Genome-Wide Analysis of NAC Transcription Factor Gene Family in Morus atropurpurea
Abstract
1. Introduction
2. Results
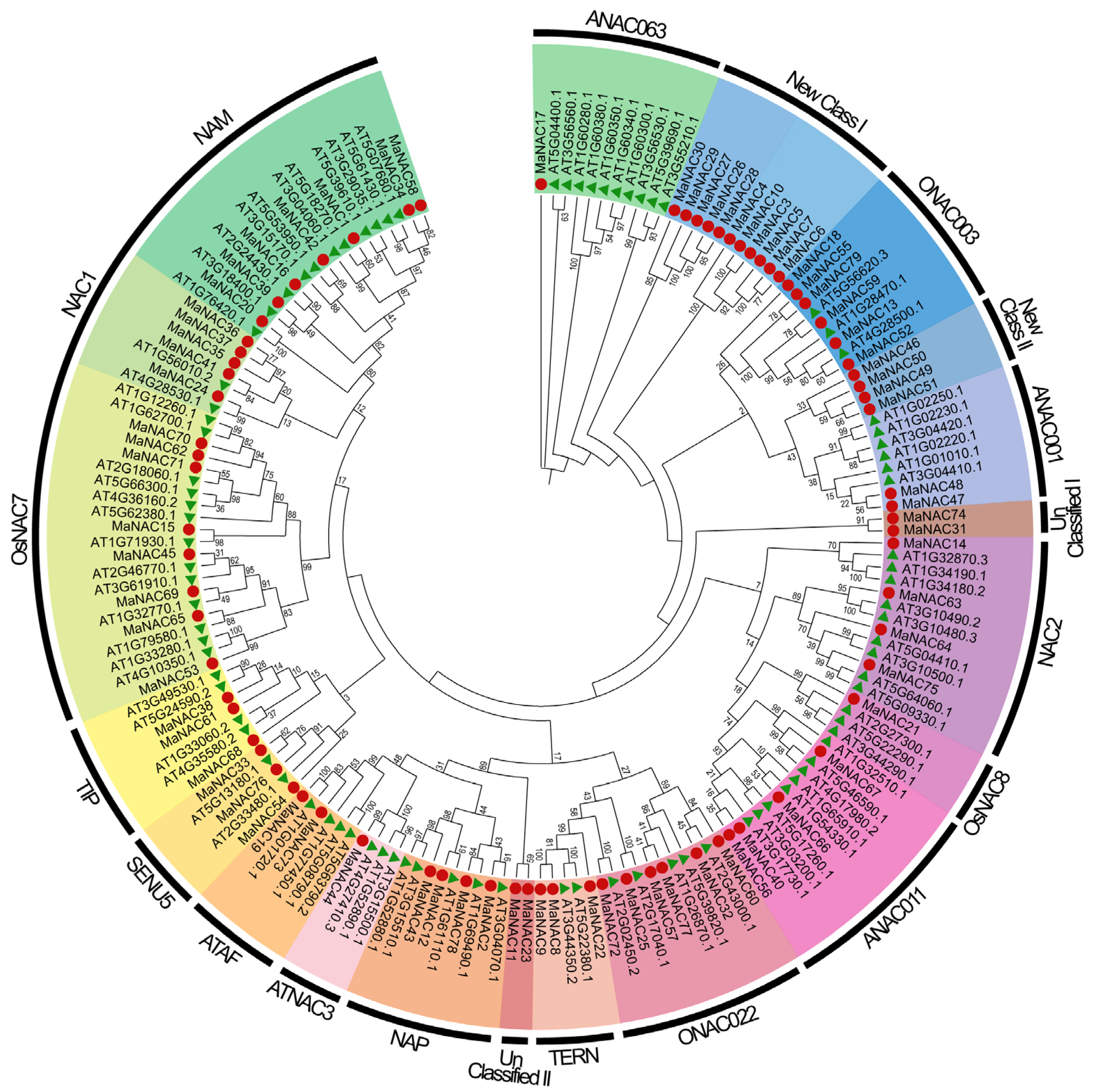
2.1. Identification and Phylogenetic Analysis of NAC Gene Family
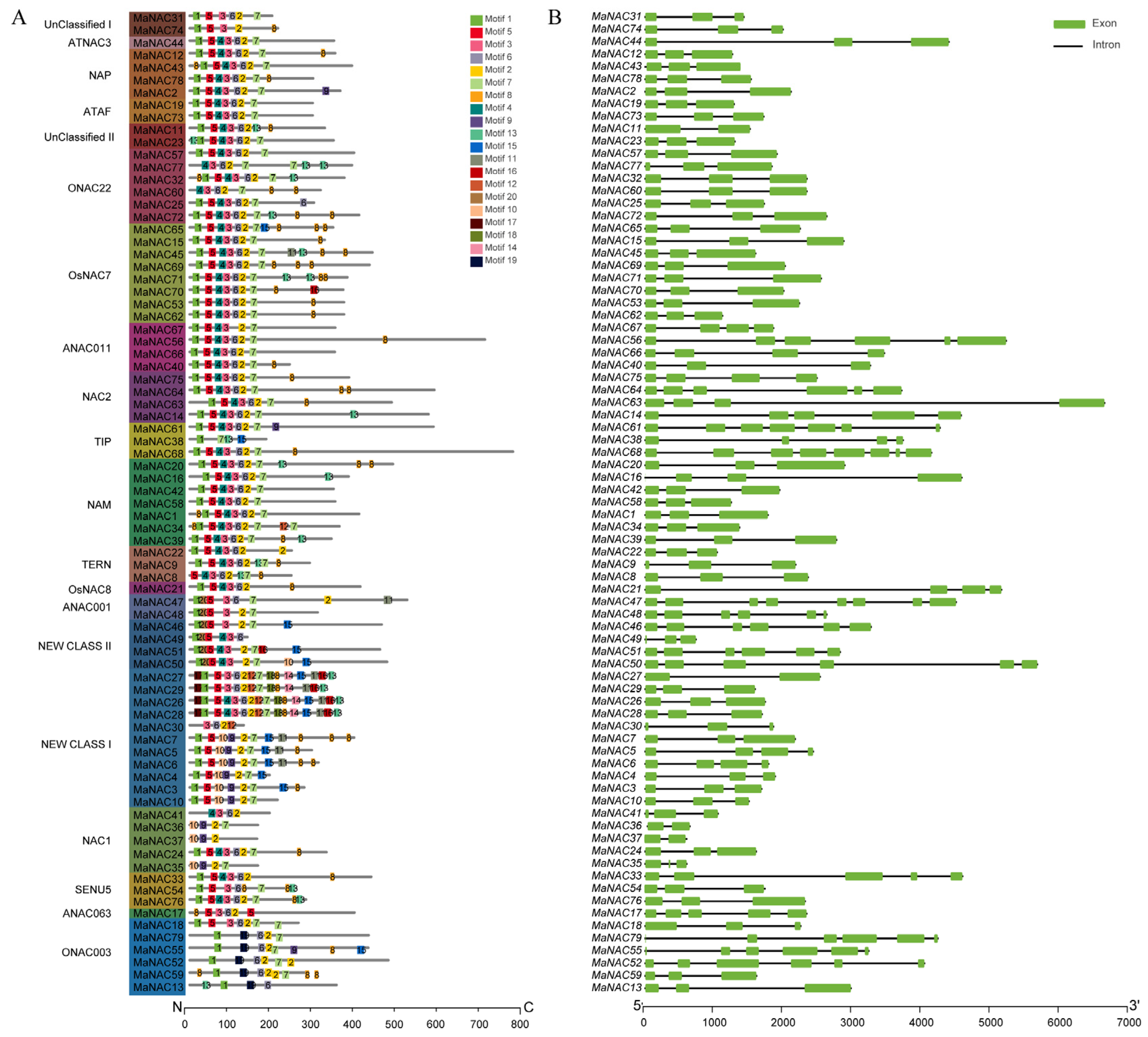
2.2. Conserved Protein Motif and Gene Structure Analysis of MaNACs
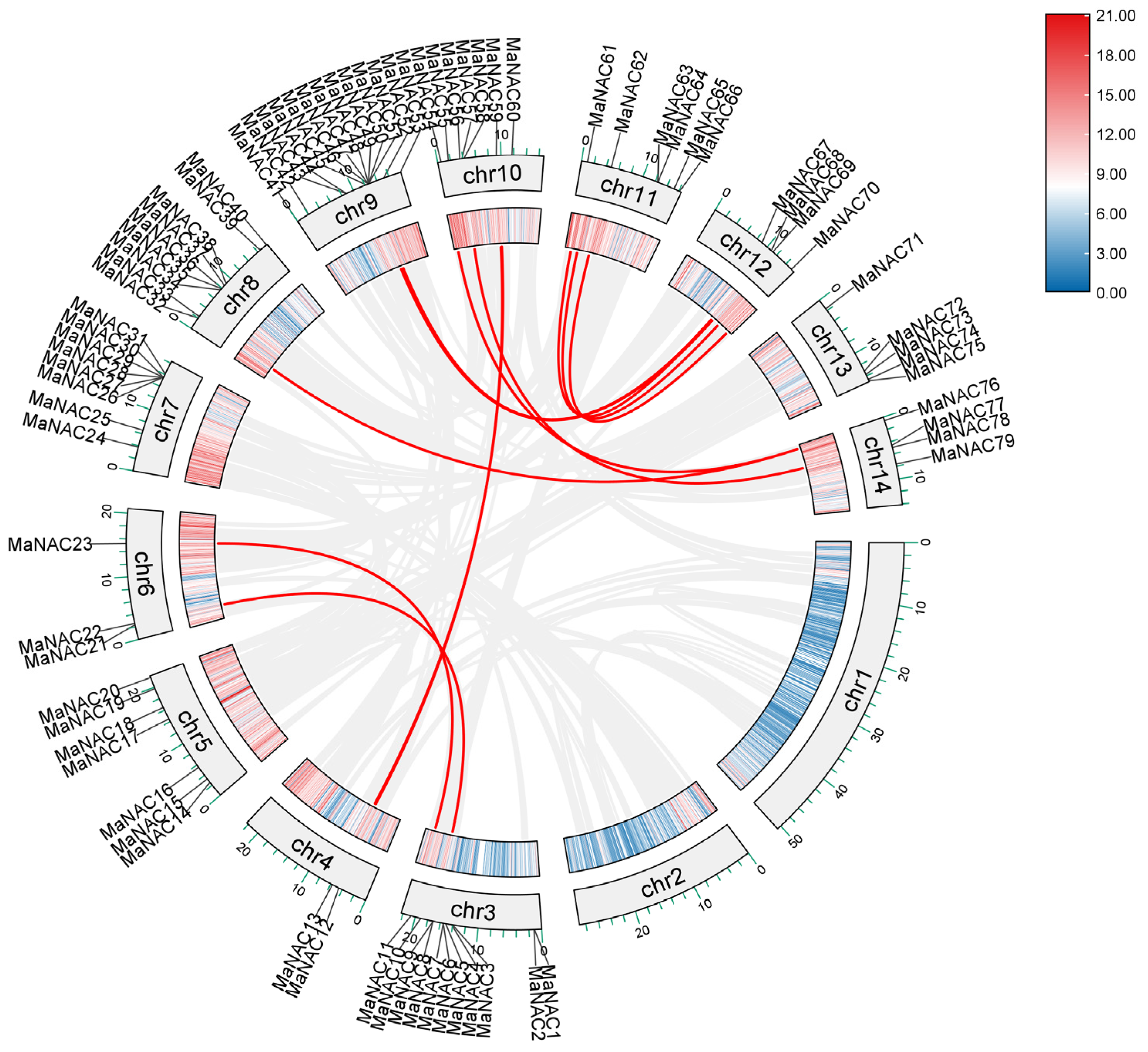
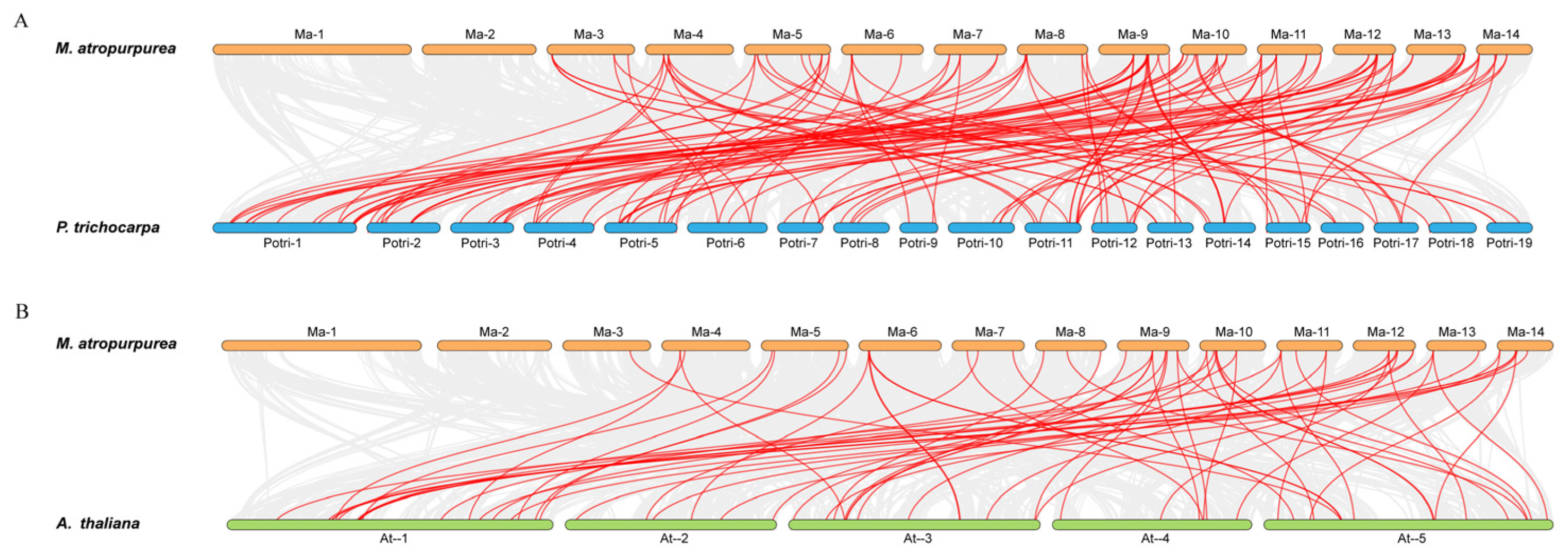
2.3. Gene Duplication and Collinearity Analysis of MaNACs
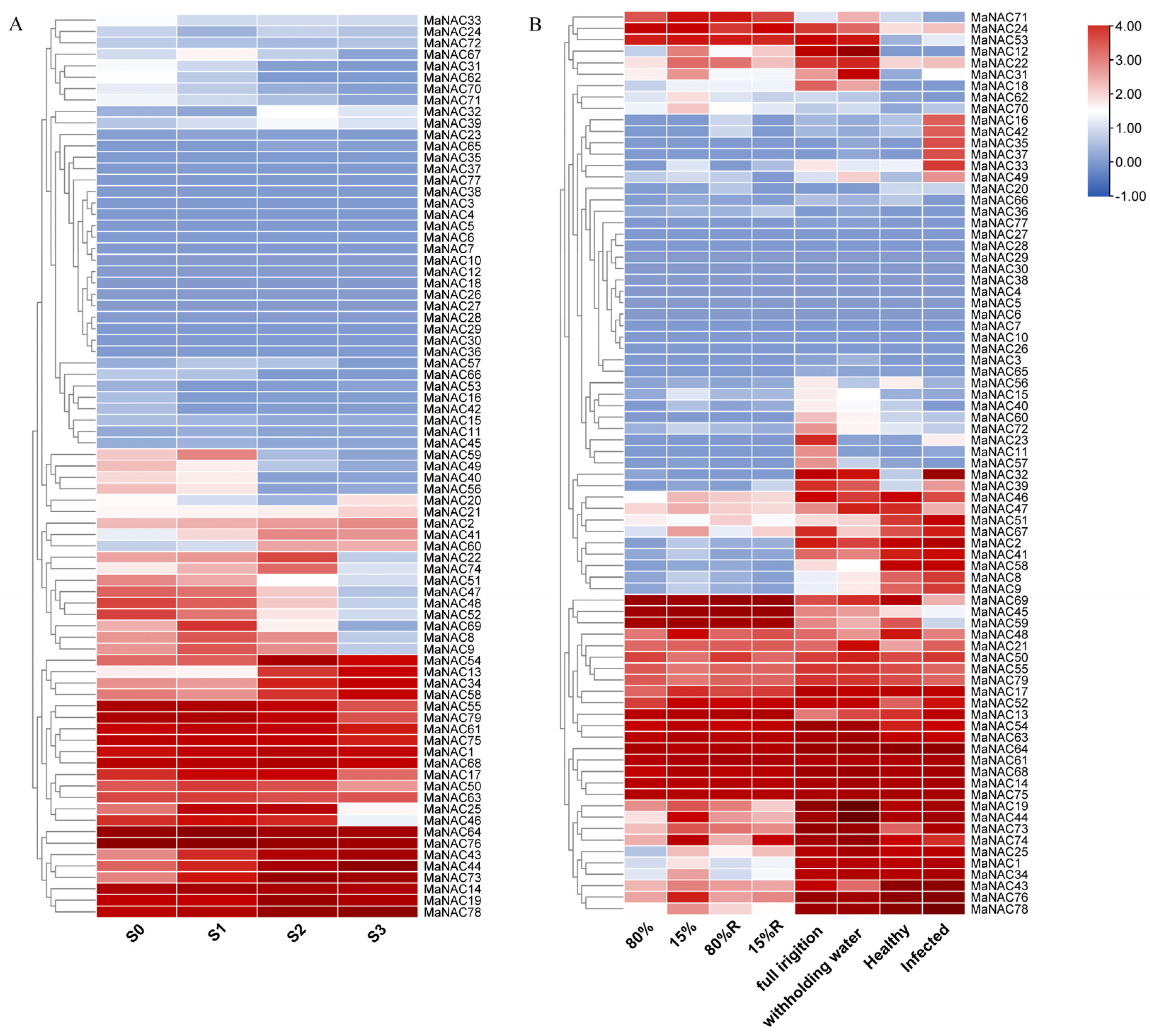
2.4. Temporal Expression and Response Profiling of MaNACs
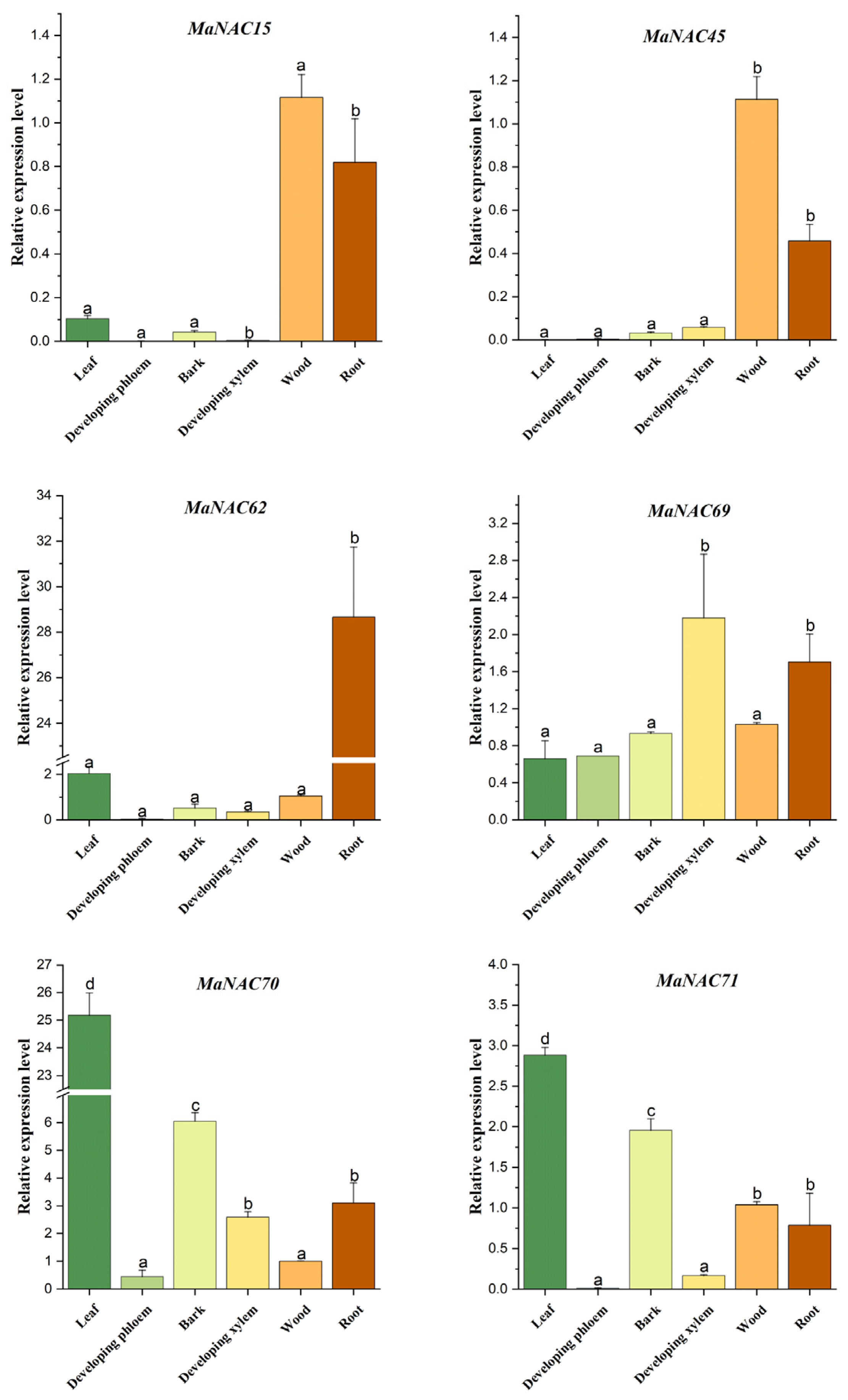
2.5. Tissue-Specific Expression Profiling of OsNAC7 Subfamily
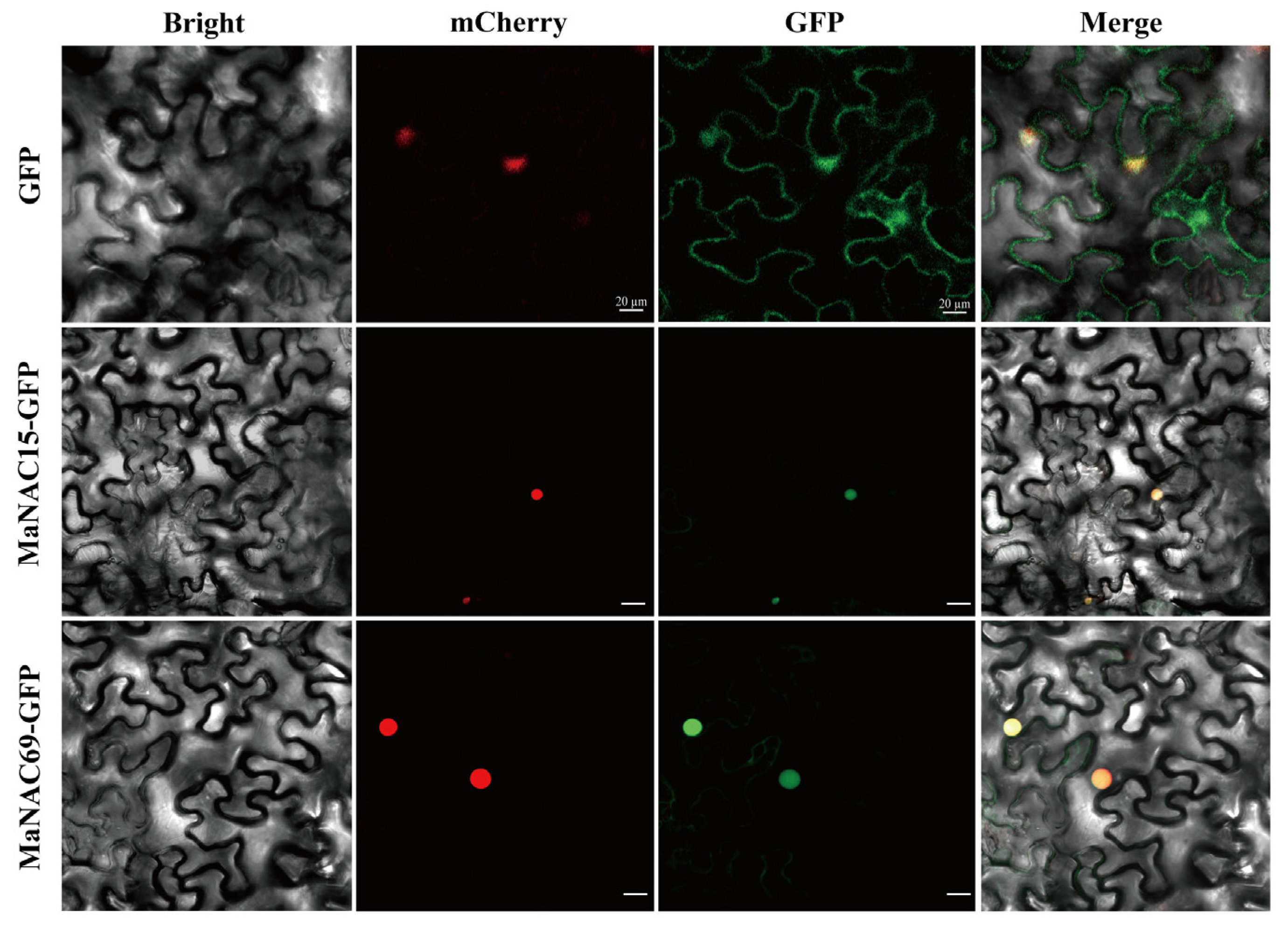
2.6. Subcellular Localization of MaNAC15 and MaNAC69
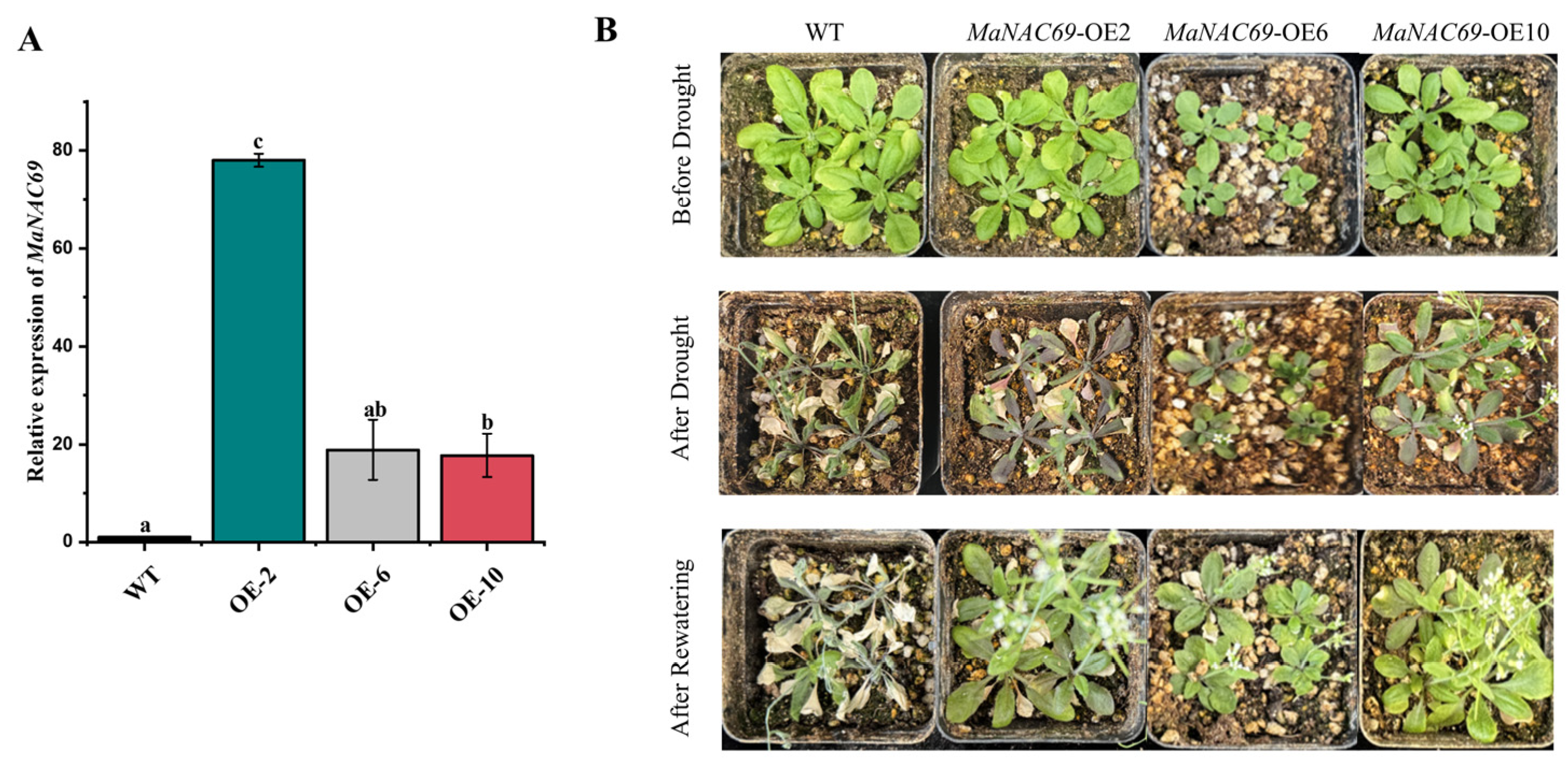
2.7. Drought Tolerance of MaNAC69-Overexpressing Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatment
4.2. Identification of Mulberry NAC
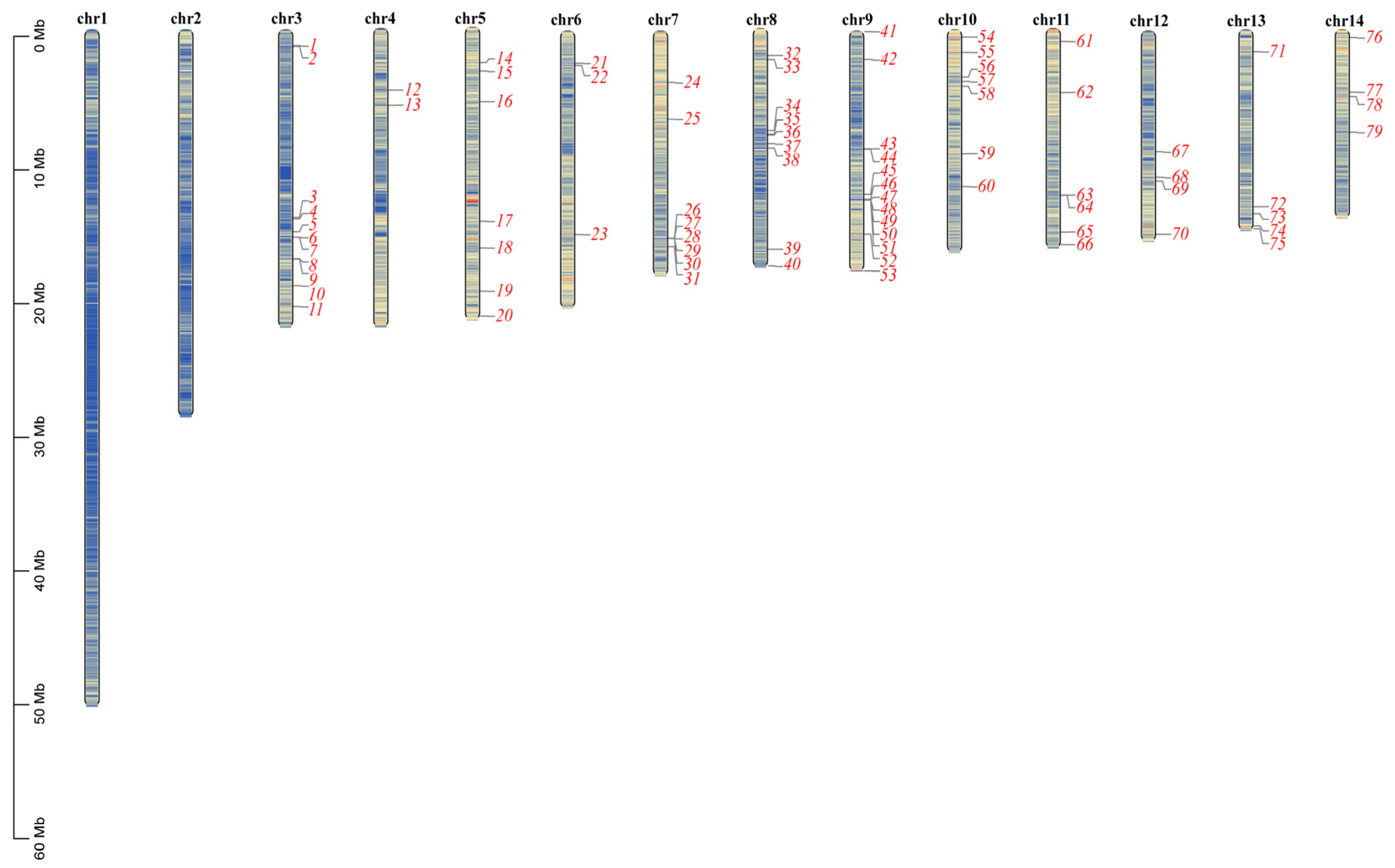
4.3. Phylogenetic Tree Construction, Chromosomal Localization and Collinearity Analysis
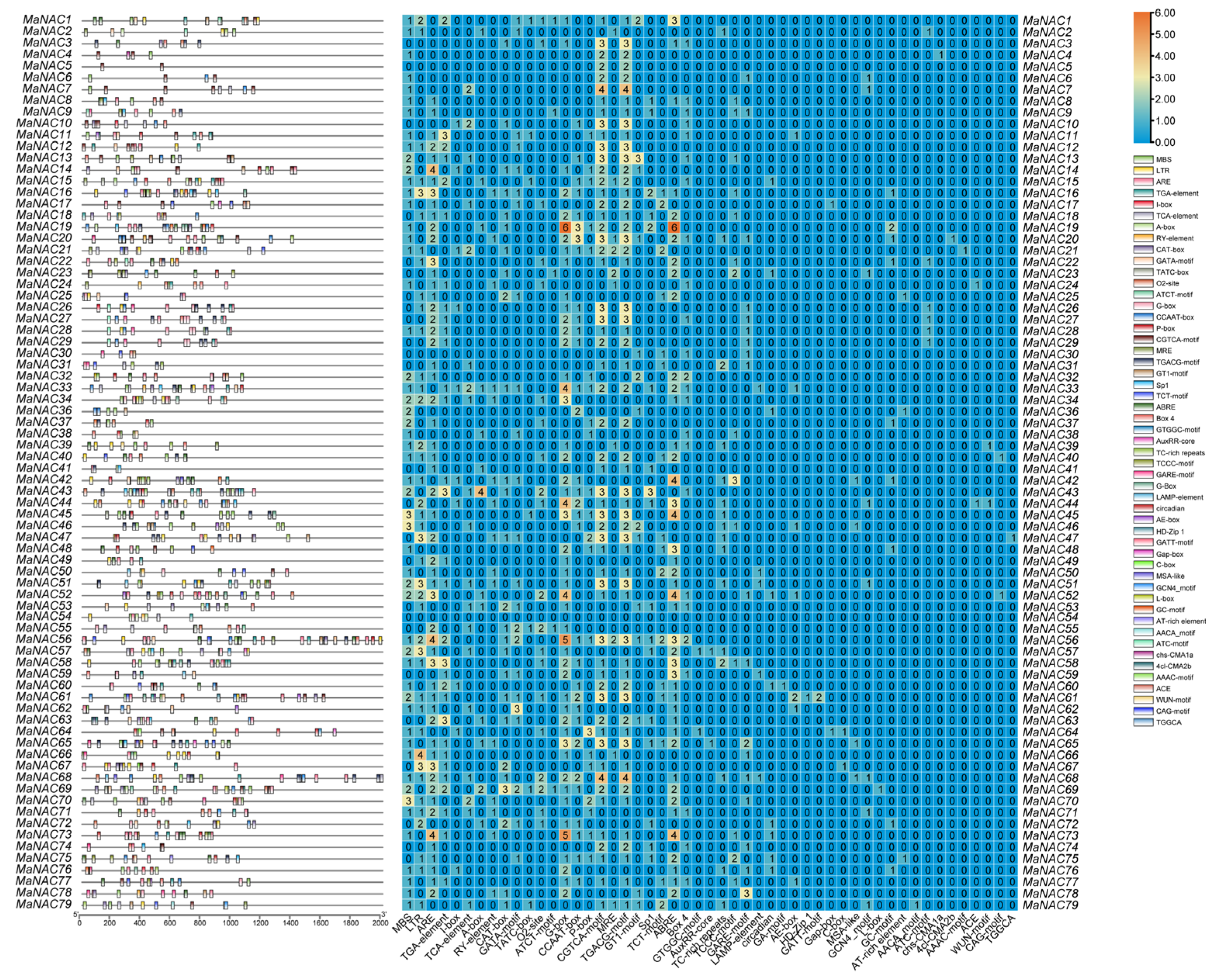
4.4. Bioinformatic Analysis of MaNACs
4.5. Expression Profiling of MaNAC Based on Transcriptome Analysis
4.6. qRT-PCR Analysis
4.7. Subcellular Localization
4.8. Functional Analysis of MaNAC69 in Arabidopsis in Response to Drought
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, D.; Chen, G.; Ma, B.; Zhong, C.; He, N. Metabolic profiling and transcriptome analysis of Mulberry leaves provide insights into flavonoid biosynthesis. J. Agric. Food Chem. 2020, 68, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Rohela, G.K.; Shukla, P.; Muttanna; Kumar, R.; Chowdhury, S.R. Mulberry (Morus spp.): An ideal plant for sustainable development. Trees For. People 2020, 2, 100011. [Google Scholar] [CrossRef]
- Zhu, Z.; Yu, C.; Dong, Z.; Mo, R.; Zhang, C.; Liu, X.; Zuo, Y.; Li, Y.; Deng, W.; Hu, X. Phylogeny and fungal community structures of helotiales associated with Sclerotial disease of Mulberry fruits in China. Plant Dis. 2024, 108, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Ernst, H.A.; Olsen, A.N.; Larsen, S.; Lo Leggio, L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Su, H.; Zhang, S.; Yuan, X.; Chen, C.; Wang, X.-F.; Hao, Y.-J. Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1,2-CUC2 transcription factor family in apple. Plant Physiol. Biochem. 2013, 71, 11–21. [Google Scholar] [CrossRef]
- Guo, L.; Liao, Y.; Deng, S.; Li, J.; Bu, X.; Zhu, C.; Zhang, W.; Cong, X.; Cheng, S.; Chen, Q.; et al. Genome-wide analysis of NAC transcription factors and exploration of candidate genes regulating selenium metabolism in Broussonetia papyrifera. Planta 2024, 260, 1. [Google Scholar] [CrossRef]
- Li, Y.; Han, H.; Fu, M.; Zhou, X.; Ye, J.; Xu, F.; Zhang, W.; Liao, Y.; Yang, X. Genome-wide identification and expression analysis of NAC family genes in Ginkgo biloba L. Plant Biol. 2023, 25, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Zhao, Y.; Liu, J.; Tian, Y.; El-Kassaby, Y.A.; Qi, Y.; Ke, M.; Sun, Y.; Li, Y. Genome-wide investigation and analysis of NAC transcription factor family in Populus tomentosaand expression analysis under salt stress. Plant Biol. 2024, 26, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, F.; Yao, Z.; Zhao, X.; Chu, G.; Ye, J. Comprehensive genomic characterisation of the NAC transcription factor family and its response to drought stress in Eucommia ulmoides. PeerJ 2023, 11, e16298. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wei, C.; Duan, W.; Gao, Y.; Kuang, J.; Liu, M.; Chen, K.; Klee, H.; Zhang, B. Transcriptional and epigenetic analysis reveals that NAC transcription factors regulate fruit flavor ester biosynthesis. Plant J. 2021, 106, 785–800. [Google Scholar] [CrossRef]
- Zhong, R.; McCarthy, R.L.; Lee, C.; Ye, Z.-H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011, 157, 1452–1468. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a stress-responsive NAC transcription factor gene ONACO22 improves drought and salt tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Mei, C.; Yang, J.; Mei, Q.; Jia, D.; Yan, P.; Feng, B.; Mamat, A.; Gong, X.; Guan, Q.; Mao, K.; et al. MdNAC104 positively regulates apple cold tolerance via CBF-dependent and CBF-independent pathways. Plant Biotechnol. J. 2023, 21, 2057–2073. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Foresti, C.; Orduña, L.; Matus, J.T.; Vandelle, E.; Danzi, D.; Bellon, O.; Tornielli, G.B.; Amato, A.; Zenoni, S. NAC61 regulates late- and post-ripening osmotic, oxidative, and biotic stress responses in grapevine. J. Exp. Bot. 2024, 75, 2330–2350. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, V.K.; Khurana, P. Genome-wide analysis, expression dynamics and varietal comparison of NAC gene family at various developmental stages in Morus notabilis. Mol. Genet. Genom. MGG 2016, 291, 1305–1317. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Fang, Y.; Cheng, J.; Tian, Y.; Liu, L.; Cao, X. Intrinsic morphology and spatial distribution of non-structural carbohydrates contribute to drought resistance of two mulberry cultivars. Plant Biol. 2023, 25, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Kang, X.; Huang, Y.; Guo, Z.; Wang, Y.; Ma, S.; Li, H.; Chao, N.; Liu, L. Functional characterization of MaEXPA11 and its roles in response to biotic and abiotic stresses in mulberry. Plant Physiol. Biochem. 2024, 206, 108289. [Google Scholar] [CrossRef]
- Xia, Z.; Fan, W.; Liu, D.; Chen, Y.; Lv, J.; Xu, M.; Zhang, M.; Ren, Z.; Chen, X.; Wang, X.; et al. Haplotype-resolved chromosomal-level genome assembly reveals regulatory variations in mulberry fruit anthocyanin content. Hortic. Res. 2024, 11, uhae120. [Google Scholar] [CrossRef]
- Meng, L.; Chen, S.; Li, D.; Huang, M.; Zhu, S. Genome-Wide characterization and evolutionary expansion of poplar NAC transcription factors and their tissue-specific expression profiles under drought. Int. J. Mol. Sci. 2022, 24, 253. [Google Scholar] [CrossRef]
- Gao, Y.; Fan, Z.Q.; Zhang, Q.; Li, H.L.; Liu, G.S.; Jing, Y.; Zhang, Y.P.; Zhu, B.Z.; Zhu, H.L.; Chen, J.Y.; et al. A tomato NAC transcription factor, SlNAM1, positively regulates ethylene biosynthesis and the onset of tomato fruit ripening. Plant J. 2021, 108, 1317–1331. [Google Scholar] [CrossRef]
- Ailizati, A.; Nagahage, I.S.P.; Miyagi, A.; Ishikawa, T.; Kawai-Yamada, M.; Demura, T.; Yamaguchi, M. An Arabidopsis NAC domain transcriptional activator VND7 negatively regulates VNI2 expression. Plant Biotechnol. 2021, 38, 415–420. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.H. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal. Behav. 2015, 10, e989746. [Google Scholar] [CrossRef]
- Kamiya, M.; Higashio, S.Y.; Isomoto, A.; Kim, J.M.; Seki, M.; Miyashima, S.; Nakajima, K. Control of root cap maturation and cell detachment by BEARSKIN transcription factors in Arabidopsis. Development 2016, 143, 4063–4072. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Heal, R.; Wyler, M.; Schmid, M.W.; Jones, J.D.G. Concerted expansion and contraction of immune receptor gene repertoires in plant genomes. Nat. Plants 2022, 8, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 288. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, X.; Chao, J.; Zhang, Z.; Wang, W.; Guo, Y. NAC family transcription factors in Tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 2018, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Alshareef, N.O.; Otterbach, S.L.; Allu, A.D.; Woo, Y.H.; de Werk, T.; Kamranfar, I.; Mueller-Roeber, B.; Tester, M.; Balazadeh, S.; Schmöckel, S.M. NAC transcription factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis. Sci. Rep. 2022, 12, 11264. [Google Scholar] [CrossRef]
- Xi, D.; Chen, X.; Wang, Y.; Zhong, R.; He, J.; Shen, J.; Ming, F. Arabidopsis ANAC092 regulates auxin-mediated root development by binding to the ARF8 and PIN4 promoters. J. Integr. Plant Biol. 2019, 61, 1015–1031. [Google Scholar] [CrossRef]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef]
- Dorjee, T.; Cui, Y.; Zhang, Y.; Liu, Q.; Li, X.; Sumbur, B.; Yan, H.; Bing, J.; Geng, Y.; Zhou, Y.; et al. Characterization of NAC gene family in Ammopiptanthus mongolicus and functional analysis of AmNAC24, an osmotic and cold-stress-induced NAC gene. Biomolecules 2024, 14, 182. [Google Scholar] [CrossRef]
- Chen, J.; Gong, Y.; Gao, Y.; Zhou, Y.; Chen, M.; Xu, Z.; Guo, C.; Ma, Y. TaNAC48 positively regulates drought tolerance and ABA responses in wheat (Triticum aestivum L.). Crop J. 2021, 9, 785–793. [Google Scholar] [CrossRef]
- Huang, D.; Wang, S.; Zhang, B.; Shang-Guan, K.; Shi, Y.; Zhang, D.; Liu, X.; Wu, K.; Xu, Z.; Fu, X.; et al. A Gibberellin-Mediated DELLA-NAC signaling cascade regulates cellulose synthesis in Rice. Plant Cell 2015, 27, 1681–1696. [Google Scholar] [CrossRef]
- Forlani, S.; Mizzotti, C.; Masiero, S. The NAC side of the fruit: Tuning of fruit development and maturation. BMC Plant Biol. 2021, 21, 238. [Google Scholar] [CrossRef]
- Liu, G.S.; Li, H.L.; Grierson, D.; Fu, D.Q. NAC transcription factor family regulation of fruit ripening and quality: A Review. Cells 2022, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, Y.; Zhao, L.; Li, C.; Yu, J.; Li, T.; Yang, W.; Zhang, S.; Su, H.; Wang, L. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 2020, 40, 413–423. [Google Scholar] [CrossRef]
- Zhang, R.-X.; Liu, Y.; Zhang, X.; Chen, X.; Sun, J.; Zhao, Y.; Zhang, J.; Yao, J.-L.; Liao, L.; Zhou, H.; et al. Two adjacent NAC transcription factors regulate fruit maturity date and flavor in peach. New Phytol. 2024, 241, 632–649. [Google Scholar] [CrossRef]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef]
- Zhou, J.; Zhong, R.; Ye, Z.-H. Arabidopsis NAC Domain Proteins, VND1 to VND5, Are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS ONE 2014, 9, e105726. [Google Scholar] [CrossRef]
- Tamadaddi, C.; Choi, J.; Ghasemi, M.; Kim, S.H.; Gomez, E.D.; Gomez, E.W.; Anderson, C.T. NST3 induces ectopic transdifferentiation, forming secondary walls with diverse patterns and composition in Arabidopsis thaliana. Ann. Bot. 2024, 134, 1097–1111. [Google Scholar] [CrossRef]
- Akiyoshi, N.; Ihara, A.; Matsumoto, T.; Takebayashi, A.; Hiroyama, R.; Kikuchi, J.; Demura, T.; Ohtani, M. Functional analysis of poplar Sombrero-Type NAC Transcription Factors yields a strategy to modify woody cell wall properties. Plant Cell Physiol 2021, 62, 1963–1974. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.-H. Functional Characterization of Poplar Wood-Associated NAC Domain Transcription Factors. Plant Physiol. 2009, 152, 1044–1055. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kudapa, H.; Garg, V.; Varshney, R.K. Comprehensive analysis and identification of drought-responsive candidate NAC genes in three semi-arid tropics (SAT) legume crops. BMC Genom. 2021, 22, 289. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, N.; Lodeyro, A.F.; Carrillo, N.; Gomez, R. Meta-analysis reveals key features of the improved drought tolerance of plants overexpressing NAC transcription factors. Environ. Exp. Bot. 2021, 186, 104449. [Google Scholar] [CrossRef]
- Chang, Y.; Fang, Y.; Liu, J.; Ye, T.; Li, X.; Tu, H.; Ye, Y.; Wang, Y.; Xiong, L. Stress-induced nuclear translocation of ONAC023 improves drought and heat tolerance through multiple processes in rice. Nat. Commun. 2024, 15, 5877. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.-S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Matsuoka, K.; Sato, R.; Matsukura, Y.; Kawajiri, Y.; Iino, H.; Nozawa, N.; Shibata, K.; Kondo, Y.; Satoh, S.; Asahina, M. Wound-inducible ANAC071 and ANAC096 transcription factors promote cambial cell formation in incised Arabidopsis flowering stems. Commun. Biol. 2021, 4, 369. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Kim, S.Y.; Hyeon do, Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.H.; Kim, S.K.; Hong, J.C.; et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef]
- Alvarado, V.Y.; Tag, A.; Thomas, T.L. A cis regulatory element in the TAPNAC promoter directs tapetal gene expression. Plant Mol. Biol. 2011, 75, 129–139. [Google Scholar] [CrossRef]
- Amin, N.; Du, Y.; Lu, L.; Khalifa, M.A.S.; Ahmad, N.; Ahmad, S.; Wang, P. GmNAC3 acts as a key regulator in soybean against drought stress. Curr. Plant Biol. 2024, 38, 100346. [Google Scholar] [CrossRef]
- Sintaha, M.; Man, C.K.; Yung, W.S.; Duan, S.; Li, M.W.; Lam, H.-M. Drought stress priming improved the drought tolerance of Soybean. Plants 2022, 11, 2954. [Google Scholar] [CrossRef]
- Dong, B.; Liu, Y.; Huang, G.; Song, A.; Chen, S.; Jiang, J.; Chen, F.; Fang, W. Plant NAC transcription factors in the battle against pathogens. BMC Plant Biol. 2024, 24, 958. [Google Scholar] [CrossRef] [PubMed]
- Vranic, M.; Perochon, A.; Benbow, H.; Doohan, F.M. Comprehensive analysis of pathogen-responsive wheat NAC transcription factors: New candidates for crop improvement. G3 2022, 12, jkac247. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, D.; Kong, X.; Song, Y.; Jing, L. Pangenome characterization and analysis of the NAC gene family reveals genes for Sclerotinia sclerotiorum resistance in sunflower (Helianthus annuus). BMC Genom. Data 2024, 25, 39. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Nobmann, B.; Devi Allu, A.; Balazadeh, S. JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal. Behav. 2016, 11, e1181245. [Google Scholar] [CrossRef]
- Afreen, U.; Kumar, M. 5-mC methylation study of sORFs in 3’UTR of transcription factor JUNGBRUNNEN 1-like during leaf rust pathogenesis in wheat. Mol. Biol. Rep. 2024, 51, 801. [Google Scholar] [CrossRef]
- Cao, X.; Du, W.; Shang, C.; Shen, Q.; Liu, L.; Cheng, J. Comparative transcriptome reveals circadian and hormonal control of adventitious rooting in mulberry hardwood cuttings. Acta Physiol. Plant. 2018, 40, 197. [Google Scholar] [CrossRef]
- Kang, X.; Huang, S.; Feng, Y.; Fu, R.; Tang, F.; Zheng, L.; Li, P.; Chao, N.; Liu, L. SWEET transporters and their potential roles in response to abiotic and biotic stresses in mulberry. Beverage Plant Res. 2023, 3, 6. [Google Scholar] [CrossRef]
- Tian, Y.; Zhai, Z.; Yang, Y.; Zhang, K.; Ma, S.; Cheng, J.; Liu, L.; Cao, X. Transcriptome-based WGCNA reveals the molecular regulation of xylem plasticity in acclimation to drought and rewatering in mulberry. Front. Plant Sci. 2024, 15, 1512645. [Google Scholar] [CrossRef]
- Liu, L.; Guo, Z.; Kang, X.; Li, S.; Huang, S.; Zheng, L.; Fu, R.; Yidilisi, K.; Chao, N. Comparative transcriptome analysis of different Mulberry varieties to reveal candidate genes and small secreted peptides involved in the Sclerotiniose response. Forests 2024, 15, 1126. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: “A one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Du, W.; Ban, Y.; Nie, H.; Tang, Z.; Du, X.; Cheng, J. A comparative transcriptome analysis leads to new insights into the molecular events governing root formation in Mulberry softwood cuttings. Plant Mol. Biol. Rep. 2016, 34, 365–373. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Kang, X.; Wang, Y.; Huang, S.; Guo, Y.; Wang, R.; Chao, N.; Liu, L. Functional characterization of flavanone 3-Hydroxylase (F3H) and its role in Anthocyanin and Flavonoid biosynthesis in Mulberry. Molecules 2022, 27, 3341. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for -mediated transformation of. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; He, M.; Zhang, K.; Zhai, Z.; Cheng, J.; Tian, Y.; Cao, X.; Liu, L. Genome-Wide Analysis of NAC Transcription Factor Gene Family in Morus atropurpurea. Plants 2025, 14, 1179. https://doi.org/10.3390/plants14081179
Yang Y, He M, Zhang K, Zhai Z, Cheng J, Tian Y, Cao X, Liu L. Genome-Wide Analysis of NAC Transcription Factor Gene Family in Morus atropurpurea. Plants. 2025; 14(8):1179. https://doi.org/10.3390/plants14081179
Chicago/Turabian StyleYang, Yujie, Meiyu He, Kaixin Zhang, Zeyang Zhai, Jialing Cheng, Yue Tian, Xu Cao, and Li Liu. 2025. "Genome-Wide Analysis of NAC Transcription Factor Gene Family in Morus atropurpurea" Plants 14, no. 8: 1179. https://doi.org/10.3390/plants14081179
APA StyleYang, Y., He, M., Zhang, K., Zhai, Z., Cheng, J., Tian, Y., Cao, X., & Liu, L. (2025). Genome-Wide Analysis of NAC Transcription Factor Gene Family in Morus atropurpurea. Plants, 14(8), 1179. https://doi.org/10.3390/plants14081179




