Genome-Wide Identification of the CYP78A Gene Family in Lycium and Functional Characterization of LrCYP78A5
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Characterization of CYP78A Genes in Goji
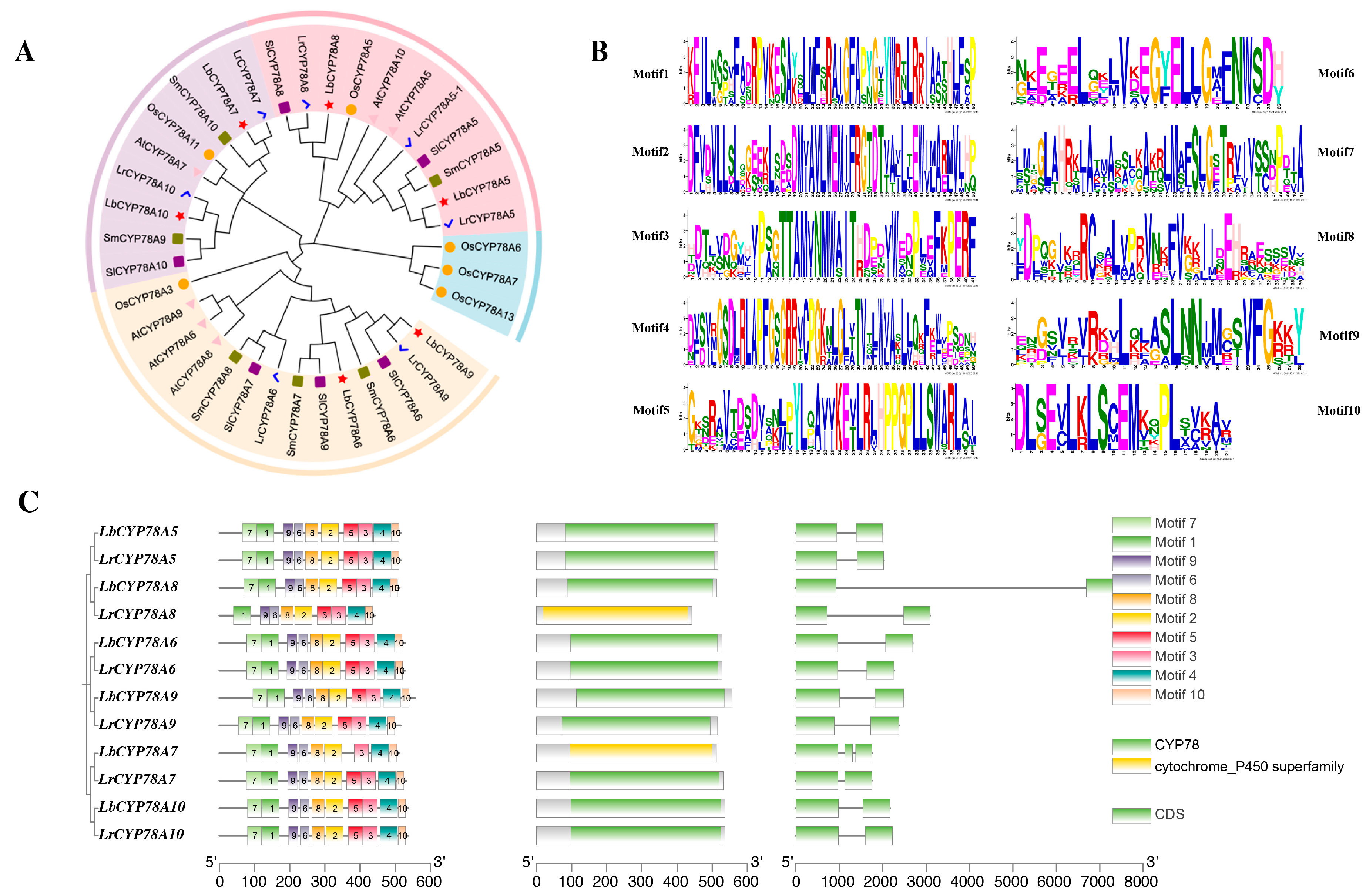
2.2. Analysis of Phylogeny, Gene Structures, and Conserved Motifs of the CYP78A Family in Lycium
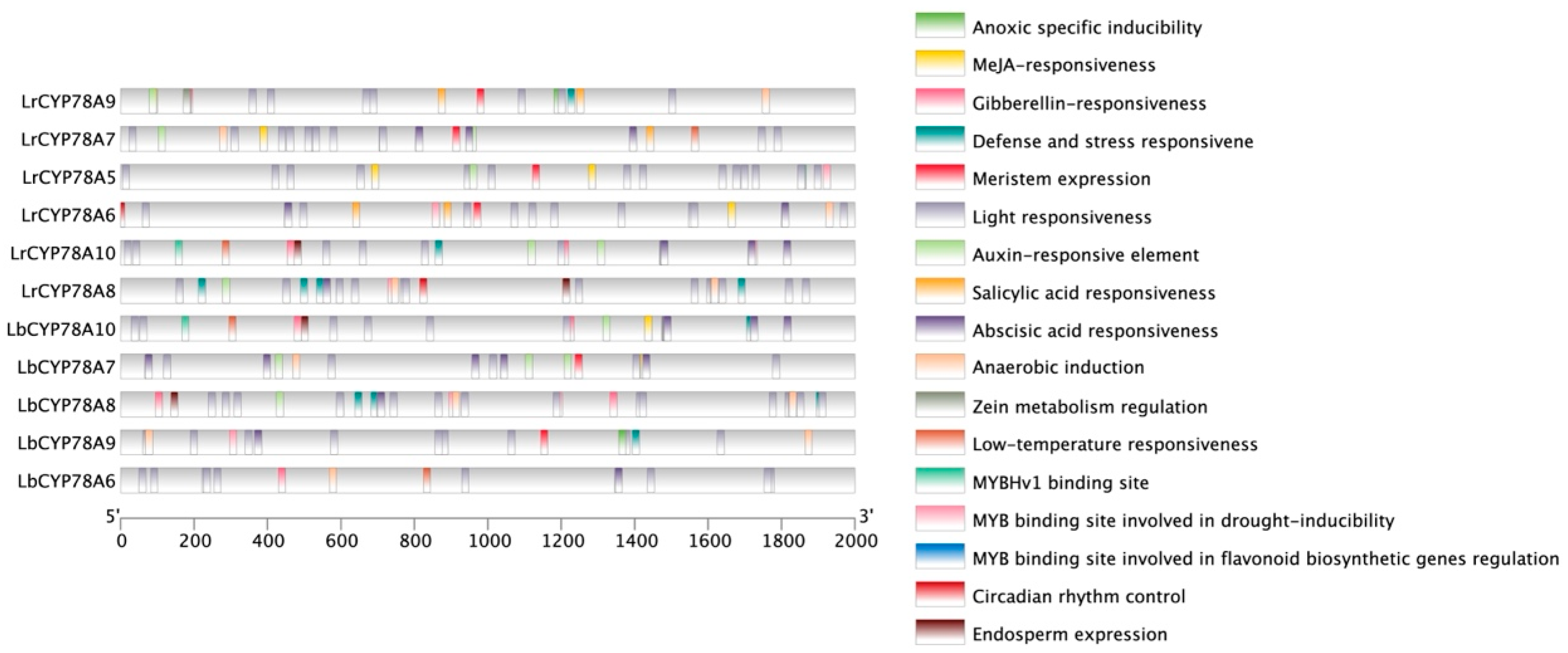
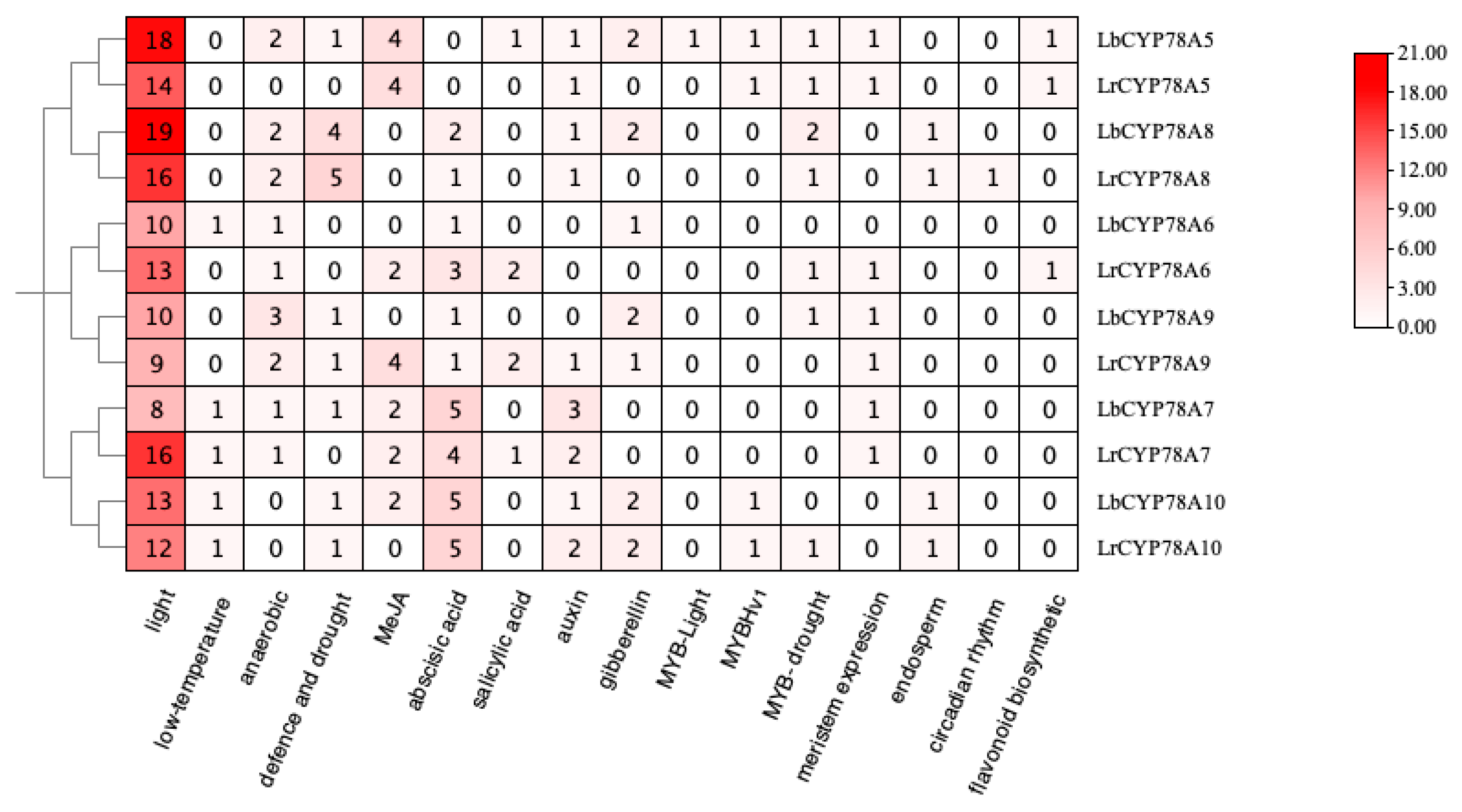
2.3. Analysis of Cis-Acting Elements in the Promoter Regions of CYP78A Genes in Lycium
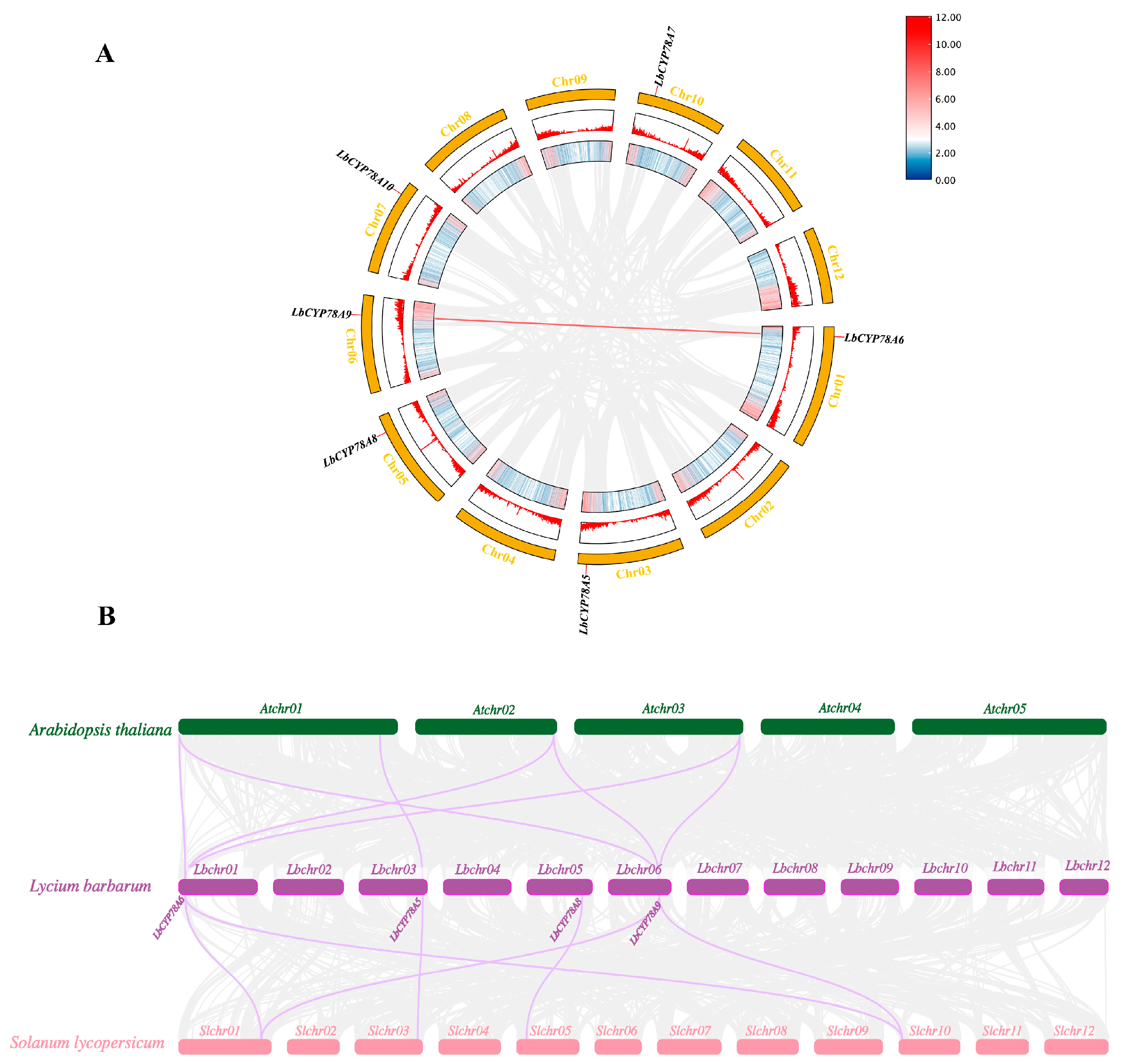
2.4. Analysis of Covariance Between L. barbarum and Other Plant Species
2.5. Expression Patterns of CYP78A Genes in L. barbarum, and L. ruthenicum
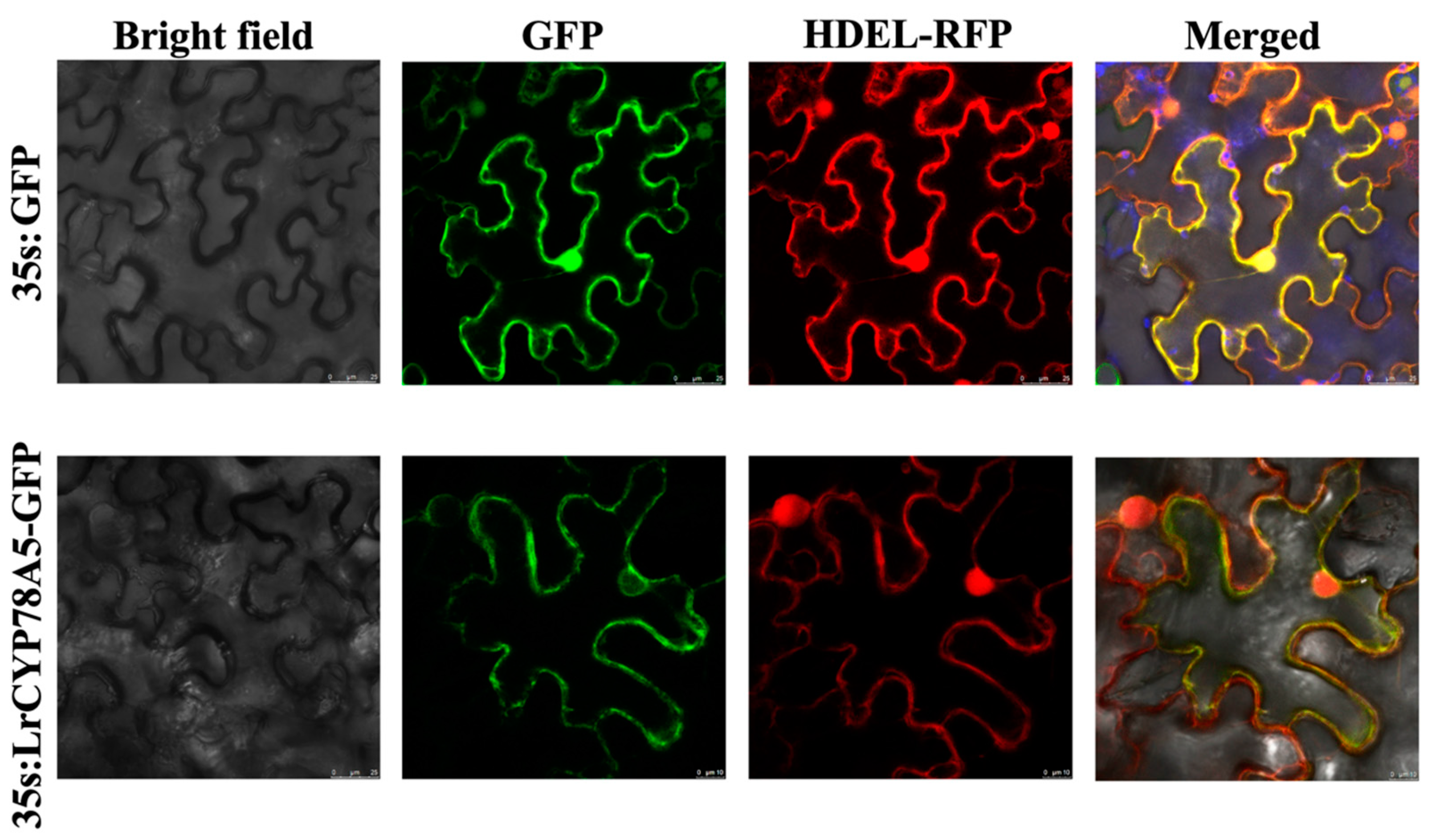
2.6. Subcellular Localization of CYP78A5
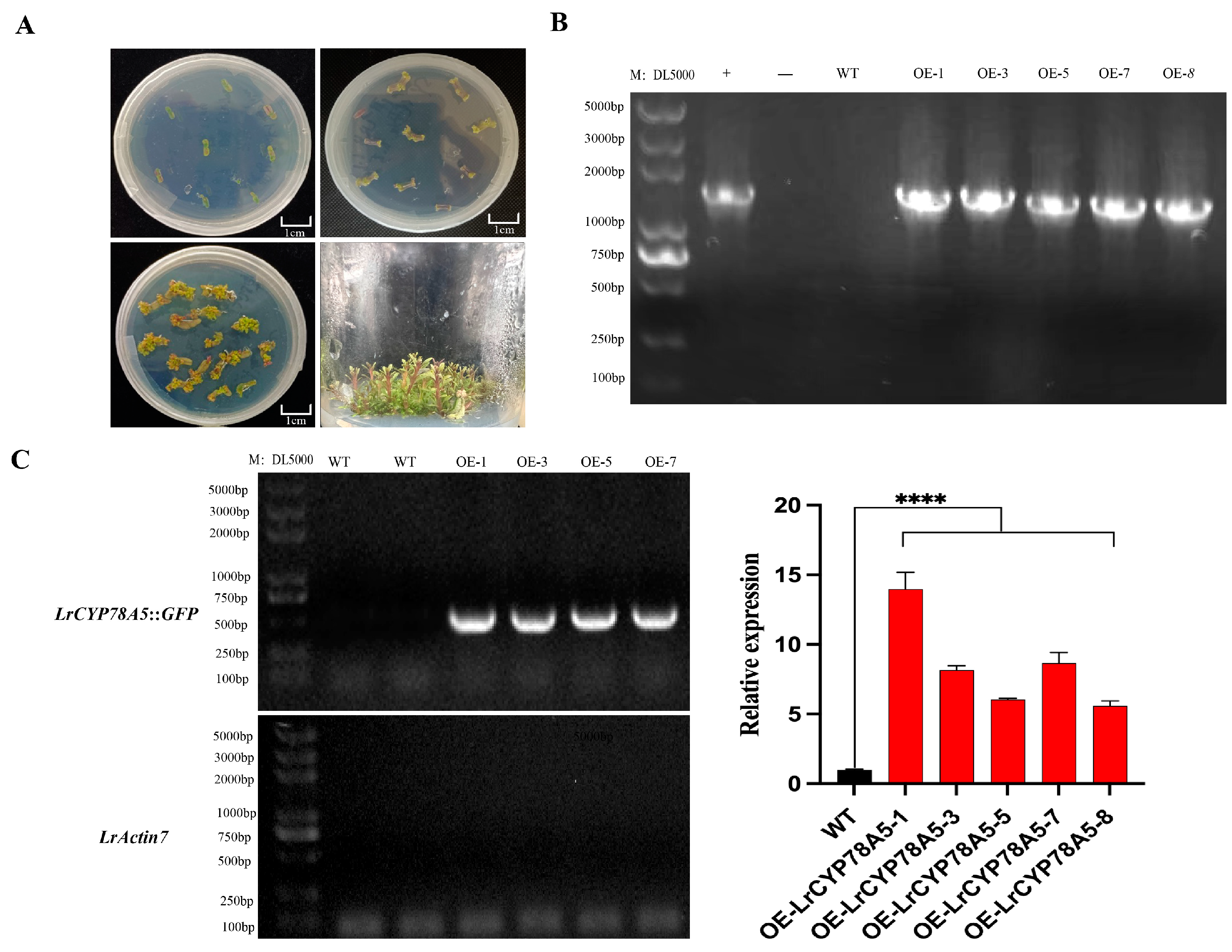
2.7. Overexpression and Identification of LrCYP78A5 in L. ruthenicum
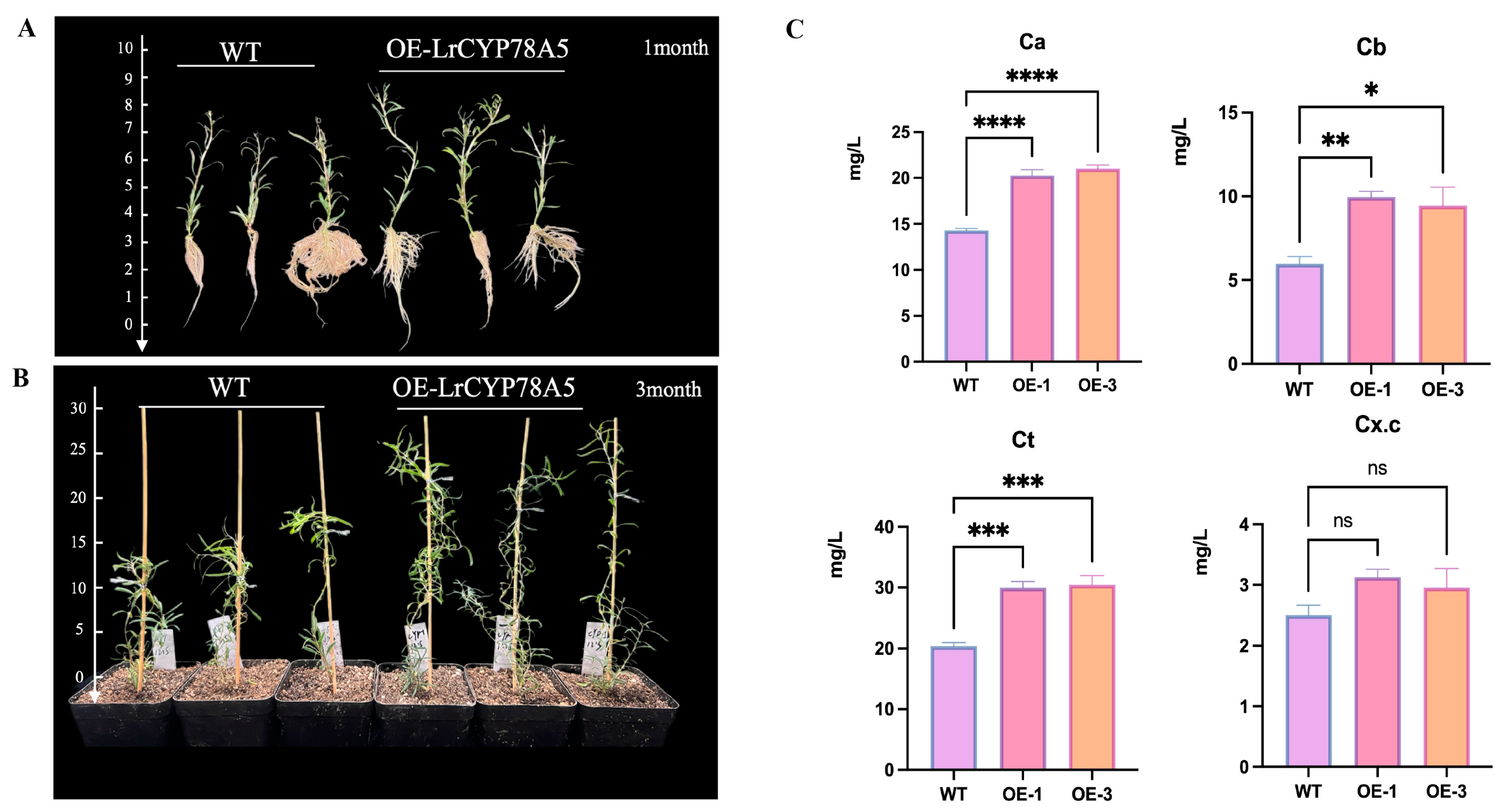
2.8. Phenotypic Observation and Chlorophyll Content Measurement of LrCYP78A5-Overexpressing Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of Members of the CYP78A Gene Family in Lycium
4.3. Analysis of CYP78A Proteins in Goji Berry
4.4. Conserved Motif and Phylogenetic Analysis of CYP78A in Lycium
4.5. Prediction of Cis-Acting Elements in the Promoter Regions of CYP78A Genes in Goji Berry
4.6. Expression Patterns of CYP78A Genes
4.7. Overexpression of LrCYP78A5 in L. ruthenicum
4.8. Obtaining and Identifying Overexpression LrCYP78A5 Plants
4.9. Subcellular Localization of LrCYP78A5
4.10. Real-Time Quantitative PCR Analysis
4.11. Determination of Photosynthetic Pigments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mizutani, M.; Ohta, D. Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 2010, 61, 291–315. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [PubMed]
- Schuler, M.A.; Werck-Reichhart, D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [PubMed]
- Adamski, N.M.; Anastasiou, E.; Eriksson, S.; O’Neill, C.M.; Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20115–20120. [Google Scholar]
- Anastasiou, E.; Kenz, S.; Gerstung, M.; MacLean, D.; Timmer, J.; Fleck, C.; Lenhard, M. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 2007, 13, 843–856. [Google Scholar]
- Kajino, T.; Yamaguchi, M.; Oshima, Y.; Nakamura, A.; Narushima, J.; Yaguchi, Y.; Yotsui, I.; Sakata, Y.; Taji, T. KLU/CYP78A5, a cytochrome P450 monooxygenase identified via fox hunting, contributes to cuticle biosynthesis and improves various abiotic stress tolerances. Front. Plant Sci. 2022, 13, 904121. [Google Scholar] [CrossRef]
- Jiang, L.; Yoshida, T.; Stiegert, S.; Jing, Y.; Alseekh, S.; Lenhard, M.; Pérez-Alfocea, F.; Fernie, A.R. Multiomics approach reveals the contribution of KLU to leaf longevity and drought tolerance. Plant Physiol. 2021, 185, 352–368. [Google Scholar]
- Zhou, C.; Lin, Q.; Ren, Y.; Lan, J.; Miao, R.; Feng, M.; Wang, X.; Liu, X.; Zhang, S.; Pan, T.; et al. A CYP78As–small grain4–coat protein complex II pathway promotes grain size in rice. Plant Cell 2023, 35, 4325–4346. [Google Scholar]
- Wang, J.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar]
- Nobusawa, T.; Kamei, M.; Ueda, H.; Matsushima, N.; Yamatani, H.; Kusaba, M. Highly pleiotropic functions of CYP78As and AMP1 are regulated in non-cell-autonomous/organ-specific manners. Plant Physiol. 2021, 186, 767–781. [Google Scholar] [PubMed]
- Fang, W.; Wang, Z.; Cui, R.; Li, J.; Li, Y. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 2012, 70, 929–939. [Google Scholar] [PubMed]
- Sotelo-Silveira, M.; Cucinotta, M.; Chauvin, A.L.; Chávez Montes, R.A.; Colombo, L.; Marsch-Martínez, N.; de Folter, S. Cytochrome P450 CYP78A9 is involved in Arabidopsis reproductive development. Plant Physiol. 2013, 162, 779–799. [Google Scholar] [PubMed]
- Wang, C.; Ma, S.; Sun, F.; Wei, B.; Nie, Y. Spatial genetic patterns of the medicinal and edible shrub Lycium ruthenicum (Solanaceae) in arid Xinjiang, China. Tree Genet. Genomes 2021, 17, 22. [Google Scholar]
- Rao, S.; Tian, Y.; Xia, X.; Li, Y.; Chen, J. Chromosome doubling mediates superior drought tolerance in Lycium ruthenicum via abscisic acid signaling. Hortic. Res. 2020, 7, 40. [Google Scholar]
- Ni, W.; Gao, T.; Wang, H.; Du, Y.; Li, J.; Li, C.; Wei, L.; Bi, H. Anti-fatigue activity of polysaccharides from the fruits of four Tibetan plateau indigenous medicinal plants. J. Ethnopharmacol. 2013, 150, 529–535. [Google Scholar]
- Potterat, O. Goji (Lycium barbarum and L. chinense): Phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med. 2010, 76, 7–19. [Google Scholar]
- Chen, S.; Zeng, Z.; Hu, N.; Bai, B.; Wang, H.; Suo, Y. Simultaneous optimization of the ultrasound-assisted extraction for phenolic compounds content and antioxidant activity of Lycium ruthenicum Murr. fruit using response surface methodology. Food Chem. 2018, 242, 1–8. [Google Scholar]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Munos, S.; Blanca, J.; Canizares, J.; Diez, M.J.; Schneider, D.; Mazourek, M.; McClead, J.; et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar]
- Ma, M.; Wang, Q.; Li, Z.; Cheng, H.; Li, Z.; Liu, X.; Song, W.; Appels, R.; Zhao, H. Expression of TaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size. Plant J. 2015, 83, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [PubMed]
- Xu, F.; Fang, J.; Ou, S.; Gao, S.; Zhang, F.; Du, L.; Xiao, Y.; Wang, H.; Sun, X.; Chu, J.; et al. Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 2015, 38, 800–811. [Google Scholar] [PubMed]
- Qi, X.; Liu, C.; Song, L.; Li, Y.; Li, M. PaCYP78A9, a cytochrome P450, regulates fruit size in sweet cherry (Prunus avium L.). Front. Plant Sci. 2017, 8, 2076. [Google Scholar]
- Ito, T.; Meyerowitz, E.M. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell 2000, 12, 1541–1550. [Google Scholar]
- Zhao, B.; Dai, A.; Wei, H.; Yang, S.; Wang, B.; Jiang, N.; Feng, X. Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 2016, 90, 33–47. [Google Scholar]
- Sun, X.; Cahill, J.; Van Hautegem, T.; Feys, K.; Whipple, C.; Novák, O.; Delbare, S.; Versteele, C.; Demuynck, K.; De Block, J.; et al. Altered expression of maize PLASTOCHRON1 enhances biomass and seed yield by extending cell division duration. Nat. Commun. 2017, 8, 14752. [Google Scholar]
- Mimura, M.; Itoh, J.I. Genetic interaction between rice PLASTOCHRON genes and the gibberellin pathway in leaf development. Rice 2014, 7, 25. [Google Scholar]
- Li, Q.; Chakrabarti, M.; Taitano, N.K.; Okazaki, Y.; Saito, K.; Al-Abdallat, A.M.; van der Knaap, E. Differential expression of SlKLUH controlling fruit and seed weight is associated with changes in lipid metabolism and photosynthesis-related genes. J. Exp. Bot. 2021, 72, 1225–1244. [Google Scholar]
- Kai, K.; Hashidzume, H.; Yoshimura, K.; Suzuki, H.; Sakurai, N.; Shibata, D.; Ohta, D. Metabolomics for the characterization of cytochromes P450-dependent fatty acid hydroxylation reactions in Arabidopsis. Plant Biotechnol. 2009, 26, 175–182. [Google Scholar]
- Shi, L.; Song, J.; Guo, C.; Wang, B.; Guan, Z.; Yang, P.; Chen, X.; Zhang, Q.; King, G.J.; Wang, J.; et al. A CACTA-like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 2019, 98, 524–539. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Ma, M.; Wu, L.; Zhou, M.; Li, M.; Wu, B.; Li, L.; Liu, X.; Jing, R.; Chen, W.; et al. Modified expression of TaCYP78A5 enhances grain weight with yield potential by accumulating auxin in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2022, 20, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Vercruysse, J.; Baekelandt, A.; Gonzalez, N.; Inzé, D. Molecular networks regulating cell division during Arabidopsis leaf growth. J. Exp. Bot. 2020, 71, 2365–2378. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Wang, C.; Dong, Y.; Zhu, L.; Wang, L.; Yan, L.; Wang, M.; Zhu, Q.; Nan, X.; Li, Y.; Li, J. Comparative transcriptome analysis of two contrasting wolfberry genotypes during fruit development and ripening and characterization of the LrMYB1 transcription factor that regulates flavonoid biosynthesis. BMC Genom. 2020, 21, 295. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.Y.; Song, S.S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef]
- Zhao, J.; Li, A.; Xu, M.; Dai, G.; Chen, J. Genome-wide analysis of the TIFY family in Lycium and the negative regulation of stomatal development by LrJAZ2 gene. Plant Physiol. Biochem. 2024, 206, 108285. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Amino Acid | Protein Molecular | Isoelectirc Point (pI) | Instability Index | Aliphatic Index | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Length/aa | Mass/kDa | ||||||
| LbCYP78A5 | Lba03g02369 | 515 | 58.12 | 6.69 | 30.08 | 94.45 | Endoplasmic reticulum. |
| LbCYP78A6 | Lba01g00553 | 527 | 59.5 | 8.94 | 24.98 | 94.5 | Endoplasmic reticulum. |
| LbCYP78A7 | Lba10g00932 | 511 | 56.4 | 5.75 | 35.26 | 97.1 | Endoplasmic reticulum. |
| LbCYP78A8 | Lba05g01623 | 512 | 58.3 | 9.17 | 43.62 | 94.63 | Endoplasmic reticulum. |
| LbCYP78A9 | Lba06g01865 | 555 | 62.4 | 8.81 | 31.98 | 93.41 | Endoplasmic reticulum. |
| LbCYP78A10 | Lba07g01446 | 536 | 60.2 | 6.8 | 32.08 | 94.07 | Endoplasmic reticulum. |
| LrCYP78A5 | Lru01G036233 | 515 | 58.25 | 6.93 | 37.6 | 93.88 | Endoplasmic reticulum. |
| LrCYP78A6 | Lru01G045721 | 527 | 60.51 | 8.2 | 34.82 | 92.05 | Endoplasmic reticulum. |
| LrCYP78A7 | Lru01G004383 | 531 | 58.74 | 6.13 | 33.42 | 96.93 | Endoplasmic reticulum. |
| LrCYP78A8 | Lru01G025311 | 441 | 49.97 | 8.76 | 44.46 | 96.83 | Endoplasmic reticulum. |
| LrCYP78A9 | Lru01G034235 | 514 | 57.94 | 9.32 | 32.77 | 90.45 | Endoplasmic reticulum. |
| LrCYP78A10 | Lru01G043957 | 536 | 60.22 | 7.69 | 32.62 | 94.61 | Endoplasmic reticulum. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Rao, S.; Dai, G.; Chen, J. Genome-Wide Identification of the CYP78A Gene Family in Lycium and Functional Characterization of LrCYP78A5. Plants 2025, 14, 1152. https://doi.org/10.3390/plants14081152
Zhao Y, Rao S, Dai G, Chen J. Genome-Wide Identification of the CYP78A Gene Family in Lycium and Functional Characterization of LrCYP78A5. Plants. 2025; 14(8):1152. https://doi.org/10.3390/plants14081152
Chicago/Turabian StyleZhao, Yiru, Shupei Rao, Guoli Dai, and Jinhuan Chen. 2025. "Genome-Wide Identification of the CYP78A Gene Family in Lycium and Functional Characterization of LrCYP78A5" Plants 14, no. 8: 1152. https://doi.org/10.3390/plants14081152
APA StyleZhao, Y., Rao, S., Dai, G., & Chen, J. (2025). Genome-Wide Identification of the CYP78A Gene Family in Lycium and Functional Characterization of LrCYP78A5. Plants, 14(8), 1152. https://doi.org/10.3390/plants14081152






