Characterization and Early Response of the DEAD Gene Family to Heat Stress in Tomato
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of SlDEAD Proteins
2.2. Collinearity Analysis and Chromosome Localization of SlDEAD Family Genes
2.3. Gene Structure and Conserved Motif Analysis of Tomato SlDEAD Genes
2.4. Subcellular Localization of SlDEAD Proteins
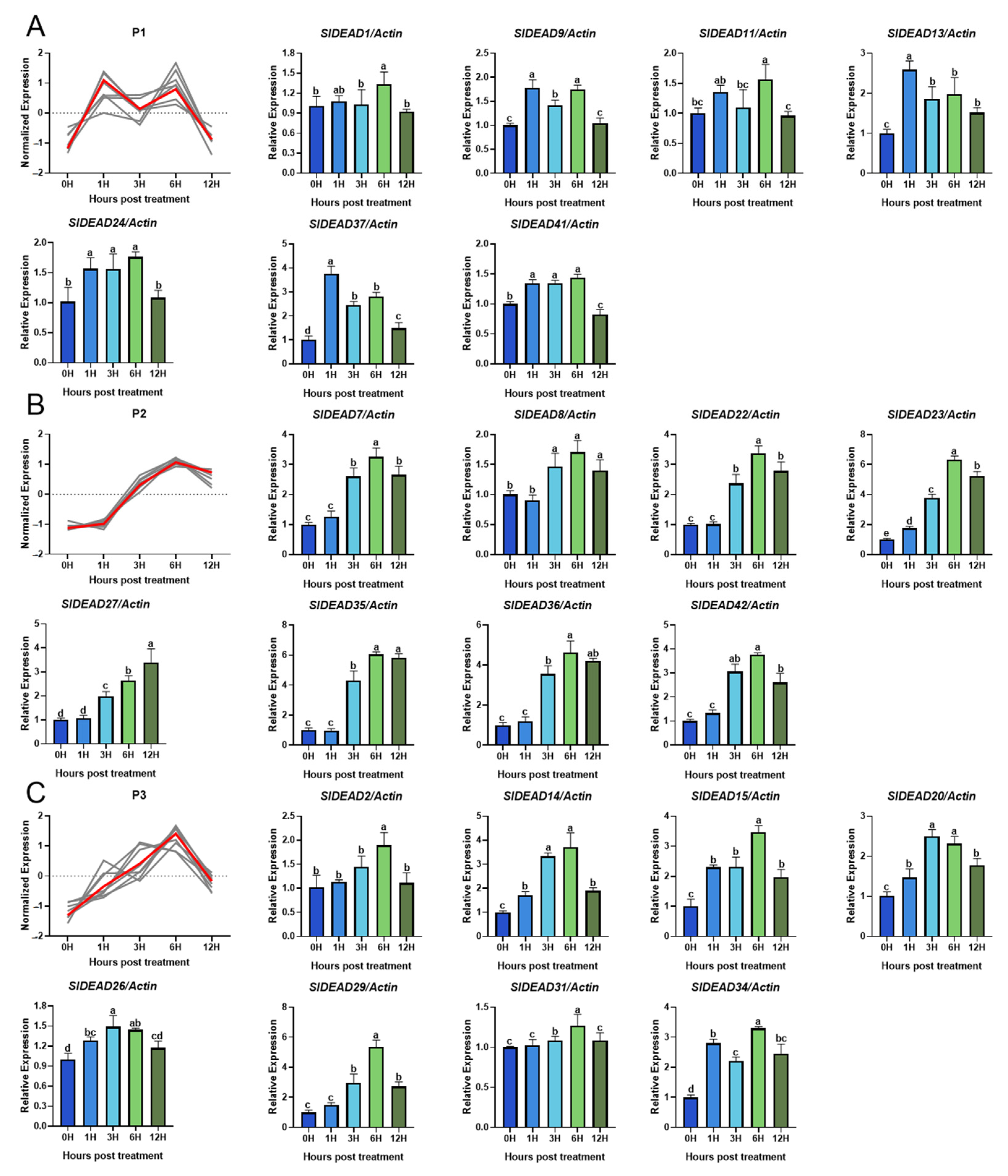
2.5. Expression Pattern of SlDEAD Genes in Response to High-Temperature Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Heat Stress Treatment
4.2. Analysis of DEAD Family Gene Structures
4.3. Analysis of SlDEAD Protein Properties
4.4. RT-qPCR Analysis
4.5. Subcellular Localization Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, Y.; Yang, S. Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 2022, 57, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lee, K.P.; Liu, Y.; Kim, C. Temperature-driven changes in membrane fluidity differentially impact FILAMENTATION TEMPERATURE-SENSITIVE H2-mediated photosystem II repair. Plant Cell 2025, 37, koae323. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Long-term mild heat causes post-mitotic pollen abortion through a local effect on flowers. Front. Plant Sci. 2022, 13, 925754. [Google Scholar] [CrossRef]
- Katano, K.; Honda, K.; Suzuki, N. Integration between ROS regulatory systems and other signals in the regulation of various types of heat responses in plants. Int. J. Mol. Sci. 2018, 19, 3370. [Google Scholar] [CrossRef]
- Schroda, M.; Vallon, O.; Wollman, F.-A.; Beck, C.F. A chloroplast-targeted Heat Shock Protein 70 (HSP70) contributes to the photoprotection and repair of Photosystem II during and after Photoinhibition. Plant Cell 1999, 11, 1165–1187. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; Koskull-Döring, P.V. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Aldubai, A.A.; Alsadon, A.A.; Migdadi, H.H.; Alghamdi, S.S.; Al-Faifi, S.A.; Afzal, M. Response of tomato (Solanum lycopersicum L.) genotypes to heat stress using morphological and expression study. Plants 2022, 11, 615. [Google Scholar] [CrossRef]
- Marondedze, C. The increasing diversity and complexity of the RNA-binding protein repertoire in plants. Proc. R. Soc. B Biol. Sci. 2020, 287, 20201397. [Google Scholar] [CrossRef]
- Yan, Y.; Gan, J.; Tao, Y.; Okita, T.W.; Tian, L. RNA-Binding Proteins: The Key Modulator in Stress Granule Formation and Abiotic Stress Response. Front. Plant Sci. 2022, 13, 882596. [Google Scholar] [CrossRef]
- Muthusamy, M.; Kim, J.H.; Kim, J.A.; Lee, S.I. Plant RNA binding proteins as critical modulators in drought, high salinity, heat, and cold stress responses: An updated overview. Int. J. Mol. Sci. 2021, 22, 6731. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Song, Y.; Wang, L.; Yang, D.; Yan, J.; Sun, Y.; Hsu, Y.F. CaRH57, a RNA helicase, contributes pepper tolerance to heat stress. Plant Physiol. Biochem. 2023, 205, 108202. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Kamphuis, L.G.; Hane, J.K.; Oñate-Sánchez, L.; Singh, K.B. The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS ONE 2015, 10, e0126978. [Google Scholar] [CrossRef] [PubMed]
- Mu, F.; Zheng, H.; Zhao, Q.; Zhu, M.; Dong, T.; Kai, L.; Li, Z. Genome-wide systematic survey and analysis of the RNA helicase gene family and their response to abiotic stress in sweetpotato. BMC Plant Biol. 2024, 24, 193. [Google Scholar] [CrossRef]
- You, L.; Shi, C.; Wang, D.; Fu, Z.Q. Helicases clear hurdles during plant defense protein translation. Trends Biochem. Sci. 2024, 49, 192–194. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Zhu, J.; Zhong, S.; Zhu, H.; Zhang, X. Functions and mechanisms of RNA helicases in plants. J. Exp. Bot. 2023, 74, 2295–2310. [Google Scholar] [CrossRef]
- Umate, P.; Tuteja, R.; Tuteja, N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: A comparison with yeast and human. Plant Mol. Biol. 2010, 73, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhang, S.; Huang, J.; Zheng, C. Genome-wide comparative in silico analysis of the RNA helicase gene family in Zea mays and Glycine max: A comparison with Arabidopsis and Oryza sativa. PLoS ONE 2013, 8, e78982. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Liu, J.; Xia, M.; Wang, W.; Shen, F. Genome-wide analysis of the RNA helicase gene family in Gossypium raimondii. Int. J. Mol. Sci. 2014, 15, 4635–4656. [Google Scholar] [CrossRef]
- Linder, P.; Owttrim, G.W. Plant RNA helicases: Linking aberrant and silencing RNA. Trends Plant Sci. 2009, 14, 344–352. [Google Scholar] [CrossRef]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Jankowsky, A.; Guenther, U.P.; Jankowsky, E. The RNA helicase database. Nucleic Acids Res. 2011, 39, D338–D341. [Google Scholar] [CrossRef]
- Tanner, N.K. The newly identified Q motif of DEAD box helicases is involved in adenine recognition. Cell Cycle 2003, 2, 18–19. [Google Scholar] [CrossRef] [PubMed]
- Rocak, S.; Linder, P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004, 5, 232–241. [Google Scholar] [CrossRef]
- Guan, Q.; Wu, J.; Zhang, Y.; Jiang, C.; Liu, R.; Chai, C.; Zhu, J. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 2013, 25, 342–356. [Google Scholar] [CrossRef]
- Lorsch, J.R. RNA chaperones exist and DEAD box proteins get a life. Cell 2002, 109, 797–800. [Google Scholar] [CrossRef]
- Wang, H.; Ye, T.; Guo, Z.; Yao, Y.; Tu, H.; Wang, P.; Zhang, Y.; Wang, Y.; Li, X.; Li, B.; et al. A double-stranded RNA binding protein enhances drought resistance via protein phase separation in rice. Nat. Commun. 2024, 15, 2514. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Bohnsack, M.T. Unravelling the Mechanisms of RNA Helicase Regulation. Trends Biochem. Sci. 2018, 43, 237–250. [Google Scholar] [CrossRef]
- Pandey, S.; Muthamilarasan, M.; Sharma, N.; Chaudhry, V.; Dulani, P.; Shweta, S.; Jha, S.; Mathur, S.; Prasad, M. Characterization of DEAD-box family of RNA helicases in tomato provides insights into their roles in biotic and abiotic stresses. Environ. Exp. Bot. 2019, 158, 107–116. [Google Scholar] [CrossRef]
- Hou, X.L.; Chen, W.Q.; Hou, Y.; Gong, H.Q.; Sun, J.; Wang, Z.; Zhao, H.; Cao, X.; Song, X.F.; Liu, C.M. DEAD-BOX RNA HELICASE 27 regulates microRNA biogenesis, zygote division, and stem cell homeostasis. Plant Cell 2021, 33, 66–84. [Google Scholar] [CrossRef]
- Iserman, C.; Desroches Altamirano, C.; Jegers, C.; Friedrich, U.; Zarin, T.; Fritsch, A.W.; Mittasch, M.; Domingues, A.; Hersemann, L.; Jahnel, M.; et al. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 2020, 181, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, H.; Zhang, H.; Wang, X.; Song, F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J. Exp. Bot. 2008, 59, 2133–2146. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.A.; Huang, C.K.; Huang, W.S.; Huang, T.S.; Liu, H.Y.; Chen, Y.F. DEAD-Box RNA helicase 42 plays a critical role in pre-mRNA splicing under cold stress. Plant Physiol. 2020, 182, 255–271. [Google Scholar] [CrossRef]
- Baek, W.; Lim, C.W.; Lee, S.C. A DEAD-box RNA helicase, RH8, is critical for regulation of ABA signalling and the drought stress response via inhibition of PP2CA activity. Plant Cell Environ. 2018, 41, 1593–1604. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lim, C.W.; Lan, W.Z.; He, K.; Luan, S. ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 2013, 6, 528–538. [Google Scholar] [CrossRef]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Env. 2012, 35, 53–60. [Google Scholar] [CrossRef]
- Huang, C.K.; Shen, Y.L.; Huang, L.F.; Wu, S.J.; Yeh, C.H.; Lu, C.A. The DEAD-Box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in Arabidopsis. Plant Cell Physiol. 2016, 57, 174–191. [Google Scholar] [CrossRef]
- Xiaomei, W.; Rongrong, K.; Ting, Z.; Yuanyuan, G.; Jianlong, X.; Zhongze, P.; Gangseob, L.; Dongzhi, L.; Yanjun, D. A DEAD-box RNA helicase TCD33 that confers chloroplast development in rice at seedling stage under cold stress. J. Plant Physiol. 2020, 248, 153138. [Google Scholar] [CrossRef]
- José, O.G.A.; Nieves, F.G.; Carmen, L.B.; Isabel, E.; Francisco, B.F.; Trinidad, A.; Juan, C.; Rafael, L.; Benito, P.; Vicente, M.; et al. The tomato res mutant which accumulates JA in roots in non-stressed conditions restores cell structure alterations under salinity. Physiol. Plant 2015, 155, 296–314. [Google Scholar] [CrossRef]
- Albaladejo, I.; Egea, I.; Morales, B.; Flores, F.B.; Capel, C.; Lozano, R.; Bolarin, M.C. Identification of key genes involved in the phenotypic alterations of res (restored cell structure by salinity) tomato mutant and its recovery induced by salt stress through transcriptomic analysis. BMC Plant Biol. 2018, 18, 213. [Google Scholar] [CrossRef]
- Capel, C.; Albaladejo, I.; Egea, I.; Massaretto, I.L.; Yuste-Lisbona, F.J.; Pineda, B.; Garcia-Sogo, B.; Angosto, T.; Flores, F.B.; Moreno, V.; et al. The res (restored cell structure by salinity) tomato mutant reveals the role of the DEAD-box RNA helicase SlDEAD39 in plant development and salt response. Plant Cell Environ. 2020, 43, 1722–1739. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Dong, T.; Wang, L.; Zhang, J.; Zhao, Z.; Hu, Z. SlDEAD31, a Putative DEAD-Box RNA helicase gene, regulates salt and drought tolerance and stress-related genes in tomato. PLoS ONE 2015, 10, e0133849. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhang, S.; Lu, L.; Cao, H.; Zheng, C. A genome-wide analysis of the RNA helicase gene family in Solanum lycopersicum. Gene 2013, 513, 128–140. [Google Scholar] [CrossRef]
- Cui, L.; Wall, P.K.; Leebens-Mack, J.H.; Lindsay, B.G.; Soltis, D.E.; Doyle, J.J.; Soltis, P.S.; Carlson, J.E.; Arumuganathan, K.; Barakat, A.; et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006, 16, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Guo, J.; Sun, W.; Wang, Y. Whole genome duplication of intra- and inter-chromosomes in the tomato genome. J. Genet. Genom. 2012, 39, 361–368. [Google Scholar] [CrossRef]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef]
- Caruthers, J.M.; McKay, D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002, 12, 123–133. [Google Scholar] [CrossRef]
- Cordin, O.; Tanner, N.K.; Monique, D.; Linder, P.; Banroques, J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004, 23, 2478–2487. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Pause, A.; Haghighat, A.; Pyronnet, S.; Witherell, G.; Belsham, G.J.; Sonenberg, N. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 2001, 7, 382–394. [Google Scholar] [CrossRef]
- Rocak, S.; Emery, B.; Tanner, N.K.; Linder, P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 2005, 33, 999–1009. [Google Scholar] [CrossRef]
- Xing, Y.H.; Yao, R.W.; Zhang, Y.; Guo, C.J.; Chen, L.L. SLERT regulates DDX21 rings associated with Pol I transcription. Cell 2017, 169, 664–678. [Google Scholar] [CrossRef]
- Andreou, A.Z.; Klostermeier, D. The DEAD-box helicase eIF4A: Paradigm or the odd one out? RNA Biol. 2013, 10, 19–32. [Google Scholar] [CrossRef]
- Choe, J.; Ryu, I.; Park, O.H.; Park, J.; Cho, H.; Yoo, J.S.; Chi, S.W.; Kim, M.K.; Song, H.K.; Kim, Y.K. eIF4AIII enhances translation of nuclear cap-binding complex-bound mRNAs by promoting disruption of secondary structures in 5′UTR. Proc. Natl. Acad. Sci. USA 2014, 111, 4577–4586. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Tuteja, N. microRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 183. [Google Scholar] [CrossRef]
- Shivakumara, T.N.; Sreevathsa, R.; Dash, P.K.; Sheshshayee, M.S.; Papolu, P.K.; Rao, U.; Tuteja, N.; Udayakumar, M. Overexpression of Pea DNA Helicase 45 (PDH45) imparts tolerance to multiple abiotic stresses in chili (Capsicum annuum L.). Sci. Rep. 2017, 7, 2760. [Google Scholar] [CrossRef]
- Ma, L.; Yang, Y.; Wang, Y.; Cheng, K.; Zhou, X.; Li, J.; Zhang, J.; Li, R.; Zhang, L.; Wang, K.; et al. SlRBP1 promotes translational efficiency via SleIF4A2 to maintain chloroplast function in tomato. Plant Cell 2022, 34, 2747–2764. [Google Scholar] [CrossRef]
- Wang, S.; Meng, Y.; Ding, F.; Yang, K.; Wang, C.; Zhang, H.; Jin, H. Comparative Analysis of TPR Gene Family in Cucurbitaceae and Expression Profiling under Abiotic Stress in Cucumis melo L. Horticulturae 2024, 10, 83. [Google Scholar] [CrossRef]
- Tong, J.J.; Ren, Z.T.; Linhua, S.; Zhou, S.X.; Yuan, W.; Hui, Y.F.; Ci, D.; Wang, W.; Fan, L.M.; Wu, Z.; et al. ALBA proteins confer thermotolerance through stabilizing HSF messenger RNAs in cytoplasmic granules. Nat. Plants 2022, 8, 778–791. [Google Scholar] [CrossRef]
- Samanta, N.; Ribeiro, S.; Becker, M.; Laborie, E.; Pollak, R.; Timr, S.; Sterpone, F.; Ebbinghaus, S. Sequestration of proteins in stress granules relies on the in-cell but not the in Vitro folding stability. J. Am. Chem. Soc. 2021, 143, 19909–19918. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gu, J.; Yao, J.; Li, Y.; Zhang, Z.; Xia, W.; Wang, Z.; Gui, X.; Li, L.; Li, D.; et al. Liquid-liquid phase separation of RBGD2/4 is required for heat stress resistance in Arabidopsis. Dev. Cell 2022, 57, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, A.A.; Tuteja, N. Stress responsive DEAD-box helicases: A new pathway to engineer plant stress tolerance. J. Photochem. Photobiol. B Biol. 2006, 84, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Kant, P.; Kant, S.; Gordon, M.; Shaked, R.; Barak, S. STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 2007, 145, 814–830. [Google Scholar] [CrossRef] [PubMed]
- Toribio, R.; Muñoz, A.; Castro-Sanz, A.B.; Merchante, C.; Castellano, M.M. A novel eIF4E-interacting protein that forms non-canonical translation initiation complexes. Nat. Plants 2019, 5, 1283–1296. [Google Scholar] [CrossRef]
- Weber, C.; Nover, L.; Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008, 56, 517–530. [Google Scholar] [CrossRef]
- Wang, D.; Qin, B.; Li, X.; Tang, D.; Zhang, Y.e.; Cheng, Z.; Xue, Y. Nucleolar DEAD-Box RNA helicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice. PLoS Genet. 2016, 12, 23. [Google Scholar] [CrossRef]
- Nawaz, G.; Sai, T.Z.T.; Lee, K.; Kim, Y.O.; Kang, H.S. Rice DEAD-box RNA helicase OsRH53 has negative impact on Arabidopsis response to abiotic stresses. Plant Growth Regul. 2018, 85, 153–163. [Google Scholar] [CrossRef]
- Bortesi, L. and Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Tanner, N.K.; Cordin, O.; Banroques, J.; Doère, M.; Linder, P. The Q Motif: A newly identified motif in DEAD Box helicases may regulate ATP binding and hydrolysis. Mol. Cell 2003, 11, 127–138. [Google Scholar] [CrossRef]
- Si, Z.; Wang, L.; Ji, Z.; Zhao, M.; Zhang, K.; Qiao, Y. Comparative analysis of the MYB gene family in seven Ipomoea species. Front. Plant Sci. 2023, 14, 1155018. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma Biomath. 2013, 3, 71–85. [Google Scholar]
- Guo, Y.; Bao, Z.; Deng, Y.; Li, Y.; Wang, P. Protein subcellular localization and functional studies in horticultural research: Problems, solutions, and new approaches. Hortic. Res. 2023, 10, uhac271. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-Y.; Chen, H.J. Single-stage analysis of variance under heteroscedasticity. Commun. Stat.-Simul. Comput. 2010, 27, 641–666. [Google Scholar] [CrossRef]







| Gene Name | Gene ID | Genomic Position | Predicted Location | Amino Acid | Mass (Da) | Isoelectric Point | Coefficient of Instability | Number of Exons | Homologous Gene |
|---|---|---|---|---|---|---|---|---|---|
| SlDEAD1 | solyc01g005960 | 626,619–632,574 | cyto | 604 | 65,709.62 | 8.16 | 41.9 | 6 | AT2G42520.1 |
| SlDEAD2 | solyc01g057760 | 56,514,938–56,523,137 | nucl | 1221 | 135,061.40 | 9.96 | 63.63 | 11 | AT3G06480.1 |
| SlDEAD3 | solyc01g079330 | 70,910,189–70,918,827 | nucl | 774 | 84,440.06 | 6.06 | 39.78 | 5 | AT2G47330.1 |
| SlDEAD4 | solyc01g094350 | 77,623,834–77,631,727 | nucl | 498 | 56,905.28 | 8.73 | 45.84 | 9 | AT4G00660.2 |
| SlDEAD5 | solyc01g095740 | 78,699,069–78,705,889 | chlo | 870 | 96,191.09 | 9.17 | 51.97 | 10 | AT5G08610.1 |
| SlDEAD6 | solyc02g068190 | 32,785,826–32,787,814 | nucl | 662 | 75,669.38 | 9.27 | 40.07 | 1 | AT2G33730.1 |
| SlDEAD7 | solyc02g070100 | 34,512,603–34,519,106 | cyto | 585 | 65,234.03 | 8.95 | 39.07 | 15 | AT4G34910.1 |
| SlDEAD8 | solyc02g078880 | 38,098,218–38,103,721 | nucl | 594 | 66,304.59 | 9.04 | 41.17 | 11 | AT5G05450.1 |
| SlDEAD9 | solyc02g081290 | 39,890,841–39,892,976 | nucl | 653 | 76,086.82 | 8.93 | 44.51 | 1 | AT2G33730.1 |
| SlDEAD10 | solyc02g086660 | 43,885,348–43,891,471 | nucl | 706 | 77,150.90 | 9.33 | 52.1 | 8 | AT2G33730.1 |
| SlDEAD11 | solyc03g052980 | 19,500,306–19,506,839 | chlo | 612 | 66,413.24 | 7.28 | 39.88 | 6 | AT2G42520.1 |
| SlDEAD12 | solyc03g112350 | 56,827,775–56,836,207 | nucl | 651 | 70,294.39 | 9.85 | 46.22 | 10 | AT5G63120.2 |
| SlDEAD13 | solyc03g114370 | 58,425,702–58,432,243 | nucl | 566 | 64,392.81 | 8.97 | 45.68 | 12 | AT3G18600.1 |
| SlDEAD14 | solyc03g117440 | 60,661,466–60,664,137 | cyto | 595 | 66,647.42 | 6.11 | 45.02 | 27 | AT5G51280.1 |
| SlDEAD15 | solyc04g081580 | 63,117,504–63,123,510 | nucl | 744 | 84,016.41 | 8.35 | 45.75 | 19 | AT4G16630.1 |
| SlDEAD16 | solyc04g082790 | 63,926,584–63,931,290 | nucl | 499 | 55,268.09 | 5.26 | 36.21 | 8 | AT3G53110.1 (LOS4) |
| SlDEAD17 | solyc05g006130 | 839,121–843,666 | chlo | 559 | 62,335.58 | 6.5 | 46.64 | 7 | AT1G59990.1 |
| SlDEAD18 | solyc05g048850 | 58,709,311–58,724,460 | nucl | 537 | 61,075.08 | 8.76 | 38.23 | 10 | AT4G00660.2 |
| SlDEAD19 | solyc06g062800 | 36,012,777–36,014,915 | nucl | 413 | 46,827.74 | 5.58 | 43.31 | 4 | AT1G54270.1 |
| SlDEAD20 | solyc06g068280 | 38,699,501–38,702,707 | cyto | 595 | 66,829.38 | 6.09 | 48.06 | 3 | AT5G51280.1 |
| SlDEAD21 | solyc07g040750 | 46,856,884–46,859,084 | nucl | 413 | 46,659.80 | 5.21 | 47.44 | 4 | AT1G54270.1 |
| SlDEAD22 | solyc07g042270 | 52,721,025–52,727,332 | nucl | 597 | 67,629.27 | 9.54 | 38.7 | 10 | AT2G40700.1 |
| SlDEAD23 | solyc07g044760 | 55,132,119–55,137,026 | chlo | 630 | 67,146.29 | 9.43 | 33.14 | 7 | AT3G22330.1 |
| SlDEAD24 | solyc07g064520 | 63,853,690–63,858,103 | nucl | 754 | 85,281.23 | 8.97 | 42.64 | 8 | AT5G54910.1 |
| SlDEAD25 | solyc08g042050 | 28924,652–28934,324 | chlo | 746 | 81,518.47 | 6.26 | 46.94 | 10 | AT5G26742.2 |
| SlDEAD26 | solyc08g062800 | 49,300,134–49,301,991 | nucl | 413 | 46,887.76 | 5.54 | 48.39 | 4 | AT1G54270.1 |
| SlDEAD27 | solyc08g076200 | 57,382,754–57,390,648 | nucl | 553 | 61,228.04 | 8.75 | 33.96 | 13 | AT1G31970.1 (STRS1) |
| SlDEAD28 | solyc09g015930 | 11,382,872–11,400,498 | chlo | 558 | 60,842.18 | 6.62 | 46.01 | 14 | AT3G58570.1 |
| SlDEAD29 | solyc09g090740 | 65,522,411–65,531,624 | nucl | 799 | 90,445.84 | 9.06 | 51.31 | 15 | AT3G16840.1 |
| SlDEAD30 | solyc10g005520 | 426,164–431,795 | nucl | 488 | 54,692.30 | 8.53 | 38.21 | 6 | AT1G16280.1 |
| SlDEAD31 | solyc10g007550 | 1,854,816–1,862,972 | nucl | 439 | 48,897.70 | 9.16 | 44.83 | 13 | AT5G60990.1 |
| SlDEAD32 | solyc10g009070 | 3,093,425–3,105,024 | cyto | 785 | 87,843.93 | 9.9 | 36.06 | 11 | AT1G77030.1 |
| SlDEAD33 | solyc10g017530 | 5,410,295–5,419,584 | chlo | 791 | 55,720.00 | 8.66 | 40.29 | 9 | AT4G00660.2 |
| SlDEAD34 | solyc10g085790 | 64,194,367–64,199,371 | nucl | 508 | 56,255.08 | 5.22 | 36.31 | 8 | AT3G53110.1 (LOS4) |
| SlDEAD35 | solyc12g006320 | 843,041–848,893 | mito | 643 | 69,762.97 | 9.56 | 43.14 | 7 | AT3G22330.1 |
| SlDEAD36 | solyc12g035130 | 23,234,550–23,240,564 | nucl | 612 | 66,663.87 | 9.7 | 47.16 | 8 | AT3G01540.2 |
| SlDEAD37 | solyc12g044860 | 45,651,195–45,658,063 | nucl | 479 | 53,288.40 | 8.75 | 48.47 | 10 | AT1G55150.1 |
| SlDEAD38 | solyc12g056340 | 47,655,899–47,661,775 | chlo | 805 | 92,434.54 | 8.89 | 48.82 | 10 | AT1G63250.1 |
| SlDEAD39 | solyc12g056740 | 48,175,584–48,181,629 | chlo | 636 | 70,077.55 | 9.88 | 49.59 | 9 | AT4G09730.1 |
| SlDEAD40 | solyc12g095990 | 63,417,531–63,419,666 | nucl | 413 | 46,843.66 | 5.46 | 47.67 | 4 | AT1G54270.1 |
| SlDEAD41 | solyc12g096000 | 63,425,877–63,428,469 | nucl | 394 | 44,850.52 | 5.64 | 46.25 | 4 | AT1G54270.1 |
| SlDEAD42 | solyc12g098700 | 64,461,108–64,464,551 | nucl | 1147 | 130,890.60 | 6 | 38.69 | 1 | AT1G20920.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Yu, C.; Xie, B.; Zhou, H.; Zhang, C.; Tian, L. Characterization and Early Response of the DEAD Gene Family to Heat Stress in Tomato. Plants 2025, 14, 1172. https://doi.org/10.3390/plants14081172
Yan Y, Yu C, Xie B, Zhou H, Zhang C, Tian L. Characterization and Early Response of the DEAD Gene Family to Heat Stress in Tomato. Plants. 2025; 14(8):1172. https://doi.org/10.3390/plants14081172
Chicago/Turabian StyleYan, Yanyan, Chao Yu, Bolun Xie, Hui Zhou, Caiyu Zhang, and Li Tian. 2025. "Characterization and Early Response of the DEAD Gene Family to Heat Stress in Tomato" Plants 14, no. 8: 1172. https://doi.org/10.3390/plants14081172
APA StyleYan, Y., Yu, C., Xie, B., Zhou, H., Zhang, C., & Tian, L. (2025). Characterization and Early Response of the DEAD Gene Family to Heat Stress in Tomato. Plants, 14(8), 1172. https://doi.org/10.3390/plants14081172







