Genome-Wide Identification of the GS3 Gene Family and the Influence of Natural Variations in BnGS3-3 on Salt and Cold Stress Tolerance in Brassica napus
Abstract
1. Introduction
2. Results
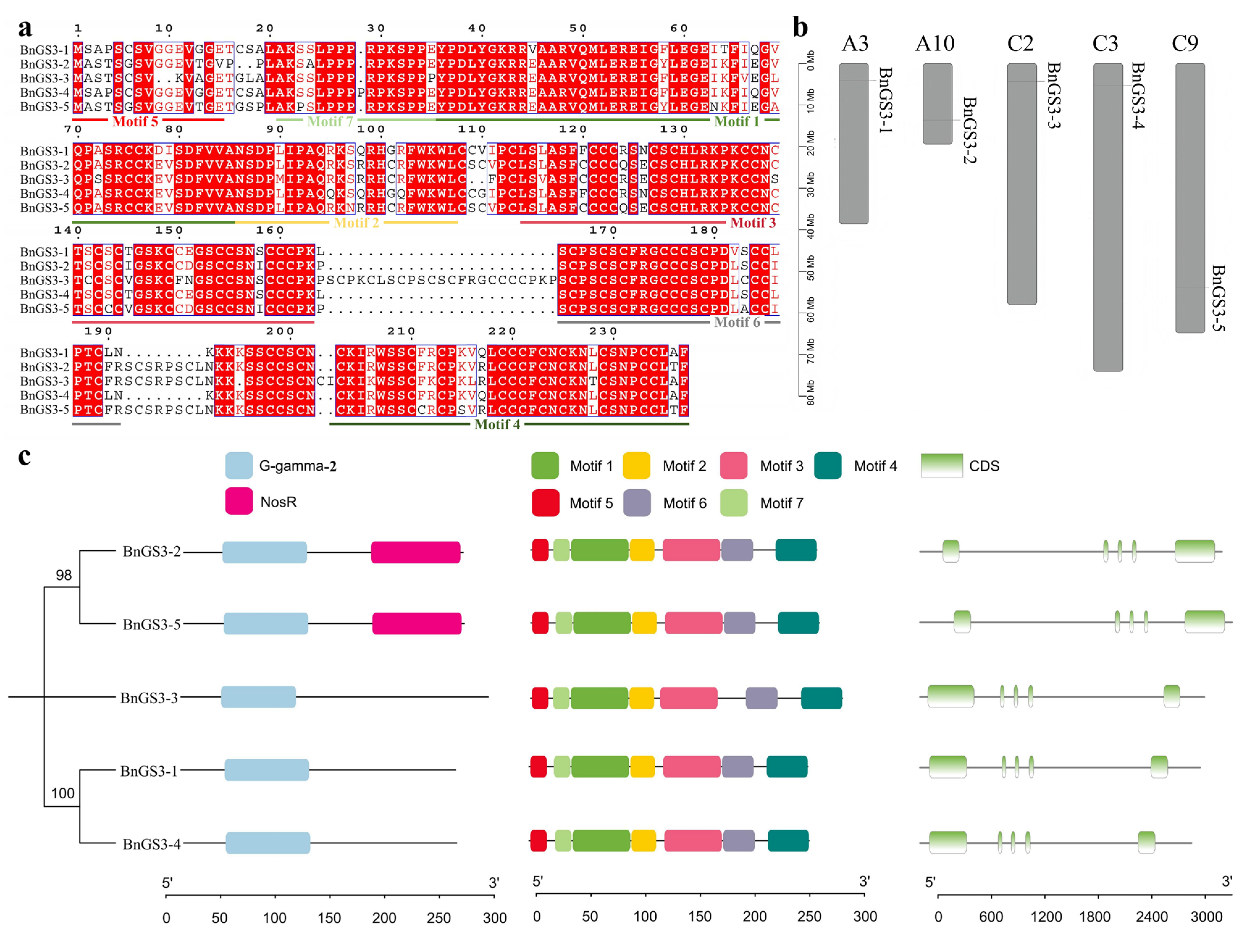
2.1. Identification and Characterization Analysis of the GS3 Gene Family in Brassica napus
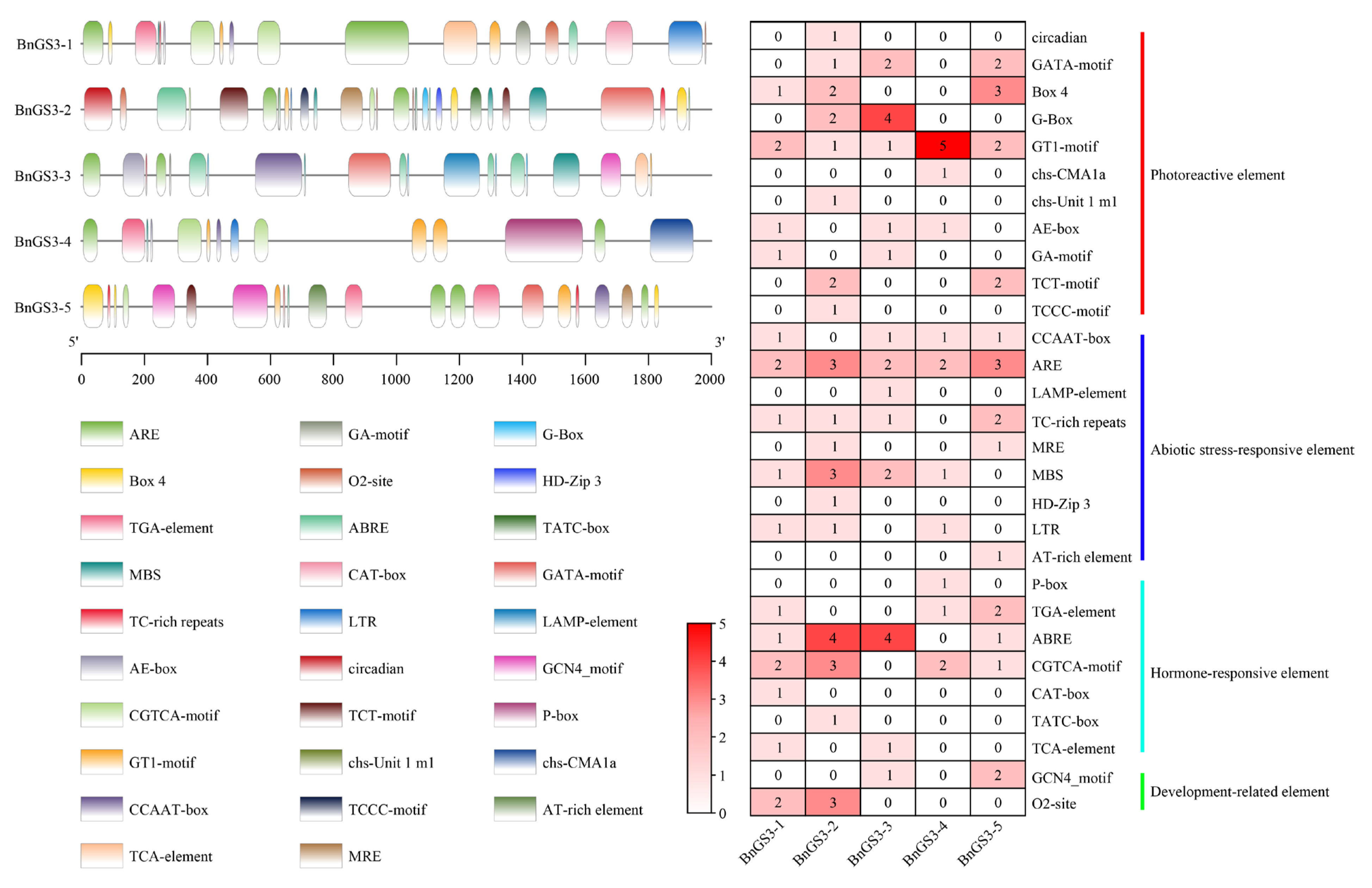
2.2. Analysis of Cis-Acting Elements in the Promoter Region of the GS3 Gene in Brassica napus
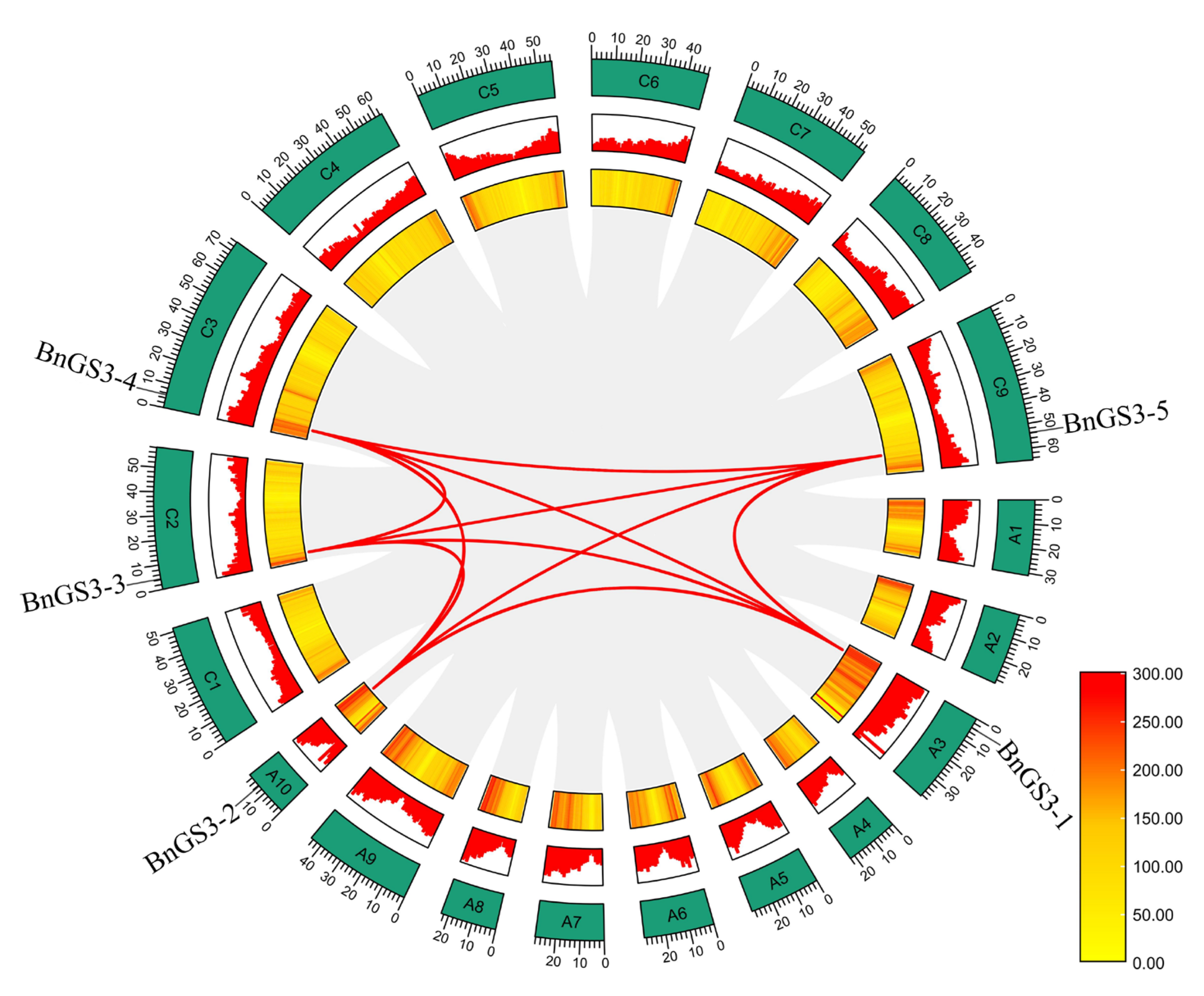
2.3. Expansion Pattern of the GS3 Genes in Brassica napus
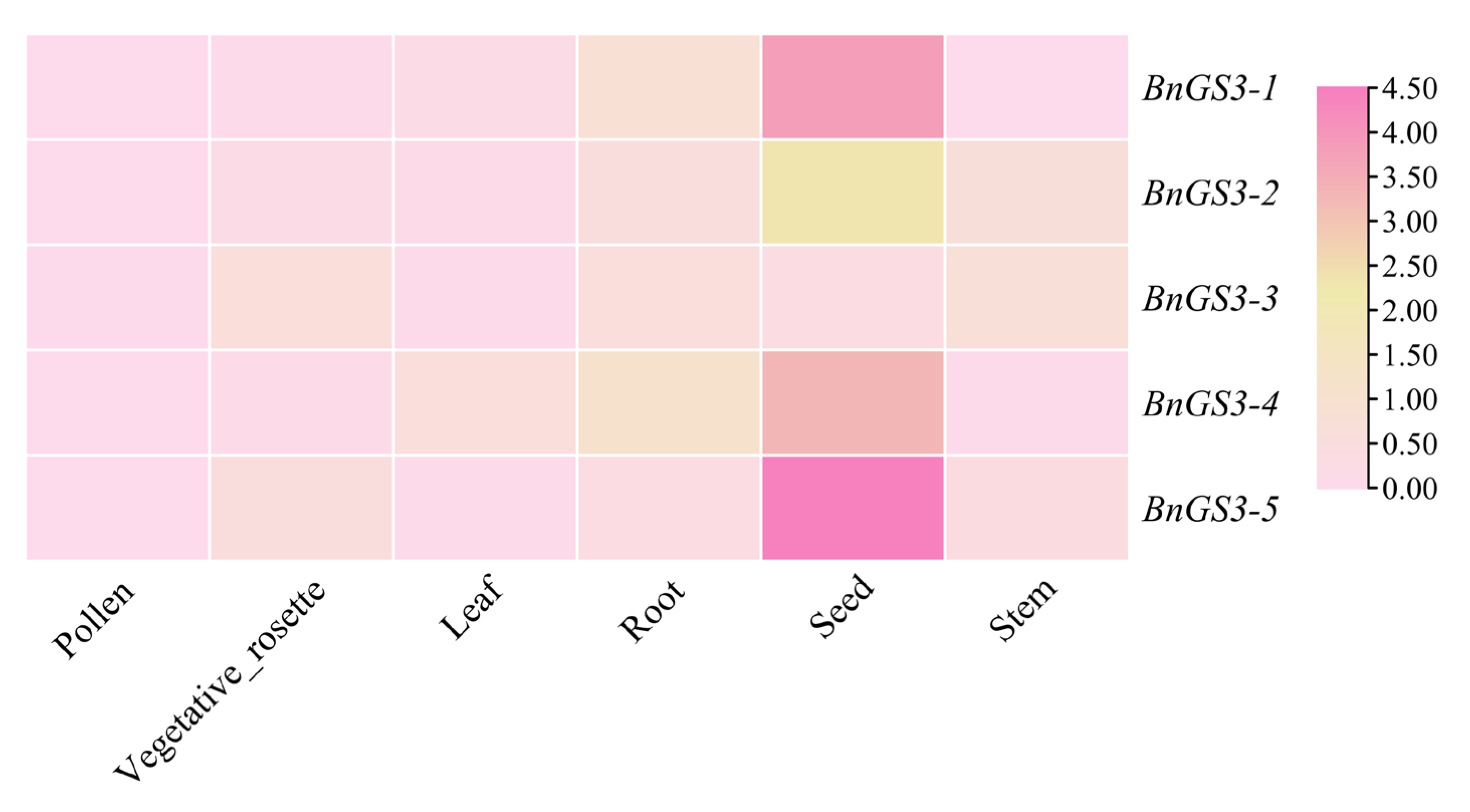
2.4. Tissue Expression Analysis of GS3 Genes in Brassica napus
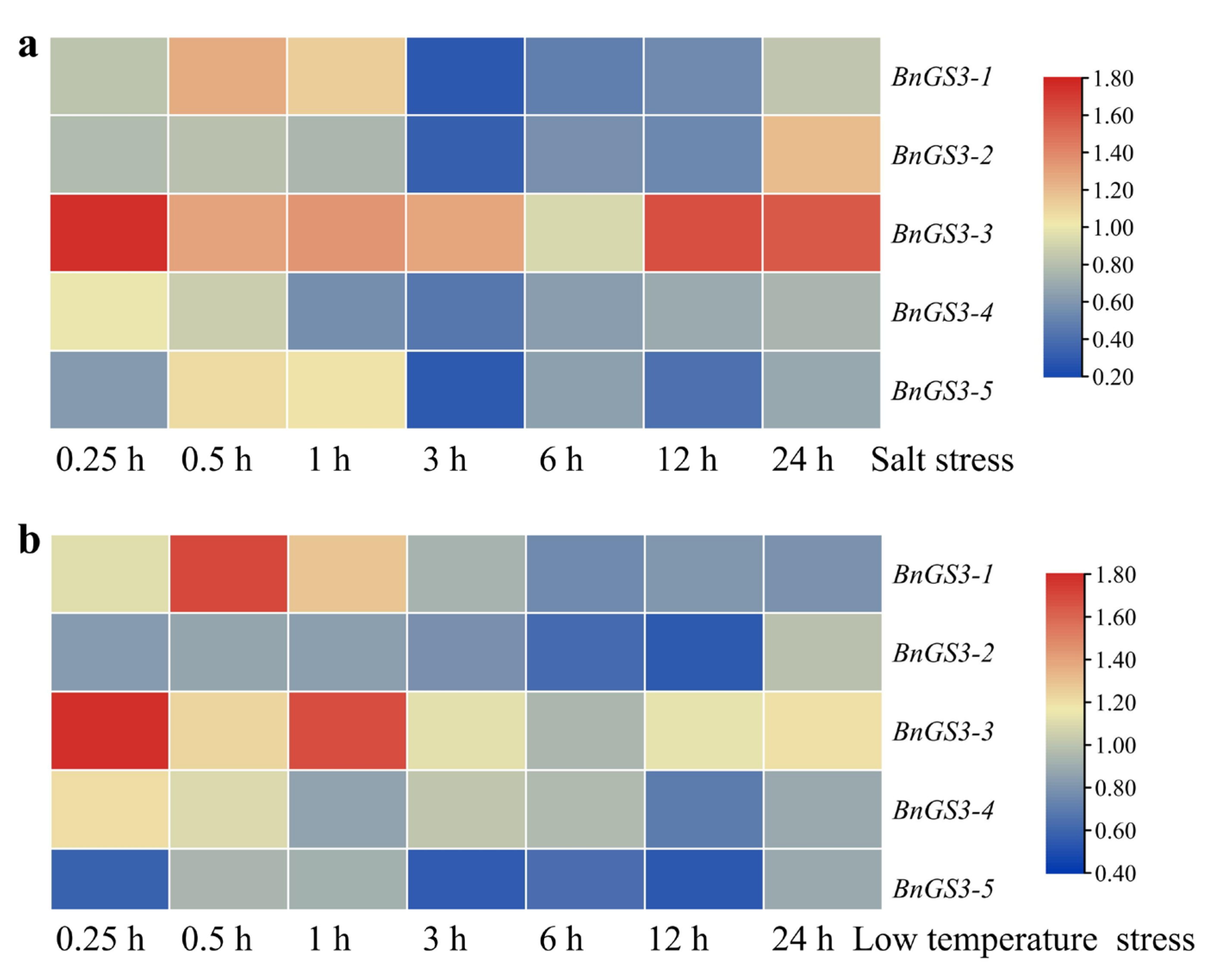
2.5. Expression Pattern of Brassica napus GS3 Genes in Response to Low Temperature and Salt Stress Treatments
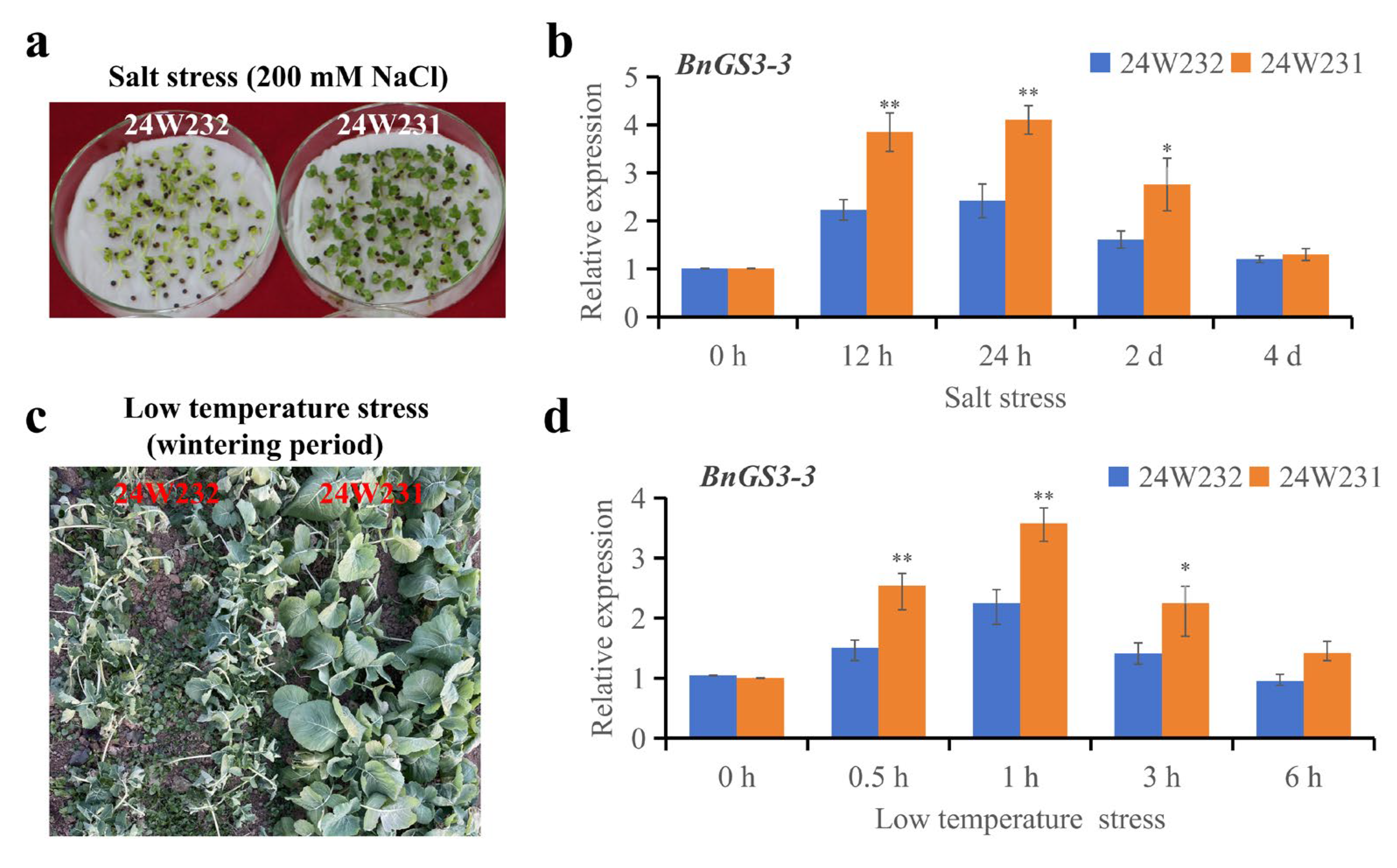
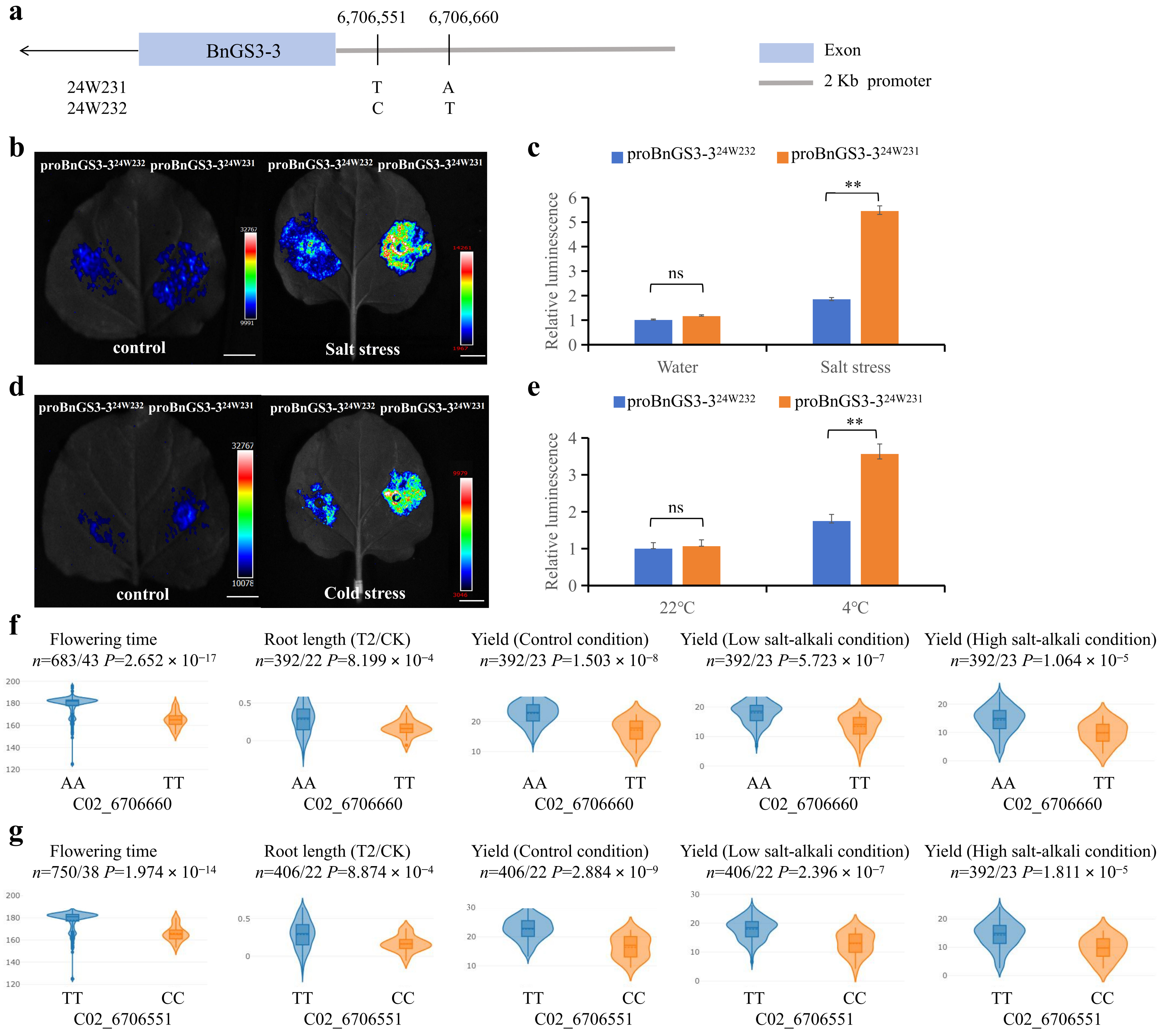
2.6. Natural Variations in the BnGS3-3 Promoter Influence Salt and Cold Stress Tolerance in Brassica napus
3. Discussion
4. Materials and Methods
4.1. Identification of GS3 Family Members in Brassica napus
4.2. Chromosome Localization and Gene Duplication Analysis of Brassica napus GS3 Gene
4.3. BnGS3 Gene Structure Analysis
4.4. BnGS3 Promoter Sequence Analysis
4.5. Analysis of Expression Patterns of the GS3 Family Genes in Brassica napus
4.6. Salt Tolerance Test in Brassica napus Seedlings
4.7. Material Treatment and RT-qPCR Analysis
4.8. Cloning and Sequencing of the BnGS3-3 Promoter
4.9. Analysis of BnGS3-3 Promoter Activity
4.10. Data Acquisition and Analysis of SNP Phenotypic Values
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| GPA | G-protein subunit alpha |
| GPCRs | G protein-coupled receptors |
| LUC | Luciferase |
| MeJA | Methyl jasmonate |
| ROS | Reactive oxygen species |
| RT-PCR | Reverse transcription polymerase chain reaction |
| SA | Salicylic acid |
References
- Garcia-Marcos, M. Stickier G-protein conformations. Nat. Chem. Biol. 2023, 19, 665–666. [Google Scholar] [CrossRef] [PubMed]
- Duc, N.M.; Kim, H.R.; Chung, K.Y. Structural mechanism of G protein activation by G protein-coupled receptor. Eur. J. Pharmacol. 2015, 763, 214–222. [Google Scholar] [PubMed]
- Syrovatkina, V.; Alegre, K.O.; Dey, R.; Huang, X.Y. Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 2016, 428, 3850–3868. [Google Scholar] [CrossRef] [PubMed]
- Maruta, N.; Trusov, Y.; Jones, A.M.; Botella, J.R. Heterotrimeric G proteins in plants: Canonical and atypical Gα subunits. Int. J. Mol. Sci. 2021, 22, 11841. [Google Scholar] [CrossRef]
- Sprang, S.R. G protein mechanisms: Insights from structural analysis. Annu. Rev. Biochem. 1997, 66, 639–678. [Google Scholar] [CrossRef]
- Weiss, C.A.; Garnaat, C.W.; Mukai, K.; Hu, Y.; Ma, H. Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc. Natl. Acad. Sci. USA 1994, 91, 9554–9558. [Google Scholar] [CrossRef]
- Xiong, X.X.; Liu, Y.; Zhang, L.L.; Li, X.J.; Zhao, Y.; Zheng, Y.; Yang, Q.H.; Yang, Y.; Min, D.H.; Zhang, X.H. G-protein β-subunit gene TaGB1-B enhances drought and salt resistance in wheat. Int. J. Mol. Sci. 2023, 24, 7337. [Google Scholar] [CrossRef]
- Torres Rodríguez, M.D.; Bhatnagar, N.; Pandey, S. Overexpression of a plant-specific Gγ protein, AGG3, in the model monocot Setaria viridis confers tolerance to heat stress. Plant Cell Physiol. 2023, 64, 1243–1256. [Google Scholar]
- Mason, M.G.; Botella, J.R. Completing the heterotrimer: Isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc. Natl. Acad. Sci. USA 2000, 97, 14784–14788. [Google Scholar]
- Delgado-Cerezo, M.; Sánchez-Rodríguez, C.; Escudero, V.; Miedes, E.; Fernández, P.V.; Jordá, L.; Hernández-Blanco, C.; Sánchez-Vallet, A.; Bednarek, P.; Schulze-Lefert, P.; et al. Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol. Plant 2012, 5, 98–114. [Google Scholar]
- Chang, Z.; Xu, C.; Huang, X.; Yan, W.; Qiu, S.; Yuan, S.; Ni, H.; Chen, S.; Xie, G.; Chen, Z.; et al. The plant-specific ABERRANT GAMETOGENESIS 1 gene is essential for meiosis in rice. J. Exp. Bot. 2020, 71, 204–218. [Google Scholar] [CrossRef] [PubMed]
- Chaya, G.; Segami, S.; Fujita, M.; Morinaka, Y.; Iwasaki, Y.; Miura, K. OsGGC2, Gγ subunit of heterotrimeric G protein, regulates plant height by functionally overlapping with DEP1 in rice. Plants 2022, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, F.; Xie, P.; Sun, S.; Qiao, X.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; Yang, D.; et al. A Gγ protein regulates alkaline sensitivity in crops. Science 2023, 379, eade8416. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, Y.; Yu, H.; Hu, P.; Wen, Y.; Wang, J.; Tan, Y.; Wu, H.; Zhu, L.; Wu, K.; et al. GR5 acts in the G protein pathway to regulate grain size in rice. Plant Commun. 2024, 5, 100673. [Google Scholar] [CrossRef]
- Olsen, R.H.J.; English, J.G. Advancements in G protein-coupled receptor biosensors to study GPCR-G protein coupling. Br. J. Pharmacol. 2023, 180, 1433–1443. [Google Scholar] [CrossRef]
- Masuho, I.; Balaji, S.; Muntean, B.S.; Skamangas, N.K.; Chavali, S.; Tesmer, J.J.G.; Babu, M.M.; Martemyanov, K.A. A global map of G protein signaling regulation by RGS proteins. Cell 2020, 183, 503–521.e519. [Google Scholar] [CrossRef]
- Mohanasundaram, B.; Pandey, S. Moving beyond the arabidopsis-centric view of G-protein signaling in plants. Trends Plant Sci. 2023, 28, 1406–1421. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Pandey, S. SymRK regulates G-protein signaling during nodulation in soybean (Glycine max) by modifying RGS phosphorylation and activity. Mol. Plant Microbe Interact. 2024, 37, 765–775. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, F.; Liu, L.; Char, S.N.; Ding, Y.; Je, B.I.; Schmelz, E.; Yang, B.; Jackson, D. The maize heterotrimeric G protein β subunit controls shoot meristem development and immune responses. Proc. Natl. Acad. Sci. USA 2020, 117, 1799–1805. [Google Scholar] [CrossRef]
- Xie, Y.; Nan, Y.; Atif, A.; Hu, W.; Zhang, Y.; Tian, H.; Gao, Y. Molecular Identification of the G-protein genes and their expression profiles in response to nitrogen deprivation in Brassica napus. Int. J. Mol. Sci. 2022, 23, 8151. [Google Scholar] [CrossRef]
- Yang, S.; Yan, N.; Bouwmeester, K.; Na, R.; Zhang, Z.; Zhao, J. Genome-wide identification of small G protein ROPs and their potential roles in Solanaceous family. Gene 2020, 753, 144809. [Google Scholar] [PubMed]
- Tian, Z.; Zhang, Z.; Kang, L.; Li, M.; Zhang, J.; Feng, Y.; Yin, J.; Gong, X.; Zhao, J. Small G Protein StRab5b positively regulates potato resistance to Phytophthora infestans. Front. Plant Sci. 2022, 13, 1065627. [Google Scholar]
- Bhagat, P.K.; Sharma, D.; Verma, D.; Singh, K.; Sinha, A.K. Arabidopsis MPK3 and MPK6 regulates D-glucose signaling and interacts with G-protein, RGS1. Plant Sci. 2022, 325, 111484. [Google Scholar] [PubMed]
- Wang, K.; Xu, F.; Yuan, W.; Zhang, D.; Liu, J.; Sun, L.; Cui, L.; Zhang, J.; Xu, W. Rice G protein γ subunit qPE9-1 modulates root elongation for phosphorus uptake by involving 14-3-3 protein OsGF14b and plasma membrane H+-ATPase. Plant J. 2021, 107, 1603–1615. [Google Scholar]
- Kansup, J.; Tsugama, D.; Liu, S.; Takano, T. Arabidopsis G-protein β subunit AGB1 interacts with NPH3 and is involved in phototropism. Biochem. Biophys. Res. Commun. 2014, 445, 54–57. [Google Scholar]
- Yang, S.; Jung, S.; Lee, H. Heterotrimeric G protein-mediated signaling is involved in stress-mediated growth inhibition in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 11027. [Google Scholar] [CrossRef]
- Wu, T.Y.; Krishnamoorthi, S.; Boonyaves, K.; Al-Darabsah, I.; Leong, R.; Jones, A.M.; Ishizaki, K.; Liao, K.L.; Urano, D. G protein controls stress readiness by modulating transcriptional and metabolic homeostasis in Arabidopsis thaliana and Marchantia polymorpha. Mol. Plant 2022, 15, 1889–1907. [Google Scholar]
- Brunetti, S.C.; Arseneault, M.K.M.; Gulick, P.J. The caleosin CLO7 and its role in the heterotrimeric G-protein signalling network. J. Plant Physiol. 2022, 279, 153841. [Google Scholar]
- Lease, K.A.; Wen, J.; Li, J.; Doke, J.T.; Liscum, E.; Walker, J.C. A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 2001, 13, 2631–2641. [Google Scholar]
- Liu, X.; Zhao, C.X.; Wang, D.; Pan, G.; Ji, X.A.; Gao, S.; Du, T.X.; Feng, Y.T.; Chen, W.J. DDG1 and G protein α subunit RGA1 interaction regulates plant height and senescence in rice (Oryza sativa). J. Exp. Bot. 2023, 92, 2051–2064. [Google Scholar]
- Sun, W.; Zhang, H.; Yang, S.; Liu, L.; Xie, P.; Li, J.; Zhu, Y.; Ouyang, Y.; Xie, Q.; Zhang, H.; et al. Genetic modification of Gγ subunit AT1 enhances salt-alkali tolerance in main graminaceous crops. Natl. Sci. Rev. 2023, 10, nwad075. [Google Scholar] [PubMed]
- Ju, C.; Wang, C. Gγ subunit AT1/GS3-the “code” of alkaline tolerance in main graminaceous crops. Stress Biol. 2023, 3, 9. [Google Scholar]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Hussain, M.A.; Wei, S.; He, H.; Zaman, Q.U.; Xuekun, Z.; Hasanuzzaman, M. Omics: The way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crops Food 2021, 12, 251–281. [Google Scholar] [PubMed]
- Jankovska-Bortkevič, E.; Jurkonienė, S.; Gavelienė, V.; Šveikauskas, V.; Mockevičiūtė, R.; Vaseva, I.; Todorova, D.; Žižytė-Eidetienė, M.; Šneideris, D.; Prakas, P. Dynamics of polyamines, proline, and ethylene metabolism under increasing cold in winter oilseed rape. Int. J. Mol. Sci. 2023, 24, 11402. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Balfagón, D.; Gómez-Cadenas, A.; Mittler, R. Plant responses to climate change: Metabolic changes under combined abiotic stresses. J. Exp. Bot. 2022, 73, 3339–3354. [Google Scholar]
- Raboanatahiry, N.; Li, H.X.; Yu, L.J.; Li, M.T. Rapeseed (Brassica napus): Processing, utilization, and genetic improvement. Agronomy 2021, 11, 1776. [Google Scholar] [CrossRef]
- Jain, S.; Farnoud, F.; Bruck, J. Capacity and expressiveness of genomic tandem duplication. IEEE Trans. Inf. Theory 2017, 63, 6129–6138. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Fan, S.; He, Y.; Wei, M.; Wang, J.; Yin, Y.; Liu, Y. Combined genomic and transcriptomic analysis reveals the contribution of tandem duplication genes to low-temperature adaptation in perennial ryegrass. Front. Plant Sci. 2023, 14, 1216048. [Google Scholar]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar]
- Li, M.; Liu, T.; Cao, R.; Cao, Q.; Tong, W.; Song, W. Evolution and expression of the expansin genes in emmer wheat. Int. J. Mol. Sci. 2023, 24, 14120. [Google Scholar] [CrossRef]
- Androsiuk, P.; Paukszto, Ł.; Jastrzębski, J.P.; Milarska, S.E.; Okorski, A.; Pszczółkowska, A. Molecular diversity and phylogeny reconstruction of genus Colobanthus (Caryophyllaceae) based on mitochondrial gene sequences. Genes 2022, 13, 1060. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.K.; Islam, S.M.; Tuteja, N. Rice heterotrimeric G-protein gamma subunits (RGG1 and RGG2) are differentially regulated under abiotic stress. Plant Signal. Behav. 2012, 7, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Kan, Y.; Mu, X.R.; Zhang, H.; Gao, J.; Shan, J.X.; Ye, W.W.; Lin, H.X. TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 2022, 8, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K.M.; Beeby, R.; Penfield, S. Yield instability of winter oilseed rape modulated by early winter temperature. Sci. Rep. 2019, 9, 6953. [Google Scholar] [CrossRef]
- Vollrath, P.; Chawla, H.S.; Schiessl, S.V.; Gabur, I.; Lee, H.; Snowdon, R.J.; Obermeier, C. A novel deletion in FLOWERING LOCUS T modulates flowering time in winter oilseed rape. Theor. Appl. Genet. 2021, 134, 1217–1231. [Google Scholar] [CrossRef]
- Schiessl, S. Regulation and subfunctionalization of flowering time genes in the allotetraploid oil crop Brassica napus. Front. Plant Sci. 2020, 11, 605155. [Google Scholar] [CrossRef]
- Tudor, E.H.; Jones, D.M.; He, Z.; Bancroft, I.; Trick, M.; Wells, R.; Irwin, J.A.; Dean, C. QTL-seq identifies BnaFT.A02 and BnaFLC.A02 as candidates for variation in vernalization requirement and response in winter oilseed rape (Brassica napus). Plant Biotechnol. J. 2020, 18, 2466–2481. [Google Scholar] [CrossRef]
- Gao, Y.; Gu, H.; Leburu, M.; Li, X.; Wang, Y.; Sheng, J.; Fang, H.; Gu, M.; Liang, G. The heterotrimeric G protein β subunit RGB1 is required for seedling formation in rice. Rice 2019, 12, 53. [Google Scholar] [CrossRef]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef]
- Duan, P.; Li, Y. Size matters: G protein signaling is crucial for grain size control in rice. Mol. Plant 2021, 14, 1618–1620. [Google Scholar] [CrossRef]
- Wu, W.H.; Assmann, S.M. A membrane-delimited pathway of G-protein regulation of the guard-cell inward K+ channel. Proc. Natl. Acad. Sci. USA 1994, 91, 6310–6314. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, D.; Trusov, Y.; Zhang, W.; Acharya, B.R.; Sheahan, M.B.; McCurdy, D.W.; Assmann, S.M.; Botella, J.R. An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K⁺-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 2011, 67, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Mostofa, M.G.; Rahman, M.M.; Ghosh, T.K.; Kabir, A.H.; Abdelrahman, M.; Khan, M.A.R.; Mochida, K.; Tran, L.P. Potassium in plant physiological adaptation to abiotic stresses. Plant Physiol. Biochem. 2022, 186, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Gorelova, V. Mind the context: K+ channel subunit AtKC1 tunes local osmotic environment to adjust stomatal movement. Plant Cell 2022, 34, 1884–1885. [Google Scholar] [CrossRef]
- Lefoulon, C. The bare necessities of plant K+ channel regulation. Plant Physiol. 2021, 187, 2092–2109. [Google Scholar] [CrossRef]
- Muraoka, Y.; Yang, G.; Munemasa, S.; Takeuchi, Y.; Ishimaru, Y.; Murata, Y.; Uozumi, N.; Ueda, M. An outward-rectifying plant K+ channel SPORK2 exhibits temperature-sensitive ion-transport activity. Curr. Biol. 2023, 33, 5488–5494.e7. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Jeon, B.W.; Acharya, B.R.; Assmann, S.M. The Arabidopsis heterotrimeric G-protein β subunit, AGB1, is required for guard cell calcium sensing and calcium-induced calcium release. Plant J. 2019, 99, 231–244. [Google Scholar] [CrossRef]
- Zhang, W.; Jeon, B.W.; Assmann, S.M. Heterotrimeric G-protein regulation of ROS signalling and calcium currents in Arabidopsis guard cells. J. Exp. Bot. 2011, 62, 2371–2379. [Google Scholar] [CrossRef]
- Leong, R.; Tan, J.J.; Koh, S.S.; Wu, T.Y.; Ishizaki, K.; Urano, D. G protein signaling and metabolic pathways as evolutionarily conserved mechanisms to combat calcium deficiency. New Phytol. 2023, 237, 615–630. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, H.; Zhang, S.; Zhang, J.; Di, H.; Zhang, L.; Dong, L.; Lu, Q.; Zeng, X.; Liu, X.; et al. The G protein-coupled receptor COLD1 promotes chilling tolerance in maize during germination. Int. J. Biol. Macromol. 2023, 253, 126877. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S. Plant receptor-like kinase signaling through heterotrimeric G-proteins. J. Exp. Bot. 2020, 71, 1742–1751. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A multi-omics database with various tools for Brassica napus research and breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef]
- Liu, D.; Yu, L.; Wei, L.; Yu, P.; Wang, J.; Zhao, H.; Zhang, Y.; Zhang, S.; Yang, Z.; Chen, G.; et al. BnTIR: An online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 2021, 19, 1895–1897. [Google Scholar] [CrossRef]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Shi, Y.; Lang, L.; Tao, S.; Zhang, Q.; Qin, M.; Wang, K.; Xu, Y.; Zheng, L.; et al. The B-box transcription factor BnBBX22.A07 enhances salt stress tolerance by indirectly activating BnWRKY33.C03. Plant Cell Environ. 2024, 47, 5424–5442. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xu, J.; Tao, X.; Wu, J.; Wang, W.; Pu, Y.; Yang, G.; Fang, Y.; Liu, L.; Li, X.; et al. Genome-wide identification of C2H2 ZFPs and functional analysis of BRZAT12 under low-temperature stress in winter rapeseed (Brassica rapa). Int. J. Mol. Sci. 2022, 23, 12218. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Sun, Y.; Shan, Z.; He, J.; Wang, N.; Gai, J.; Li, Y. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 1155–1169. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, T.; Hao, X.; Huang, Z.; Zhang, X.; Li, S.; Yang, Y.; Long, W. Genome-Wide Identification of the GS3 Gene Family and the Influence of Natural Variations in BnGS3-3 on Salt and Cold Stress Tolerance in Brassica napus. Plants 2025, 14, 1145. https://doi.org/10.3390/plants14071145
Jin T, Hao X, Huang Z, Zhang X, Li S, Yang Y, Long W. Genome-Wide Identification of the GS3 Gene Family and the Influence of Natural Variations in BnGS3-3 on Salt and Cold Stress Tolerance in Brassica napus. Plants. 2025; 14(7):1145. https://doi.org/10.3390/plants14071145
Chicago/Turabian StyleJin, Ting, Xiaoshuai Hao, Zhen Huang, Xingguo Zhang, Shimeng Li, Ying Yang, and Weihua Long. 2025. "Genome-Wide Identification of the GS3 Gene Family and the Influence of Natural Variations in BnGS3-3 on Salt and Cold Stress Tolerance in Brassica napus" Plants 14, no. 7: 1145. https://doi.org/10.3390/plants14071145
APA StyleJin, T., Hao, X., Huang, Z., Zhang, X., Li, S., Yang, Y., & Long, W. (2025). Genome-Wide Identification of the GS3 Gene Family and the Influence of Natural Variations in BnGS3-3 on Salt and Cold Stress Tolerance in Brassica napus. Plants, 14(7), 1145. https://doi.org/10.3390/plants14071145





