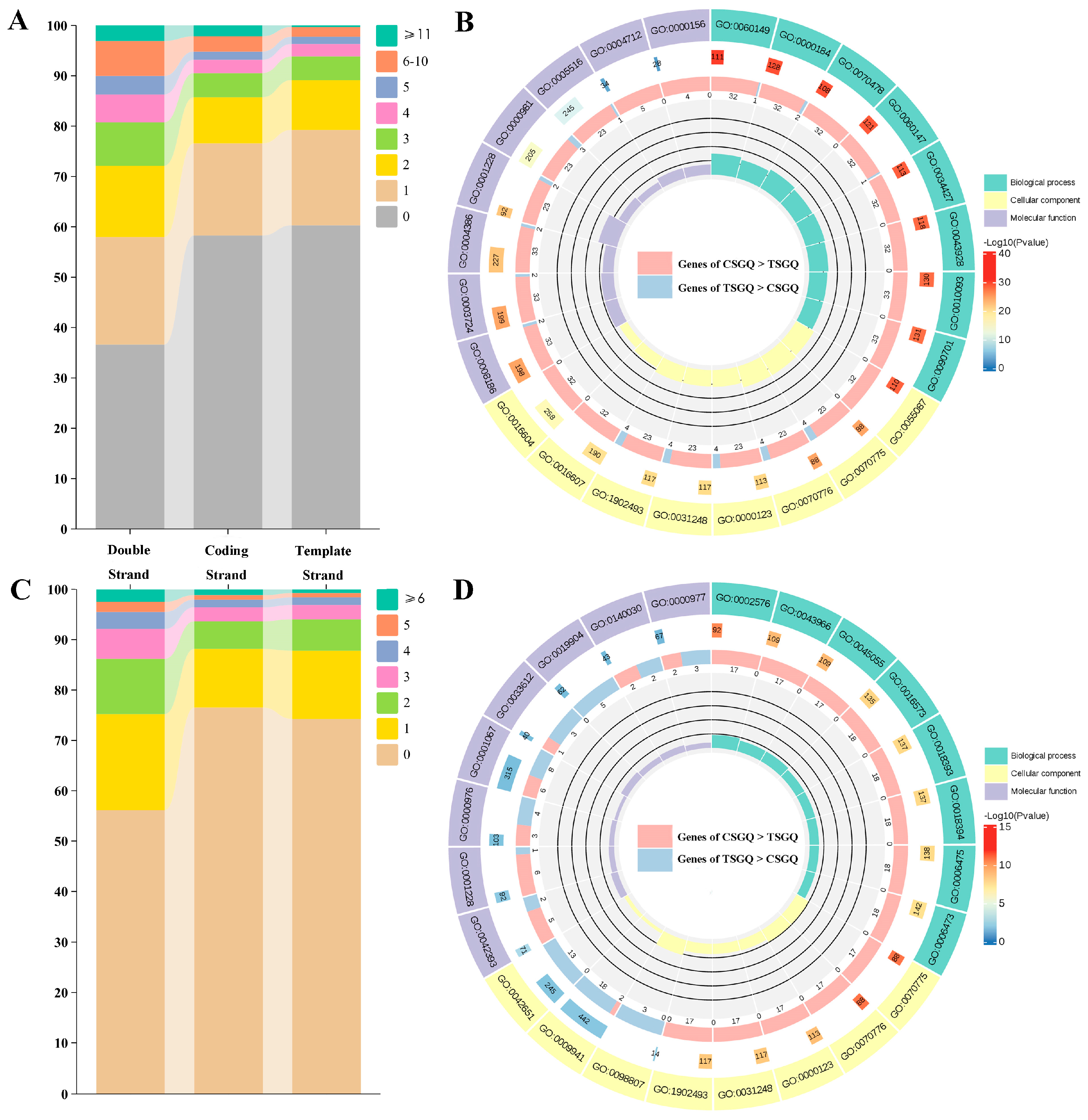
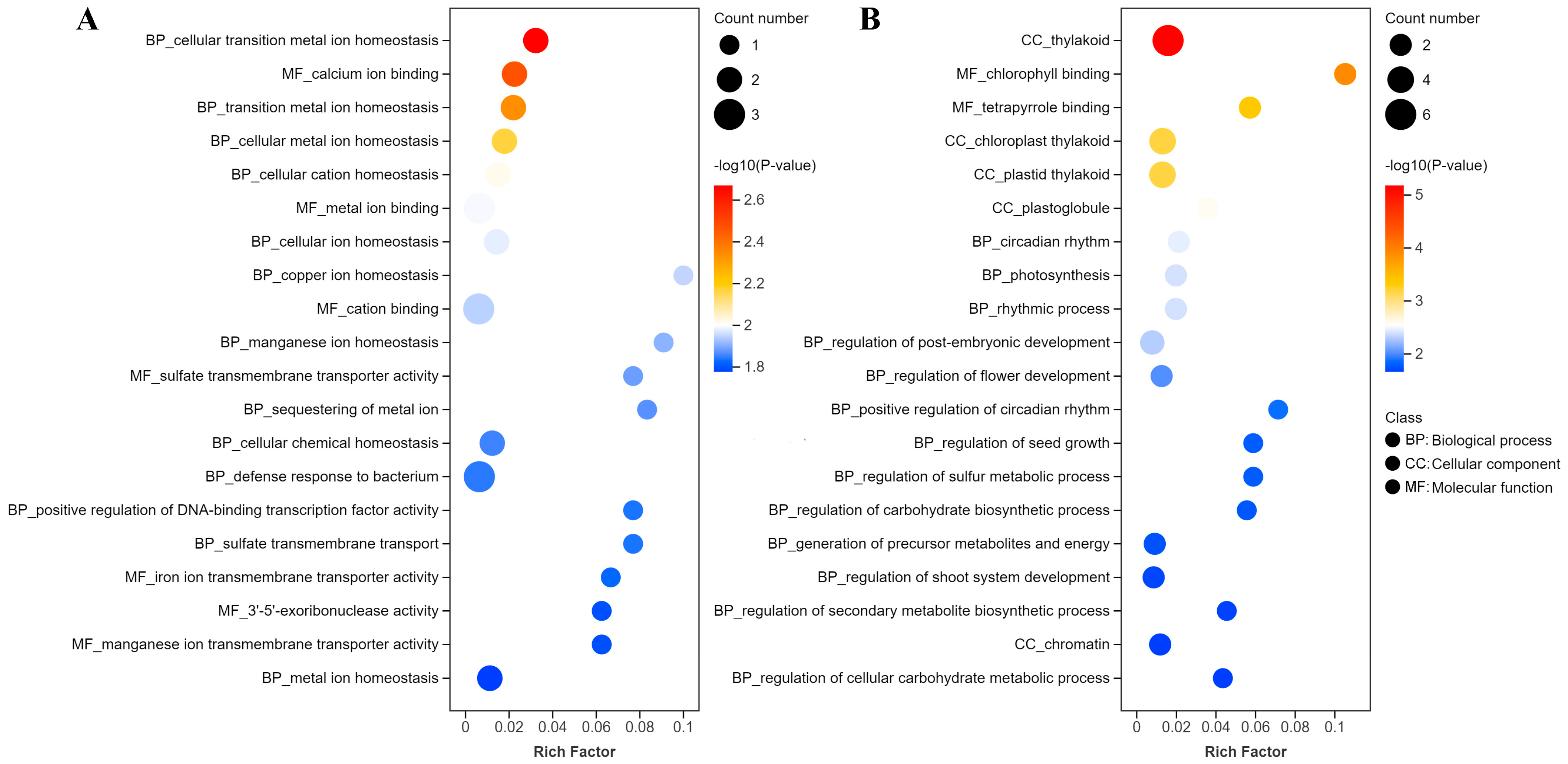
2.3. Functions of Genes with Highly Enriched G-Quadruplexes in Promoters and TSSs
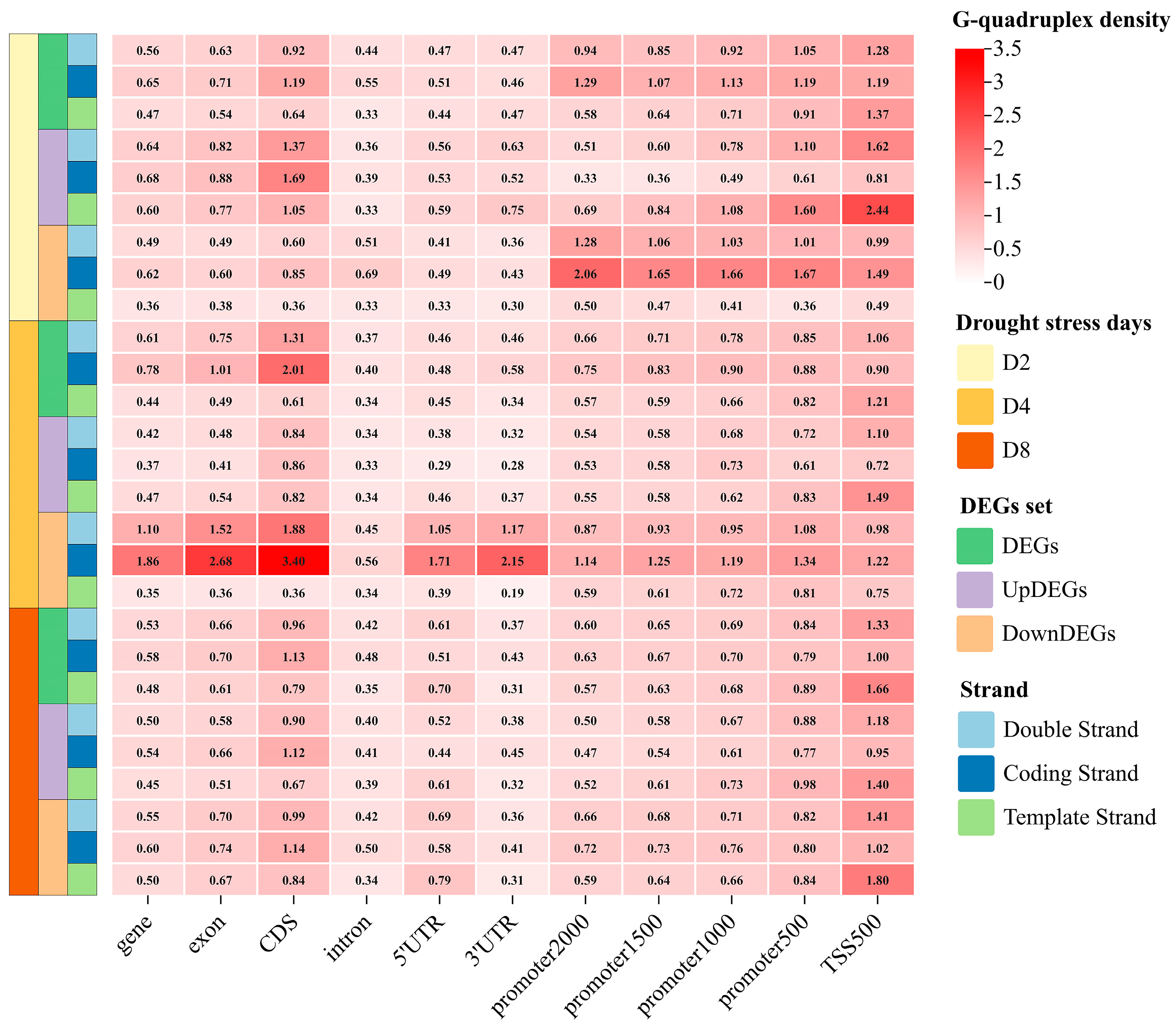
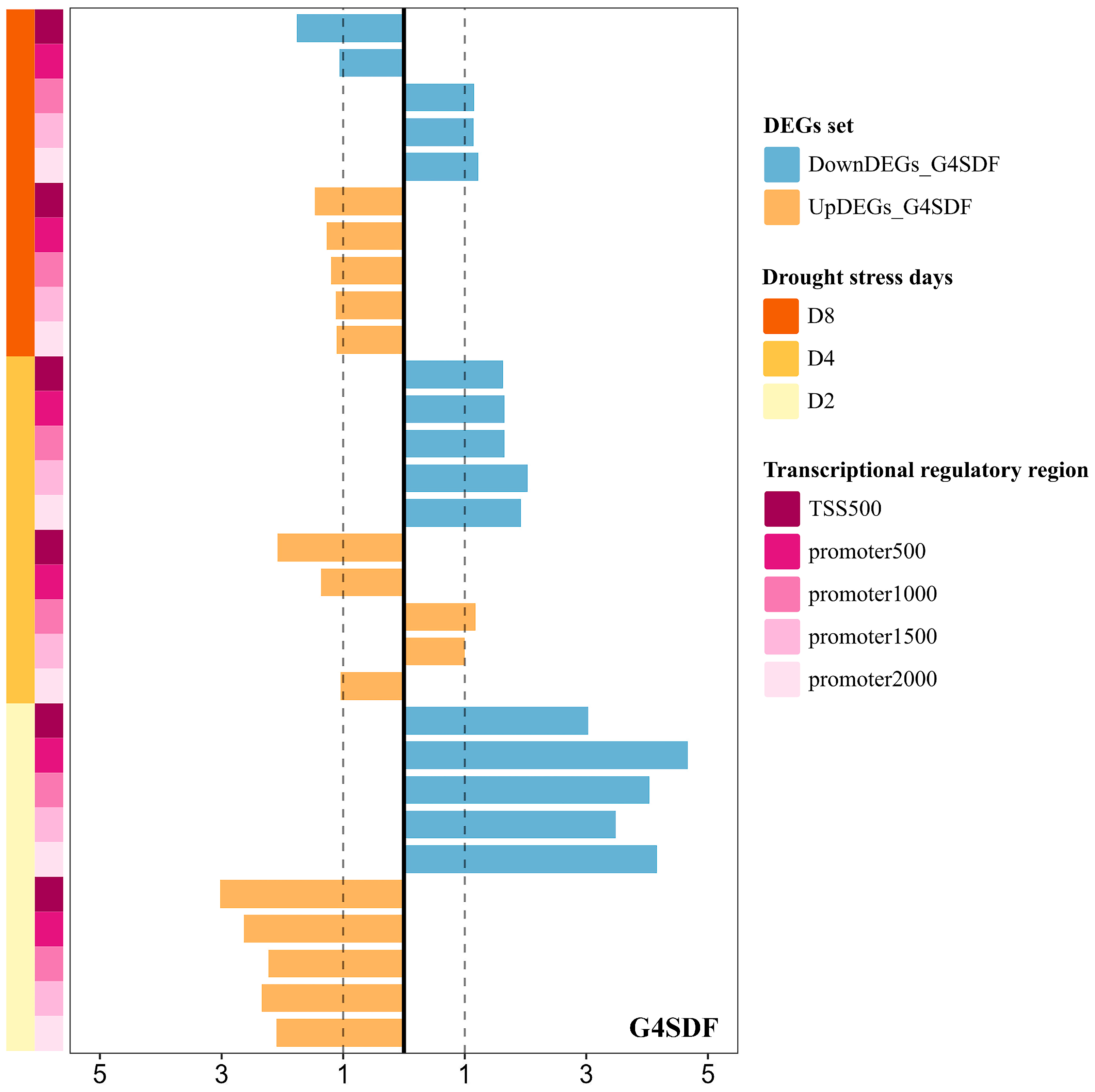
2.4. Prevalence of G-Quadruplexes in DEGs Under Drought Stress
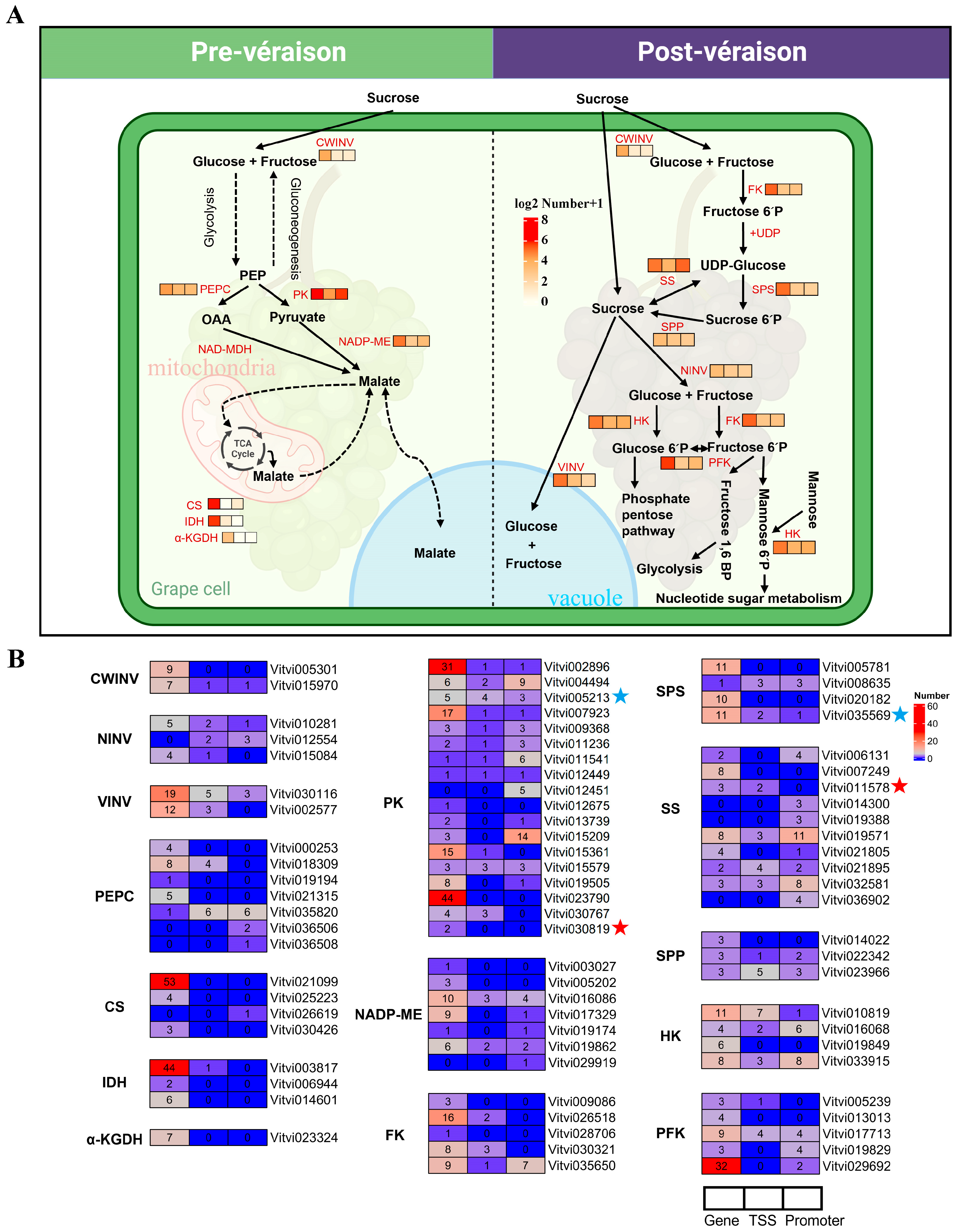
2.5. G-Quadruplexes in the Grape Berry Sugar–Acid Metabolism Pathway
3. Discussion
3.1. High-Density G-Quadruplexes May Facilitate Grapevine Adaptive Evolution
3.2. G-Quadruplexes Enriched in Transcriptional Regulatory Regions with Strand Bias
3.3. G-Quadruplex Might Participate in the Rapid Response to Drought Stress in Grapevine
3.4. G-Quadruplexes as Targets of Sugar–Acid Regulation of Grape Berry
4. Materials and Methods
4.1. Identification of G-Quadruplexes in the Vitis vinifera Genome
4.2. G-Quadruplex Analysis of Genomic Feature Regions in the Grapevine Genome
4.3. Functional Enrichment Analysis of Grapevine G-Quadruplex-Rich Genes
4.4. G-Quadruplex Analysis of DEGs Under Drought Stress at Different Stages
4.5. G-Quadruplex Analysis of Key Genes in the Sugar–Acid Metabolism Pathway of Grape Berry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Crespi, M.; Ariel, F. Non-B DNA structures emerging from plant genomes. Trends Plant Sci. 2022, 27, 624–626. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef]
- Guo, J.U.; Bartel, D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 2016, 353, aaf5371. [Google Scholar] [CrossRef]
- Garcia, I.E.; Kirchner, A.; Melidis, L.; Tavares, R.D.A.; Dhir, S.; Simeone, A.; Yu, Z.T.; Madden, S.K.; Hermann, R.; Tannahill, D.; et al. G-quadruplex DNA structure is a positive regulator of MYC transcription. Proc. Natl. Acad. Sci. USA 2024, 121, e2320240121. [Google Scholar] [CrossRef]
- Pramanik, S.; Chen, Y.L.; Song, H.Y.; Khutsishvili, I.; Marky, L.A.; Ray, S.; Natarajan, A.; Singh, P.K.; Bhakat, K.K. The human AP-endonuclease 1 (APE1) is a DNA G-quadruplex structure binding protein and regulates KRAS expression in pancreatic ductal adenocarcinoma cells. Nucleic Acids Res. 2022, 50, 3394–3412. [Google Scholar] [CrossRef]
- Agrawal, P.; Hatzakis, E.; Guo, K.X.; Carver, M.; Yang, D.Z. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: Insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013, 41, 10584–10592. [Google Scholar] [CrossRef]
- Chen, L.Y.; Dickerhoff, J.; Sakai, S.; Yang, D.Z. DNA G-Quadruplex in Human Telomeres and Oncogene Promoters: Structures, Functions, and Small Molecule Targeting. Acc. Chem. Res. 2022, 55, 2628–2646. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Sharma, H.; Garg, R. Knot-knot chronicles: Unveiling the G-quadruplexes. Crit. Rev. Biotechnol. 2025, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tavakoli, M.; Jiang, R.; Zong, L.; Zhang, W. Identification and functional characterization of ssDNA in plants. Trends Plant Sci. 2025, 25, S1360–S1385. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Aggarwal, J.; Thakkar, B. Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci. Rep. 2016, 6, 28211. [Google Scholar] [CrossRef] [PubMed]
- Mullen, M.A.; Olson, K.J.; Dallaire, P.; Major, F.; Assmann, S.M.; Bevilacqua, P.C. RNA G-Quadruplexes in the model plant species Arabidopsis thaliana: Prevalence and possible functional roles. Nucleic Acids Res. 2010, 38, 8149–8163. [Google Scholar] [CrossRef]
- Feng, Y.L.; Tao, S.T.; Zhang, P.Y.; Sperti, F.R.; Liu, G.Q.; Cheng, X.J.; Zhang, T.; Yu, H.X.; Wang, X.E.; Chen, C.Y.; et al. Epigenomic features of DNA G-quadruplexes and their roles in regulating rice gene transcription. Plant Physiol. 2022, 188, 1632–1648. [Google Scholar] [CrossRef]
- Zhou, Y.Z.; Kishchenko, O.; Stepanenko, A.; Chen, G.M.; Wang, W.; Zhou, J.; Pan, C.Z.; Borisjuk, N. The Dynamics of NO3− and NH4+ Uptake in Duckweed Are Coordinated with the Expression of Major Nitrogen Assimilation Genes. Plants 2022, 11, 11. [Google Scholar] [CrossRef]
- Song, K.K.; Li, B.; Li, H.Z.; Zhang, R.; Zhang, X.H.; Luan, R.W.; Liu, Y.; Yang, L. The Characterization of G-Quadruplexes in Tobacco Genome and Their Function under Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 4331. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Sen, T.Z. Genome-Wide Discovery of G-Quadruplexes in Wheat: Distribution and Putative Functional Roles. G3-Genes Genomes Genet. 2020, 10, 2021–2032. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Budak, H.; Sen, T.Z. Genome-wide discovery of G-quadruplexes in barley. Sci. Rep. 2021, 11, 7876. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolná, M.; Bohálová, N.; Peska, V.; Wang, J.W.; Luo, Y.; Bartas, M.; Volná, A.; Mergny, J.L.; Brázda, V. The Newly Sequenced Genome of Pisum sativum Is Replete with Potential G-Quadruplex-Forming Sequences-Implications for Evolution and Biological Regulation. Int. J. Mol. Sci. 2022, 23, 8482. [Google Scholar] [CrossRef] [PubMed]
- Griffin, B.D.; Bass, H.W. Plant G-quadruplex (G4) motifs in DNA and RNA; abundant, intriguing sequences of unknown function. Plant Sci. 2018, 269, 143–147. [Google Scholar] [CrossRef]
- Yang, X.; Yang, M.; Deng, H.; Ding, Y. New Era of Studying RNA Secondary Structure and Its Influence on Gene Regulation in Plants. Front. Plant Sci. 2018, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Cho, H.S.; Nam, H.; Jo, H.; Yoon, J.; Park, C.; Dang, T.V.T.; Kim, E.; Jeong, J.; Park, S.; et al. Translational control of phloem development by RNA G-quadruplex-JULGI determines plant sink strength. Nat. Plants 2018, 4, 376–390. [Google Scholar] [CrossRef]
- Yang, X.F.; Cheema, J.; Zhang, Y.Y.; Deng, H.J.; Duncan, S.; Umar, M.I.; Zhao, J.Y.; Liu, Q.; Cao, X.F.; Kwok, C.K.; et al. RNA G-quadruplex structures exist and function in vivo in plants. Genome Biol. 2020, 21, 226. [Google Scholar] [CrossRef]
- Foroozani, M. Unraveling the role of DNA G-quadruplexes in transcription in the rice. Plant Physiol. 2022, 188, 1412–1414. [Google Scholar] [CrossRef]
- Yang, X.F.; Yu, H.P.; Duncan, S.; Zhang, Y.Y.; Cheema, J.; Liu, H.F.; Miller, J.B.; Zhang, J.; Kwok, C.K.; Zhang, H.K.; et al. RNA G-quadruplex structure contributes to cold adaptation in plants. Nat. Commun. 2022, 13, 6224. [Google Scholar] [CrossRef]
- Andorf, C.M.; Kopylov, M.; Dobbs, D.; Koch, K.E.; Stroupe, M.E.; Lawrence, C.J.; Bass, H.W. G-Quadruplex (G4) Motifs in the Maize (Zea mays L.) Genome Are Enriched at Specific Locations in Thousands of Genes Coupled to Energy Status, Hypoxia, Low Sugar, and Nutrient Deprivation. J. Genet. Genom. 2014, 41, 627–647. [Google Scholar] [CrossRef]
- Chakrabarti, M.; de Lorenzo, L.; Abdel-Ghany, S.E.; Reddy, A.S.N.; Hunt, A.G. Wide-ranging transcriptome remodelling mediated by alternative polyadenylation in response to abiotic stresses in Sorghum. Plant J. 2020, 102, 916–930. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.; Gupta, K.; Sharma, H.; Bady, S.; Sondhi, M.; Garg, R. Genome-Wide Identification of G-Quadruplex forming regions in Arabidopsis: Unraveling the Role of Pif like Helicase (AtPLH1) in Gene Regulation and Stress Response. bioRxiv 2024. [Google Scholar] [CrossRef]
- Dong, Y.; Duan, S.C.; Xia, Q.J.; Liang, Z.C.; Dong, X.; Margaryan, K.; Musayev, M.; Goryslavets, S.; Zdunic, G.; Bert, P.F.; et al. Dual domestications and origin of traits in grapevine evolution. Science 2023, 379, 892–901. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, K.Y.; Li, H.Y.; Kuang, Y.F.; Liang, Z.C. Biography of Vitis genomics: Recent advances and prospective. Hortic. Res. 2024, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.; Massonnet, M.; Cochetel, N. The wild side of grape genomics. Trends Genet. 2024, 40, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Pecinka, P.; Bohálová, N.; Volná, A.; Kundrátová, K.; Brázda, V.; Bartas, M. Analysis of G-Quadruplex-Forming Sequences in Drought Stress-Responsive Genes, and Synthesis Genes of Phenolic Compounds in Arabidopsis thaliana. Life 2023, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Sahayasheela, V.J.; Yu, Z.; Hidaka, T.; Pandian, G.N.; Sugiyama, H. Mitochondria and G-quadruplex evolution: An intertwined relationship. Trends Genet. 2023, 39, 15–30. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Groover, A.T.; Schmutz, J.; DiFazio, S.P.; Myburg, A.; Grattapaglia, D.; Smart, L.B.; Yin, T.M.; Aury, J.M.; Kremer, A.; et al. Hardwood Tree Genomics: Unlocking Woody Plant Biology. Front. Plant Sci. 2018, 9, 1799. [Google Scholar] [CrossRef]
- Faizullah, L.; Morton, J.A.; Hersch-Green, E.I.; Walczyk, A.M.; Leitch, A.R.; Leitch, I.J. Exploring environmental selection on genome size in angiosperms. Trends Plant Sci. 2021, 26, 1039–1049. [Google Scholar] [CrossRef]
- Guo, L.; Wang, X.; Ayhan, D.H.; Rhaman, M.S.; Yan, M.; Jiang, J.; Wang, D.; Zheng, W.; Mei, J.; Ji, W.; et al. Super pangenome of Vitis empowers identification of downy mildew resistance genes for grapevine improvement. Nat. Genet. 2025, 57, 741–753. [Google Scholar] [CrossRef]
- Liu, Z.J.; Wang, N.; Su, Y.; Long, Q.M.; Peng, Y.L.; Shangguan, L.F.; Zhang, F.; Cao, S.; Wang, X.; Ge, M.Q.; et al. Grapevine pangenome facilitates trait genetics and genomic breeding. Nat. Genet. 2024, 56, 2804–2814. [Google Scholar] [CrossRef]
- Tsukahara, S.; Bousios, A.; Perez-Roman, E.; Yamaguchi, S.; Leduque, B.; Nakano, A.; Naish, M.; Osakabe, A.; Toyoda, A.; Ito, H.; et al. Centrophilic retrotransposon integration via CENH3 chromatin in Arabidopsis. Nature 2025, 637, 744–748. [Google Scholar] [CrossRef] [PubMed]
- Wlodzimierz, P.; Rabanal, F.A.; Burns, R.; Naish, M.; Primetis, E.; Scott, A.; Mandáková, T.; Gorringe, N.; Tock, A.J.; Holland, D.; et al. Cycles of satellite and transposon evolution in Arabidopsis centromeres. Nature 2023, 618, 557–565. [Google Scholar] [CrossRef]
- Naish, M. Bridging the gap: Unravelling plant centromeres in the telomere-to-telomere era. New Phytol. 2024, 244, 2143–2149. [Google Scholar] [CrossRef] [PubMed]
- Naish, M.; Henderson, I.R. The structure, function, and evolution of plant centromeres. Genome Res. 2024, 34, 161–178. [Google Scholar] [CrossRef]
- Kasinathan, S.; Henikoff, S. Non-B-Form DNA Is Enriched at Centromeres. Mol. Biol. Evol. 2018, 35, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Su, H.D.; Liu, K.P.; Xiao, X.; Li, W.; Sun, Q.W.; Birchler, J.A.; Han, F.P. Genome-wide mapping reveals R-loops associated with centromeric repeats in maize. Genome Res. 2021, 31, 1409–1418. [Google Scholar] [CrossRef]
- Patchigolla, V.S.P.; Mellone, B.G. Enrichment of Non-B-Form DNA at D. melanogaster Centromeres. Genome Biol. Evol. 2022, 14, 5. [Google Scholar] [CrossRef]
- Liu, C.; Huang, Y.H.; Guo, X.R.; Yi, C.Y.; Liu, Q.; Zhang, K.B.; Zhu, C.L.; Liu, Y.; Han, F.P. Young retrotransposons and non-B DNA structures promote the establishment of dominant rye centromere in the 1RS.1BL fused centromere. New Phytol. 2024, 241, 607–622. [Google Scholar] [CrossRef]
- Song, S.; Liu, H.; Miao, L.; Lan, H.; Chen, C. Centromeric repeats in Citrus sinensis provide new insights into centromeric evolution and the distribution of G-quadruplex structures. Hortic. Adv. 2023, 1, 7. [Google Scholar] [CrossRef]
- Liu, Q.; Yi, C.Y.; Zhang, Z.Y.; Su, H.D.; Liu, C.; Huang, Y.H.; Li, W.; Hu, X.J.; Liu, C.; Birchler, J.A.; et al. Non-B-form DNA tends to form in centromeric regions and has undergone changes in polyploid oat subgenomes. Proc. Natl. Acad. Sci. USA 2022, 120, e2211683120. [Google Scholar] [CrossRef]
- Altemose, N.; Logsdon, G.A.; Bzikadze, A.; Sidhwani, P.; Langley, S.A.; Caldas, G.; Hoyt, S.J.; Uralsky, L.; Ryabov, F.D.; Shew, C.J.; et al. Complete genomic and epigenetic maps of human centromeres. Science 2022, 376, eabl4178. [Google Scholar] [CrossRef]
- Feng, Y.L.; Luo, Z.Y.; Huang, R.R.; Yang, X.M.; Cheng, X.J.; Zhang, W.L. Epigenomic Features and Potential Functions of K+ and Na+ Favorable DNA G-Quadruplexes in Rice. Int. J. Mol. Sci. 2022, 23, 8404. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2006, 35, 406–413. [Google Scholar] [CrossRef]
- Zhang, R.X.; Wang, Y.Q.; Wang, C.; Sun, X.; Mergny, J.L. G-quadruplexes as pivotal components of cis-regulatory elements in the human genome. BMC Biol. 2024, 22, 177. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Joshi, M.; Wang, A.; Myong, S. 5′UTR G-quadruplex structure enhances translation in size dependent manner. Nat. Commun. 2024, 15, 3963. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Dong, Q.Y.; Wallrad, L.; Almutairi, B.O.; Kudla, J. Ca2+ signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 2022, 64, 287–300. [Google Scholar] [CrossRef]
- Zhang, H.M.; Zhu, J.H.; Gong, Z.Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Mendoza, O.; Gueddouda, N.M.; Boulé, J.B.; Bourdoncle, A.; Mergny, J.L. A fluorescence-based helicase assay: Application to the screening of G-quadruplex ligands. Nucleic Acids Res. 2015, 43, e71. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Balasubramanian, S. Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat. Chem. 2021, 13, 626–633. [Google Scholar] [CrossRef]
- Batra, S.; Allwein, B.; Kumar, C.; Devbhandari, S.; Brüning, J.-G.; Bahng, S.; Lee, C.M.; Marians, K.J.; Hite, R.K.; Remus, D. G-quadruplex–stalled eukaryotic replisome structure reveals helical inchworm DNA translocation. Science 2025, 387, eadt1978. [Google Scholar] [CrossRef] [PubMed]
- Li, G.S.; Zhao, Y. The critical roles of three sugar-related proteins (HXK, SnRK1, TOR) in regulating plant growth and stress responses. Hortic. Res. 2024, 11, uhae099. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, Z.; Li, B.; Qin, G.; Tian, S. Molecular basis for optimizing sugar metabolism and transport during fruit development. Abiotech 2021, 2, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.J.; Wu, L.M.; Yang, Y.J.; Zhou, G.Z.; Ruan, Y.L. Evolution of Sucrose Metabolism: The Dichotomy of Invertases and Beyond. Trends Plant Sci. 2018, 23, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Liu, Z.Q.; Yang, J.; Liao, X.F.; Zhao, C.Q.; Ren, J.S.; Qu, X.G. Targeting specific DNA G-quadruplexes with CRISPR-guided G-quadruplex-binding proteins and ligands. Nat. Cell Biol. 2024, 26, 1212–1224. [Google Scholar] [CrossRef]
- Ren, C.; Lin, Y.; Liang, Z. CRISPR/Cas genome editing in grapevine: Recent advances, challenges and future prospects. Fruit Res. 2022, 2, 7. [Google Scholar] [CrossRef]
- Lam, H.Y.I.; Ong, X.E.; Mutwil, M. Large language models in plant biology. Trends Plant Sci. 2024, 29, 1145–1155. [Google Scholar] [CrossRef]
- Nguyen, E.; Poli, M.; Durrant, M.G.; Kang, B.; Katrekar, D.; Li, D.B.; Bartie, L.J.; Thomas, A.W.; King, S.H.; Brixi, G.; et al. Sequence modeling and design from molecular to genome scale with Evo. Science 2024, 386, eado9336. [Google Scholar] [CrossRef]
- Yu, H.; Yang, H.; Sun, W.; Yan, Z.; Yang, X.; Zhang, H.; Ding, Y.; Li, K. An interpretable RNA foundation model for exploring functional RNA motifs in plants. Nat. Mach. Intell. 2024, 6, 1616–1625. [Google Scholar] [CrossRef]
- Shi, X.Y.; Cao, S.; Wang, X.; Huang, S.Y.; Wang, Y.; Liu, Z.J.; Liu, W.W.; Leng, X.P.; Peng, Y.L.; Wang, N.; et al. The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding. Hortic. Res. 2023, 10, uhad061. [Google Scholar] [CrossRef]
- Shen, W.; Sipos, B.; Zhao, L.Y. SeqKit2: A Swiss army knife for sequence and alignment processing. Imeta 2024, 3, e191. [Google Scholar] [CrossRef] [PubMed]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. Imeta 2022, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Katz, K.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O’Sullivan, C. The Sequence Read Archive: A decade more of explosive growth. Nucleic Acids Res. 2022, 50, D387–D390. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data, P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Delrot, S.; Liang, Z. From acidity to sweetness: A comprehensive review of carbon accumulation in grape berries. Mol. Hortic. 2024, 4, 22. [Google Scholar] [CrossRef]
- Hong, P.; Sadeghnezhad, E.; Wang, J.; Yu, W.; Zheng, J.; Zhong, R.; Xu, Y.; Zhang, Y.; Dong, T.; Fang, J.; et al. VvSnRK1-VvSS3 regulates sugar accumulation during grape berry ripening in response to abscisic acid. Sci. Hortic. 2023, 320, 112208. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 2024, 53, D672–D677. [Google Scholar] [CrossRef] [PubMed]
- Thumuluri, V.; Armenteros, J.J.A.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]

| Chromosome | Length (bp) | GC Content (%) | G-Quadruplex Number | G-Quadruplex Density (per kbp) |
|---|---|---|---|---|
| PN1 | 27,822,162 | 36.0 | 26,041 | 0.94 |
| PN2 | 20,941,263 | 35.2 | 21,555 | 1.03 |
| PN3 | 21,317,290 | 34.8 | 18,839 | 0.88 |
| PN4 | 25,934,928 | 34.7 | 25,232 | 0.97 |
| PN5 | 26,899,771 | 35.3 | 27,761 | 1.03 |
| PN6 | 24,571,969 | 35.5 | 23,639 | 0.96 |
| PN7 | 31,654,362 | 34.8 | 28,552 | 0.90 |
| PN8 | 23,763,023 | 35.0 | 26,679 | 1.12 |
| PN9 | 24,372,199 | 34.1 | 22,262 | 0.91 |
| PN10 | 27,504,061 | 35.6 | 25,068 | 0.91 |
| PN11 | 20,048,508 | 35.0 | 22,253 | 1.11 |
| PN12 | 24,706,008 | 34.8 | 24,271 | 0.98 |
| PN13 | 29,842,242 | 34.5 | 28,749 | 0.96 |
| PN14 | 30,475,315 | 34.5 | 29,451 | 0.97 |
| PN15 | 23,565,456 | 35.4 | 22,321 | 0.95 |
| PN16 | 27,608,946 | 36.0 | 21,207 | 0.77 |
| PN17 | 19,942,836 | 35.4 | 20,337 | 1.02 |
| PN18 | 36,684,271 | 34.7 | 32,786 | 0.89 |
| PN19 | 27,218,600 | 35.1 | 20,810 | 0.76 |
| mitochondrion | 774,663 | 44.1 | 1092 | 1.41 |
| chloroplast | 145,134 | 36.7 | 105 | 0.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, J.; Song, K.; Qiao, G.; Wang, R.; Wu, H.; Jia, Q.; Liu, Y.; Li, Y.; Xu, M. Decoding G-Quadruplexes Sequence in Vitis vinifera: Regulatory Region Enrichment, Drought Stress Adaptation, and Sugar–Acid Metabolism Modulation. Plants 2025, 14, 1180. https://doi.org/10.3390/plants14081180
Xie J, Song K, Qiao G, Wang R, Wu H, Jia Q, Liu Y, Li Y, Xu M. Decoding G-Quadruplexes Sequence in Vitis vinifera: Regulatory Region Enrichment, Drought Stress Adaptation, and Sugar–Acid Metabolism Modulation. Plants. 2025; 14(8):1180. https://doi.org/10.3390/plants14081180
Chicago/Turabian StyleXie, Jun, Kangkang Song, Gaixia Qiao, Rong Wang, Hongyuan Wu, Qiaoxia Jia, Yujuan Liu, Yi Li, and Meilong Xu. 2025. "Decoding G-Quadruplexes Sequence in Vitis vinifera: Regulatory Region Enrichment, Drought Stress Adaptation, and Sugar–Acid Metabolism Modulation" Plants 14, no. 8: 1180. https://doi.org/10.3390/plants14081180
APA StyleXie, J., Song, K., Qiao, G., Wang, R., Wu, H., Jia, Q., Liu, Y., Li, Y., & Xu, M. (2025). Decoding G-Quadruplexes Sequence in Vitis vinifera: Regulatory Region Enrichment, Drought Stress Adaptation, and Sugar–Acid Metabolism Modulation. Plants, 14(8), 1180. https://doi.org/10.3390/plants14081180






