A Truncated Endogenous U6 Promoter Enables High-Efficiency CRISPR Editing in Flax (Linum usitatissimum L.)
Abstract
1. Introduction
2. Results
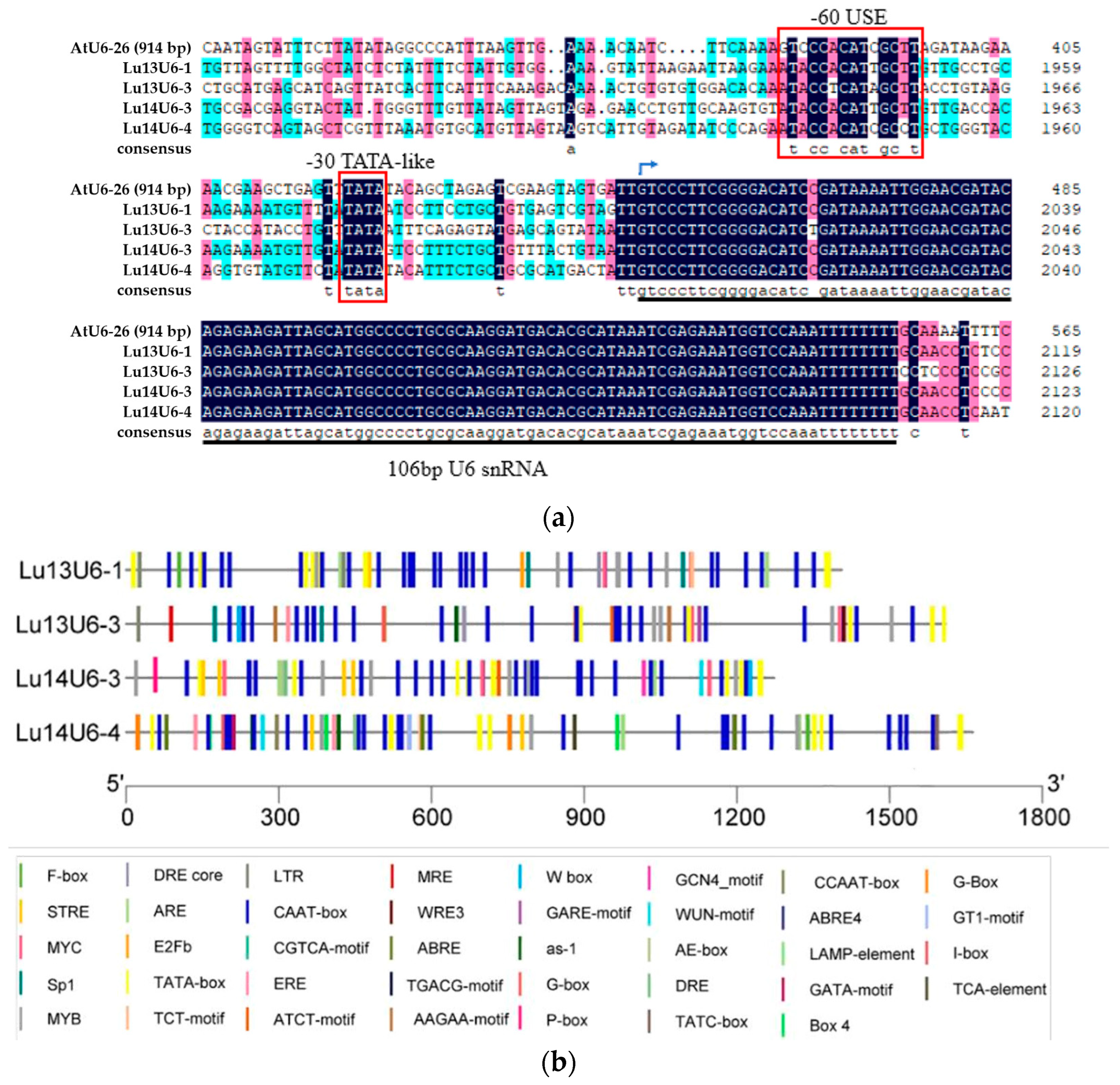
2.1. Sequence Analysis of Flax U6 Promoters: Structural Conservation and Functional Implications
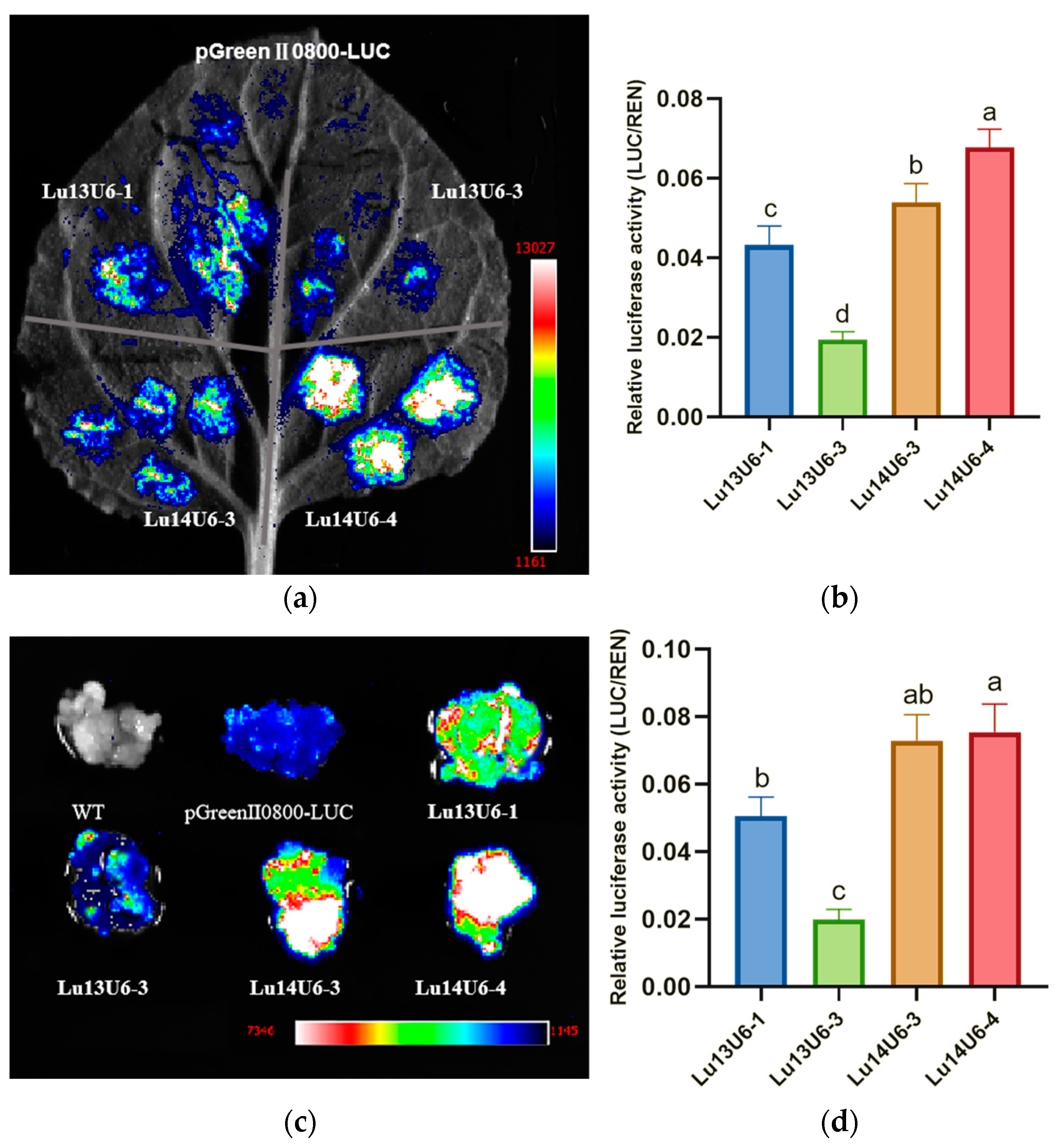
2.2. Transcriptional Validation of Endogenous Flax U6 Promoter Candidates
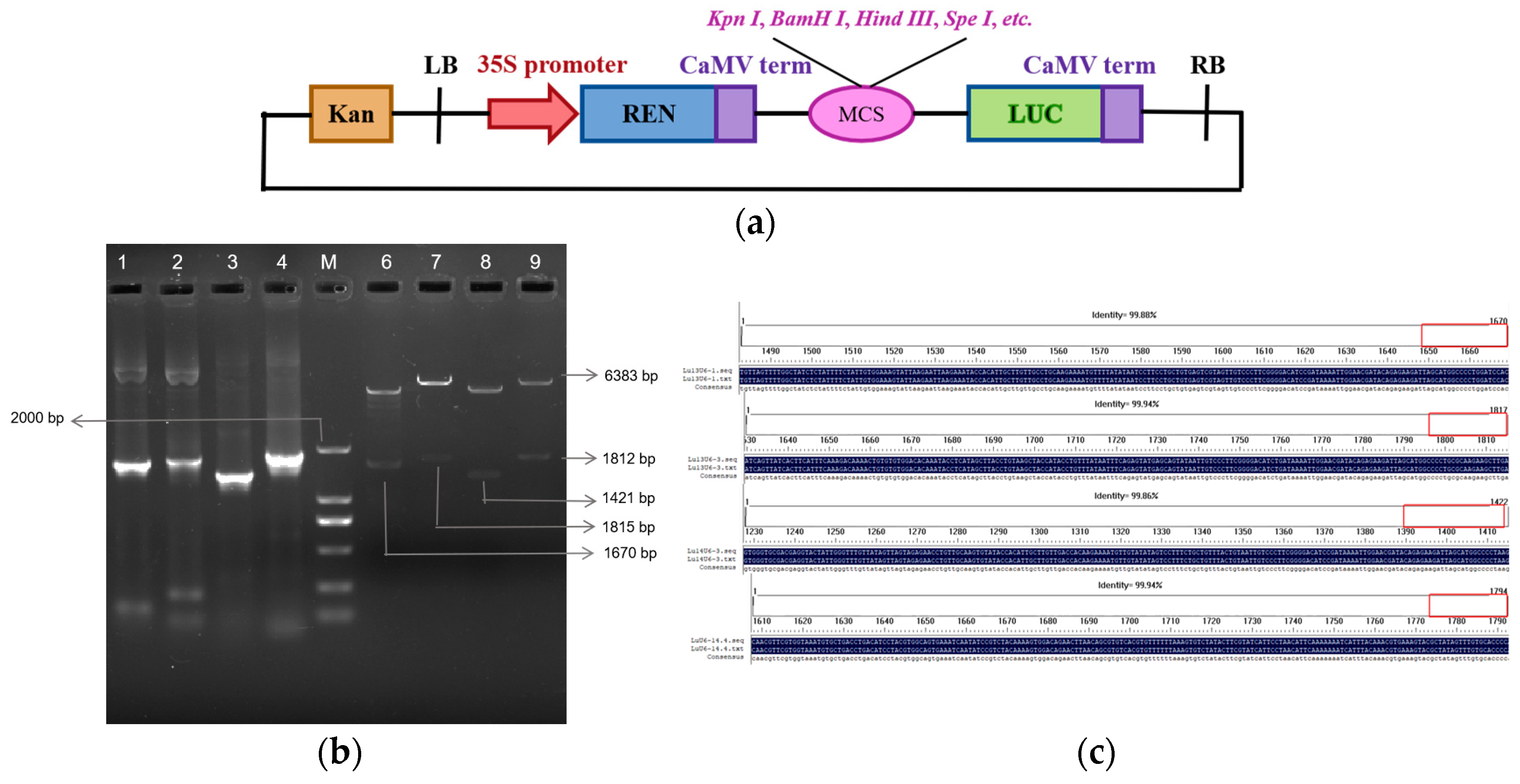
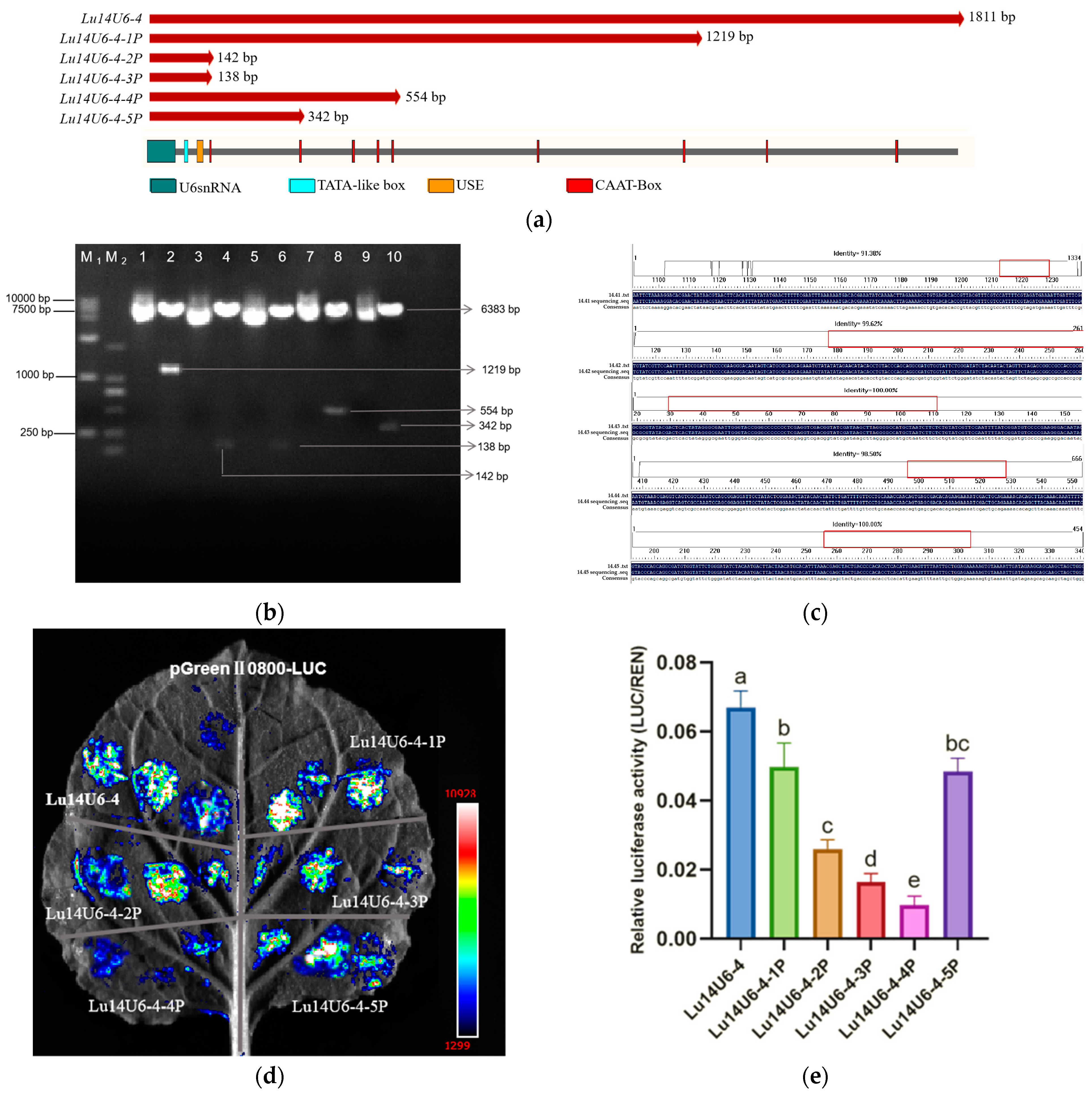
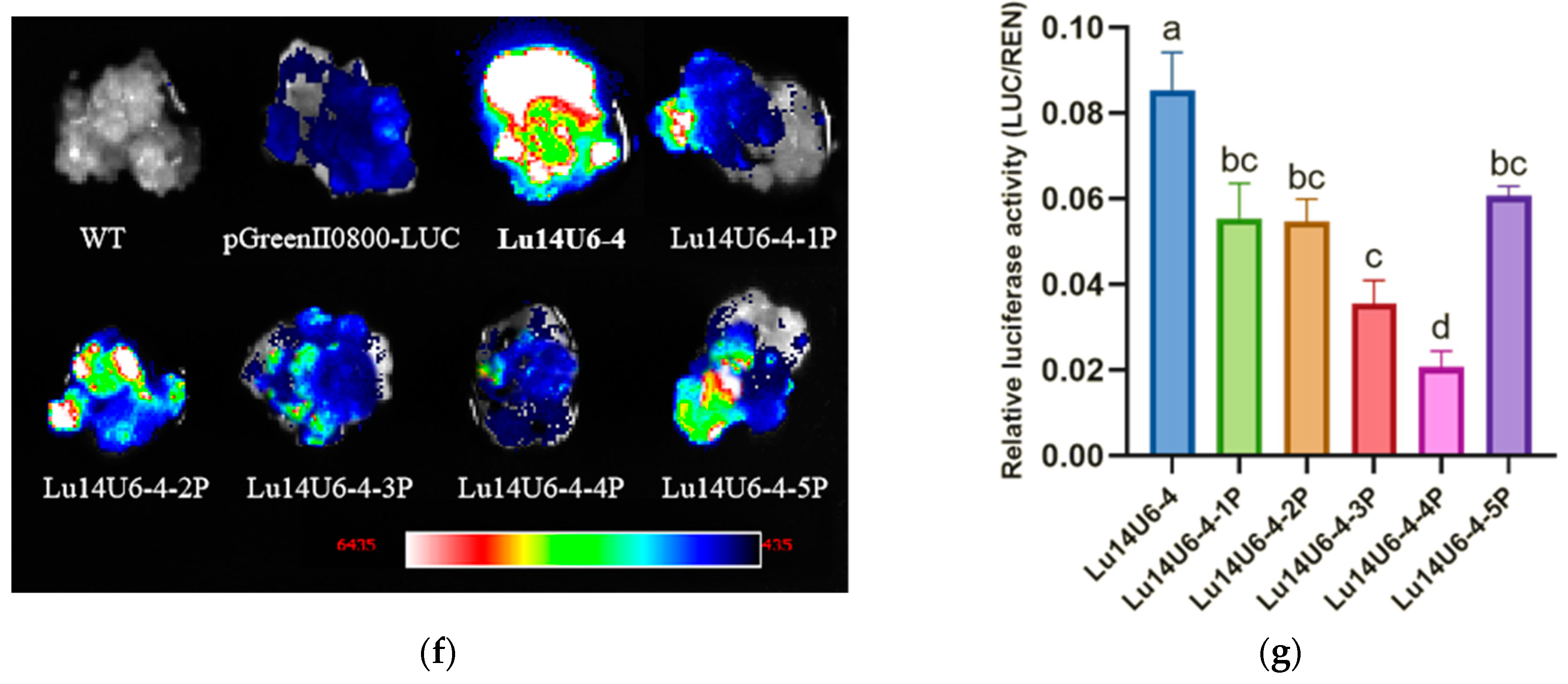
2.3. Construction and Activity Validation of Truncated Flax LuU6 Promoter Expression Vectors
2.4. Development and Genetic Transformation of CRISPR/Cas9 Gene Editing Vectors Driven by Endogenous U6 Promoters in Flax
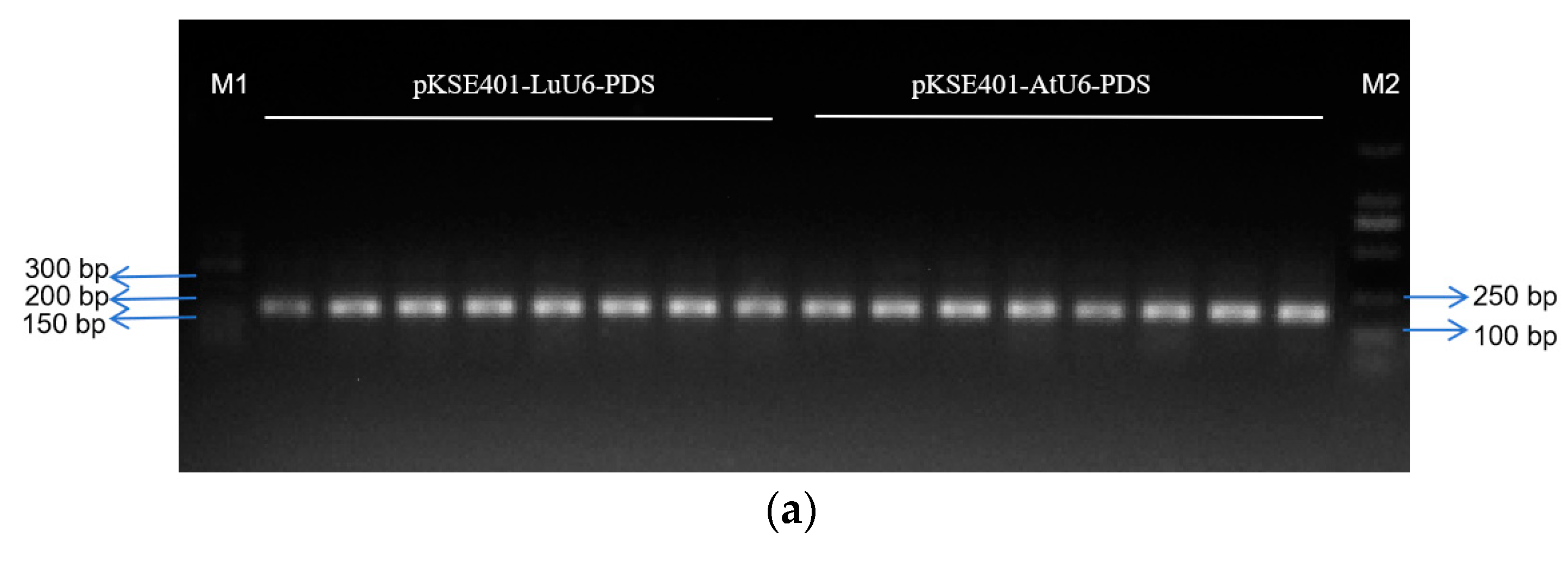
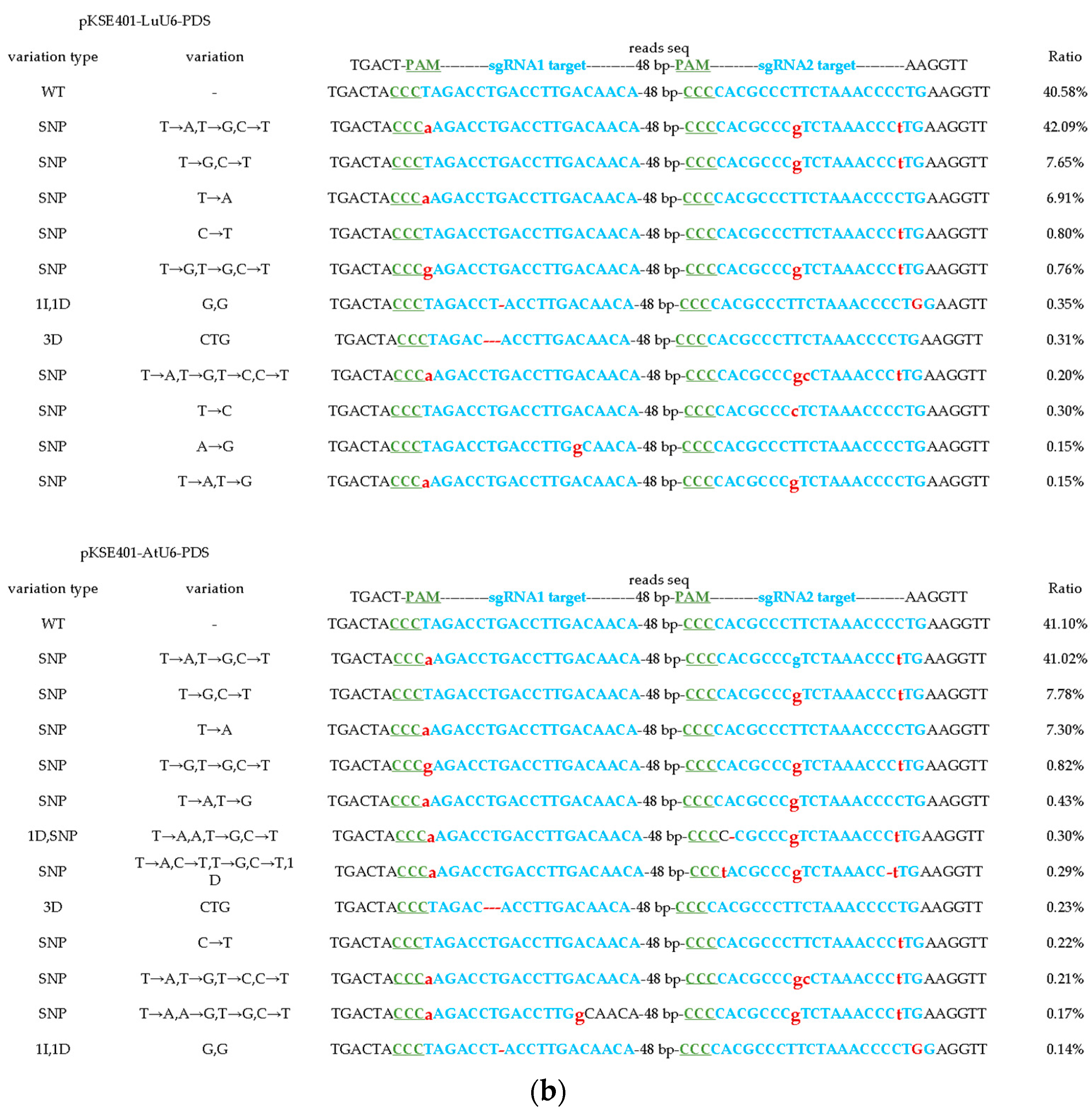
2.5. The LuU6 Promoter Demonstrates Higher Editing Efficiency in CRISPR/Cas9-Mediated Gene Editing in Flax Compared to the AtU6 Promoter
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. Cloning, Truncation, and Construction of Dual-Luciferase Reporter Gene Expression Vectors for Flax LuU6 Promoter
4.3. Transient Transformation of Flax Callus and Nicotiana benthamiana
4.4. Dual-Luciferase Reporter Gene Assay for Flax LuU6 Promoter
4.5. Construction of CRISPR/Cas9-Mediated Gene Editing Vectors in Flax
4.6. Plant Vector Construction
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primer Name | Sequence (5′-3′) | Experiment |
|---|---|---|
| M13-F | GTAAAACGACGGCCAGT | Universal Primers for Vector |
| M13-R | CAGGAAACAGCTATGAC | |
| pGreenII-LUC-F | GTTTTCCCAGTCACGACGTT | Universal Primers for pGreenII0800-LUC Vector |
| pGreenII-LUC-R | GCCTTATGCAGTTGCTCTCC | |
| Lu13U6-1F | GGGGTACCCCCTAAAGATTTGGGGGTCACT | Cloning of Candidate U6 Promoter Fragments |
| Lu13U6-1R | CGGGATCCAGGGGCCATGCTAATCTTCTCT | |
| Lu13U6-3F | GGGGTACCCGAGAACAAACCTCTAGCCG | |
| Lu13U6-3R | CCCAAGCTTCTTGCGCAGGGGCCAT | |
| Lu14U6-3F | GGGGTACCTCCCCCTGGGTTAGTAGCAATA | |
| Lu14U6-3R | CCCAAGCTTAGGGGCCATGCTAATCTTCT | |
| Lu14U6-4F | GGGGTACCTGGGGTGCACAAACTATAGCG | |
| Lu14U6-4R | CCCAAGCTTAGGGGCCATGCTAATCTTCTC | |
| SK-R | TCTAGAACTAGTGGATC | Colony PCR Screening |
| Lu14U6-4-1pF Lu14U6-4-2pF Lu14U6-4-3pF | TCGACGGTATCGATAAGCTTAGGGGCCATGCTAATCTTCTCT | Cloning and Detection of Truncated U6 Promoter Fragments |
| Lu14U6-4-1pR | GCGGCCGCTCTAGAACTAGTCAAATTCAAAAATATATACA | |
| Lu14U6-4-2pR | GCGGCCGCTCTAGAACTAGTATTGTAGATATCCCAGAATACC | |
| Lu14U6-4-3pR | GCGGCCGCTCTAGAACTAGTTAGATATCCCAGAATACCAC | |
| Lu14U6-4-4pF Lu14U6-4-5pF | TCGAGGTCGACGGTATCGATAAGCTTAGGGGCCATGCTAATCTTCT | |
| Lu14U6-4-4pR | GCGGCCGCTCTAGAACTAGTGGATCCATTGTTTGATTGAATGGAAC | |
| Lu14U6-4-5pR | GCGGCCGCTCTAGAACTAGTGGATCCATTTGGCGACTGACCTCGTT | |
| M13F (47) | CGCCAGGGTTTTCCCAGTCACGAC | Detection of Truncated U6 Promoter Fragments |
| LusPDS F | TTCAGGGGAAGTGACTCTCTCA | Detection of the 926 bp LusPDS Fragment in the Genomic DNA of Longya 10 Leaves |
| LusPDS R | TCGTACCAGTCTCCATCCTC | |
| TDNA-LusPDS F1 | CCCGCCAATATATCCTGTCAAAC | Detection of Exogenous T-DNA Insertion in Cas9 Editing Vectors |
| TDNA-LusPDS R1 | CCCTAGACCTGACCTTGACAACA | |
| Hi-TOM-LusPDS F | ggagtgagtacggtgtgcGTGTTGTGCGTTGACTACCC | Hi-TOM Assay |
| Hi-TOM-LusPDS R | gagttggatgctggatggCCTGCACCAGCAATAACAAC |
| Sample Names | Proportions of Major Mutation Types | Mutant Phenotype | ||
|---|---|---|---|---|
| Missense Mutation | Synonymous Mutation | Frameshift Mutation | ||
| pKSE401-LuU6-PDS-ref 1 | 85.57% | 14.43% | 0 | albino shoot |
| pKSE401-LuU6-PDS-ref 2 | 88.19% | 11.81% | 0 | albino shoot |
| pKSE401-LuU6-PDS-ref 3 | 89.55% | 10.45% | 0 | slightly albino shoots |
| pKSE401-LuU6-PDS-ref 4 | 87.04% | 12.96% | 0 | slightly albino shoots |
| pKSE401-LuU6-PDS-ref 5 | 86.98% | 8.52% | 4.50% | albino shoot |
| pKSE401-LuU6-PDS-ref 6 | 83.23% | 12.51% | 4.26% | albino shoot |
| pKSE401-LuU6-PDS-ref 7 | 83.57% | 16.43% | 0 | slightly albino shoots |
| pKSE401-LuU6-PDS-ref 8 | 86.44% | 13.56% | 0 | slightly albino shoots |
| pKSE401-AtU6-PDS-ref 1 | 89.28% | 10.72% | 0 | slightly albino shoots |
| pKSE401-AtU6-PDS-ref 2 | 87.70% | 12.31% | 0 | slightly albino shoots |
| pKSE401-AtU6-PDS-ref 3 | 88.25% | 11.75% | 0 | slightly albino shoots |
| pKSE401-AtU6-PDS-ref 4 | 85.95% | 14.05% | 0 | slightly albino shoots |
| pKSE401-AtU6-PDS-ref 5 | 83.56% | 11.82% | 4.62% | albino shoot |
| pKSE401-AtU6-PDS-ref 6 | 83.98% | 16.02% | 0 | slightly albino shoots |
| pKSE401-AtU6-PDS-ref 7 | 81.92% | 16.28% | 1.80% | albino shoot |
| pKSE401-AtU6-PDS-ref 8 | 85.2% | 11.43% | 3.31% | albino shoot |
| pKSE401-LuU6-PDS | |||||
|---|---|---|---|---|---|
| Sort | Reads Number | Ratio | Variation Type | Variation | Reads Seq |
| 001-ref | TGACT-PAM----------sgRNA1 target----------48 bp-PAM----------sgRNA2 target----------AAGGTT | ||||
| 1 | 1494 | 38.48% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1555 | 40.05% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 305 | 7.85% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 4 | 251 | 6.46% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 5 | 46 | 1.18% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 6 | 44 | 1.13% | SNP | T → A, T → G | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCCTGAAGGTT |
| 7 | 40 | 1.03% | SNP | C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCtTGAAGGTT |
| 002-ref | |||||
| 1 | 1645 | 42.58% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1473 | 38.13% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 262 | 6.78% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 3 | 222 | 5.75% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 5 | 54 | 1.40% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 003-ref | |||||
| 1 | 1551 | 41.22% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1501 | 39.89% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 256 | 6.80% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 205 | 5.45% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 004-ref | |||||
| 1 | 1451 | 37.52% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1558 | 40.29% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 277 | 7.17% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 4 | 201 | 5.20% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 5 | 60 | 1.55% | SNP | T → A, T → G, T → C, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgcCTAAACCCtTGAAGGTT |
| 6 | 43 | 1.11% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 005-ref | |||||
| 1 | 1483 | 39.32% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1542 | 40.88% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 361 | 9.57% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 195 | 5.17% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 103 | 2.73% | 1I,1D | G, G | TGACTACCCTAGACCT-ACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGGAaGTT |
| 6 | 88 | 2.33% | SNP | T → C | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCcTCTAAACCCCTGAAGGTT |
| 006-ref | |||||
| 1 | 1601 | 41.64% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1407 | 36.60% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 305 | 7.93% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 264 | 6.87% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 90 | 2.34% | 3D | CTG | TGACTACCCTAGAC---ACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 6 | 45 | 1.17% | SNP | A → G | TGACTACCCTAGACCTGACCTTGgCAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 007-ref | |||||
| 1 | 1140 | 29.04% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1696 | 43.21% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 409 | 10.42% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 291 | 7.41% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 131 | 3.34% | SNP | C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCtTGAAGGTT |
| 6 | 41 | 1.04% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 008-ref | |||||
| 1 | 1511 | 39.30% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1587 | 41.28% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 241 | 6.27% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 230 | 5.98% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 63 | 1.64% | SNP | C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCtTGAAGGTT |
| 6 | 40 | 1.04% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| pKSE401-AtU6-PDS | |||||
| Sort | Reads number | Ratio | variation type | variation | reads seq |
| 001-ref | TGACT-PAM----------sgRNA1 target----------48 bp-PAM----------sgRNA2 target----------AAGGTT | ||||
| 1 | 1558 | 40.64% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1482 | 38.65% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 284 | 7.41% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 224 | 5.84% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 58 | 1.51% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 6 | 41 | 1.07% | SNP | T → A, T → G | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCCTGAAGGTT |
| 002-ref | |||||
| 1 | 1549 | 40.33% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1942 | 38.84% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 296 | 7.70% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 263 | 6.85% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 47 | 1.22% | SNP | T → A, T → G | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCCTGAAGGTT |
| 6 | 40 | 1.04% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 003-ref | |||||
| 1 | 1427 | 37.75% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1558 | 41.22% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 277 | 7.33% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 258 | 6.83% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 55 | 1.46% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 6 | 49 | 1.30% | SNP | T → A, A → G, T → G, C → T | TGACTACCCaAGACCTGACCTTGgCAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 004-ref | |||||
| 1 | 1531 | 40.47% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1485 | 39.26% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 308 | 8.14% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 293 | 7.75% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 005-ref | |||||
| 1 | 1633 | 43.64% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1422 | 38.00% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 231 | 6.17% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 4 | 211 | 5.64% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 5 | 90 | 2.41% | 1D,SNP | T → A, A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCC-CGCCCgTCTAAACCCtTGAAGGTT |
| 006-ref | |||||
| 1 | 1389 | 35.63% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1449 | 37.18% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 360 | 9.23% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 3 | 352 | 9.03% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 5 | 85 | 2.18% | SNP | T → A, C → T, T → G, C → T, 1D | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCtACGCCCgTCTAAACC-tTGAAGGTT |
| 007-ref | |||||
| 1 | 1508 | 38.66% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1494 | 38.31% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 300 | 7.69% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 4 | 228 | 5.84% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 5 | 64 | 1.64% | SNP | C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCtTGAAGGTT |
| 6 | 61 | 1.56% | SNP | T → A, T → G, T → C, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgcCTAAACCCtTGAAGGTT |
| 7 | 48 | 1.23% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 8 | 40 | 1.03% | 1I,1D | G, G | TGACTACCCTAGACCT-ACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGGAaGTT |
| 008-ref | |||||
| 1 | 1580 | 41.75% | WT | - | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 2 | 1321 | 34.91% | SNP | T → A, T → G, C → T | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 3 | 349 | 9.22% | SNP | T → G, C → T | TGACTACCCTAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 4 | 235 | 6.21% | SNP | T → A | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 5 | 68 | 1.80% | 3D | CTG | TGACTACCCTAGAC---ACCTTGACAACA-48 bp-CCCCACGCCCTTCTAAACCCCTGAAGGTT |
| 6 | 43 | 1.14% | SNP | T → G, T → G, C → T | TGACTACCCgAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCtTGAAGGTT |
| 7 | 40 | 1.06% | SNP | T → A, T → G | TGACTACCCaAGACCTGACCTTGACAACA-48 bp-CCCCACGCCCgTCTAAACCCCTGAAGGTT |
Appendix B




References
- Cloutier, S.; Ragupathy, R.; Miranda, E.; Radovanovic, N.; Reimer, E.; Walichnowski, A.; Ward, K.; Rowland, G.; Duguid, S.; Banik, M. Integrated consensus genetic and physical maps of flax (linum usitatissimum L.). Theor. Appl. Genet. 2012, 125, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-B. Geographic patterns of RAPD variation in cultivated flax. Crop Sci. 2005, 45, 1084–1091. [Google Scholar] [CrossRef]
- Zuk, M.; Richter, D.; Matuła, J.; Szopa, J. Linseed, the multipurpose plant. Ind. Crops Prod. 2015, 75, 165–177. [Google Scholar] [CrossRef]
- Hall, L.M.; Booker, H.; Siloto, R.M.; Jhala, A.J.; Weselake, R.J. Flax (Linum usitatissimum L.). In Industrial Oil Crops; AOCS Press: Urbana, IL, USA, 2016; pp. 157–194. [Google Scholar]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar]
- Mishra, R.; Joshi, R.K.; Zhao, K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. J. 2020, 18, 20–31. [Google Scholar]
- Pixley, K.V.; Falck-Zepeda, J.B.; Paarlberg, R.L.; Phillips, P.W.; Slamet-Loedin, I.H.; Dhugga, K.S.; Campos, H.; Gutterson, N. Genome-edited crops for improved food security of smallholder farmers. Nat. Genet. 2022, 54, 364–367. [Google Scholar]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar]
- Zheng, N.; Li, T.; Dittman, J.D.; Su, J.; Li, R.; Gassmann, W.; Peng, D.; Whitham, S.A.; Liu, S.; Yang, B. CRISPR/Cas9-based gene editing using egg cell-specific promoters in Arabidopsis and soybean. Front. Plant Sci. 2020, 11, 800. [Google Scholar] [CrossRef]
- Riesenberg, S.; Helmbrecht, N.; Kanis, P.; Maricic, T.; Pääbo, S. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat. Commun. 2022, 13, 489. [Google Scholar]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar]
- Li, J.-F.; Zhang, D.; Sheen, J. Cas9-based genome editing in arabidopsis and tobacco. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 546, pp. 459–472. ISBN 0076-6879. [Google Scholar]
- Kumar, N.; Galli, M.; Ordon, J.; Stuttmann, J.; Kogel, K.-H.; Imani, J. Further analysis of barley MORC 1 using a highly efficient RNA-guided Cas9 gene-editing system. Plant Biotechnol. J. 2018, 16, 1892–1903. [Google Scholar] [CrossRef] [PubMed]
- Zinselmeier, M.H.; Casas-Mollano, J.A.; Cors, J.; Sychla, A.; Heinsch, S.C.; Voytas, D.F.; Smanski, M.J. Optimized dCas9 programmable transcriptional activators for plants. Plant Biotechnol. J. 2024, 22, 3202. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef] [PubMed]
- Jores, T.; Tonnies, J.; Wrightsman, T.; Buckler, E.S.; Cuperus, J.T.; Fields, S.; Queitsch, C. Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters. Nat. Plants 2021, 7, 842–855. [Google Scholar] [CrossRef]
- Basso, M.F.; Lourenço-Tessutti, I.T.; Busanello, C.; Pinto, C.E.M.; de Oliveira Freitas, E.; Ribeiro, T.P.; de Almeida Engler, J.; de Oliveira, A.C.; Morgante, C.V.; Alves-Ferreira, M. Insights obtained using different modules of the cotton uceA1. 7 promoter. Planta 2020, 251, 56. [Google Scholar] [CrossRef]
- Claeys, H.; Neyrinck, E.; Dumoulin, L.; Pharazyn, A.; Verstichele, A.; Pauwels, L.; Nuccio, M.L.; Van Ex, F. Coordinated gene upregulation in maize through CRISPR/cas-mediated enhancer insertion. Plant Biotechnol. J. 2024, 22, 16–18. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Hu, C.; Yang, Q.; Dong, T.; Sheng, O.; Deng, G.; He, W.; Dou, T.; Li, C.; et al. Increased mutation efficiency of CRISPR/Cas9 genome editing in banana by optimized construct. PeerJ 2022, 10, e12664. [Google Scholar] [CrossRef]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in arabidopsis and nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L. Targeted genome modification of crop plants using a CRISPR-cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.; Kamoun, S. Targeted mutagenesis in the model plant nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Lin, X.; Yang, J.; Chen, J.; Gunasekera, A.; Fesik, S.W.; Shen, Y. Development of a tightly regulated U6 promoter for shRNA expression. FEBS Lett. 2004, 577, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Di, Y.-H.; Sun, X.-J.; Hu, Z.; Jiang, Q.-Y.; Song, G.-H.; Zhang, B.; Zhao, S.-S.; Zhang, H. Enhancing the CRISPR/Cas9 system based on multiple GmU6 promoters in soybean. Biochem. Biophys. Res. Commun. 2019, 519, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Guo, D.-D.; Gao, W.; Yang, W.-W.; Hou, L.-P.; Ma, X.-N.; Miao, Y.-C.; Botella, J.R.; Song, C.-P. Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 2018, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Hikino, K.; Mano, S. Cloning and functional verification of endogenous U6 promoters for the establishment of efficient CRISPR/Cas9-based genome editing in castor (ricinus communis). Genes 2023, 14, 1327. [Google Scholar] [CrossRef]
- Liu, F.-H.; Chen, X.; Long, B.; Shuai, R.-Y.; Long, C.-L. Historical and botanical evidence of distribution, cultivation and utilization of Linum usitatissimum L.(flax) in China. Veg. Hist. Archaeobotany 2011, 20, 561–566. [Google Scholar]
- Baruah-Wolff, J.; Harwood, W.A.; Lonsdale, D.A.; Harvey, A.; Hull, R.; Snape, J.W. Luciferase as a reporter gene for transformation studies in rice (oryza sativa L.). Plant Cell Rep. 1999, 18, 715–720. [Google Scholar]
- Huang, Y.; Wang, Y.; Zeng, B.; Liu, Z.; Xu, X.; Meng, Q.; Huang, Y.; Yang, G.; Vasseur, L.; Gurr, G.M. Functional characterization of Pol III U6 promoters for gene knockdown and knockout in Plutella xylostella. Insect Biochem. Mol. Biol. 2017, 89, 71–78. [Google Scholar]
- Calistri, E.; Buiatti, M.; Livi, R. Variation and constraints in species-specific promoter sequences. J. Theor. Biol. 2014, 363, 357–366. [Google Scholar]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE 2015, 10, e0136064. [Google Scholar]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.; Han, X.; Yuan, G.; Zhang, L.; Tian, Y.; Zhang, C.; Cong, P. Cloning and functional analysis of U6 promoter in apple. Sci. Agric. Sin. 2019, 52, 4364–4373. [Google Scholar]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.-J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef]
- Ren, C.; Liu, Y.; Guo, Y.; Duan, W.; Fan, P.; Li, S.; Liang, Z. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic. Res. 2021, 8, 52. [Google Scholar] [CrossRef]
- Riu, Y.-S.; Kim, G.H.; Chung, K.W.; Kong, S.-G. Enhancement of the CRISPR/Cas9-based genome editing system in lettuce (lactuca sativa L.) using the endogenous U6 promoter. Plants 2023, 12, 878. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef]
- Yang, W.; Ren, J.; Liu, W.; Liu, D.; Xie, K.; Zhang, F.; Wang, P.; Guo, W.; Wu, X. An efficient transient gene expression system for protein subcellular localization assay and genome editing in citrus protoplasts. Hortic. Plant J. 2023, 9, 425–436. [Google Scholar] [CrossRef]
- Shen, M.W.; Arbab, M.; Hsu, J.Y.; Worstell, D.; Culbertson, S.J.; Krabbe, O.; Cassa, C.A.; Liu, D.R.; Gifford, D.K.; Sherwood, R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 2018, 563, 646–651. [Google Scholar] [CrossRef]
- Allen, F.; Crepaldi, L.; Alsinet, C.; Strong, A.J.; Kleshchevnikov, V.; De Angeli, P.; Palenikova, P.; Khodak, A.; Kiselev, V.; Kosicki, M.; et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2018, 37, 64–72. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [PubMed]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [PubMed]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zong, W.; Zeng, D.; Han, J.; Chen, S.; Tang, J.; Zhao, Z.; Li, X.; Ma, K.; Xie, X. Shortened snRNA promoters for efficient CRISPR/cas-based multiplex genome editing in monocot plants. Sci. China Life Sci. 2020, 63, 933–935. [Google Scholar]
- Li, X.; Jiang, D.-H.; Yong, K.; Zhang, D.-B. Varied Transcriptional Efficiencies of Multiple Arabidopsis U6 Small Nuclear RNA Genes. J. Integr. Plant Biol. 2007, 49, 222–229. [Google Scholar] [CrossRef]
- Wang, C.; Sun, C.; Shi, L.; Zhou, J.; Liu, S.; Bai, Y.; Yu, W. Establishment of a CRISPR-Cas9-mediated genome editing system in flax. CRISPR J. 2025, 8, 51–59. [Google Scholar]
- Saha, D.; Mukherjee, P.; Dutta, S.; Meena, K.; Sarkar, S.K.; Mandal, A.B.; Dasgupta, T.; Mitra, J. Genomic insights into HSFs as candidate genes for high-temperature stress adaptation and gene editing with minimal off-target effects in flax. Sci. Rep. 2019, 9, 5581. [Google Scholar]
- Aznar-Moreno, J.A.; Durrett, T.P. Simultaneous targeting of multiple gene homeologs to alter seed oil production in camelina sativa. Plant Cell Physiol. 2017, 58, 1260–1267. [Google Scholar] [CrossRef]
- Gao, Z.; Herrera-Carrillo, E.; Berkhout, B. RNA polymerase II activity of type 3 pol III promoters. Mol. Ther.-Nucleic Acids 2018, 12, 135–145. [Google Scholar] [CrossRef]
- Thorne, N.; Inglese, J.; Auld, D.S. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol. 2010, 17, 646–657. [Google Scholar] [CrossRef]
- Fuqua, T.; Sun, Y.; Wagner, A. The emergence and evolution of gene expression in genome regions replete with regulatory motifs. eLife 2024, 13, RP98654. [Google Scholar] [CrossRef] [PubMed]
- Minkenberg, B.; Wheatley, M.; Yang, Y. CRISPR/Cas9-enabled multiplex genome editing and its application. Prog. Mol. Biol. Transl. Sci. 2017, 149, 111–132. [Google Scholar] [PubMed]
- Tippens, N.D.; Vihervaara, A.; Lis, J.T. Enhancer transcription: What, where, when, and why? Genes Dev. 2018, 32, 1–3. [Google Scholar] [PubMed]
- Chen, J.; Li, S.; He, Y.; Li, J.; Xia, L. An update on precision genome editing by homology-directed repair in plants. Plant Physiol. 2022, 188, 1780–1794. [Google Scholar]
- Chen, J.; Lu, C.; Wang, G.-X.; Li, C.-G.; Li, Y.-H.; Su, F.-Y.; Wang, C.; Zhang, Y. Cloning and expression analysis of U6 promoters in panax quinquefolius. Zhongguo Zhong Yao Za Zhi 2023, 2931–2939. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar]
- Zhao, Y.; Zhu, H.; Lu, X.; Anees, M.; He, N.; Yang, D.; Chen, Z.; Hong, Z.; Zhang, J.; Liu, W. Streamlined agrobacterium rhizogenes-mediated hairy root transformation for efficient CRISPR/Cas9-based gene editing evaluation in diverse citrullus cultivars. Hortic. Plant J. 2025, 11, 816–826. [Google Scholar]
- Zhang, S.; Zhang, R.; Gao, J.; Song, G.; Li, J.; Li, W.; Qi, Y.; Li, Y.; Li, G. CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol. J. 2021, 19, 1684. [Google Scholar]
- Zhao, H.; Zhao, S.; Cao, Y.; Jiang, X.; Zhao, L.; Li, Z.; Wang, M.; Yang, R.; Zhou, C.; Wang, Z.; et al. Development of a single transcript CRISPR/Cas9 toolkit for efficient genome editing in autotetraploid alfalfa. Crop J. 2024, 12, 788–795. [Google Scholar] [CrossRef]
- Zhou, J.; He, R.; Liu, X.; Zhang, B.; Chen, G.; Wang, D.; Zhu, Y. Manipulation of plant height in garden asparagus (Asparagus officinalis L.) through CRISPR/Cas9-mediated aspSPL14 allele editing. Hortic. Res. 2023, 10, uhad096. [Google Scholar] [CrossRef]
- Alotaibi, S.S.; Sparks, C.A.; Parry, M.A.J.; Simkin, A.J.; Raines, C.A. Identification of leaf promoters for use in transgenic wheat. Plants 2018, 7, 27. [Google Scholar] [CrossRef]
- Norris, S.R.; Shen, X.; Della Penna, D. Complementation of the Arabidopsis pds1 Mutation with the Gene Encoding p-Hydroxyphenylpyruvate Dioxygenase. Plant Physiol. 1998, 117, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Krenek, P.; Samajova, O.; Luptovciak, I.; Doskocilova, A.; Komis, G.; Samaj, J. Transient plant transformation mediated by agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 2015, 33, 1024–1042. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Tian, X.; Xu, F.; Liu, J.; Singh, P.K.; Botella, J.R.; Song, C. Genome editing in cotton with the CRISPR/Cas9 system. Front. Plant Sci. 2017, 8, 1364. [Google Scholar]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar]
- Engler, C.; Youles, M.; Gruetzner, R.; Ehnert, T.-M.; Werner, S.; Jones, J.D.; Patron, N.J.; Marillonnet, S. A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 2014, 3, 839–843. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison III, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar]
- Xie, D.; Yang, X.; He, R.; Huo, H.; Ye, Z.; Ren, X.; Yuan, H.; Dai, Z.; Sun, J.; Su, J. Comprehensive analysis of the UDP-glycosyltransferase gene family in flax [Linum usitatissimum L.] and functional verification of the role of LuUGT175 in the regulation of lignin biosynthesis. Ind. Crops Prod. 2022, 188, 115720. [Google Scholar] [CrossRef]

| Gene Name | Chromosome Location | E-Value |
|---|---|---|
| Lu14U6-1 | Chr14: 10801770…10801880 | 3.00 × 10−49 |
| Lu14U6-2 | Chr14: 10810787…10810897 | 3.00 × 10−49 |
| Lu14U6-3 | Chr14: 10613237…10613349 | 1.00 × 10−48 |
| Lu14U6-4 | Chr14: 13709531…13709640 | 1.00 × 10−48 |
| Lu14U6-5 | Chr14: 13672928…13673036 | 4.00 × 10−48 |
| Lu14U6-6 | Chr14: 13685856…13685964 | 4.00 × 10−48 |
| Lu14U6-7 | Chr14: 5535449…5535484 | 6.00 × 10−6 |
| Lu13U6-1 | Chr13: 17095955…17096063 | 4.00 × 10−48 |
| Lu13U6-2 | Chr13: 14893350…14893457 | 1.00 × 10−47 |
| Lu13U6-3 | Chr13: 18154867…18154981 | 2.00 × 10−46 |
| Lu13U6-4 | Chr13: 14891387…14891491 | 6.00 × 10−46 |
| Transformation Vector | Number of Transgenic Flax Hypocotyls | Total Number of Germinated Shoots | Number of Albino Shoots | Frequency of Albino Shoot Emergence |
|---|---|---|---|---|
| pKSE401-LuU6-PDS | 80 | 45 | 26 | 32.5% |
| pKSE401-AtU6-PDS | 83 | 46 | 29 | 34.9% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Xue, M.; Guo, D.; Zhu, L.; Li, Y.; Xie, L. A Truncated Endogenous U6 Promoter Enables High-Efficiency CRISPR Editing in Flax (Linum usitatissimum L.). Plants 2025, 14, 1142. https://doi.org/10.3390/plants14071142
Li F, Xue M, Guo D, Zhu L, Li Y, Xie L. A Truncated Endogenous U6 Promoter Enables High-Efficiency CRISPR Editing in Flax (Linum usitatissimum L.). Plants. 2025; 14(7):1142. https://doi.org/10.3390/plants14071142
Chicago/Turabian StyleLi, Feifei, Min Xue, Dongliang Guo, Leilei Zhu, Yuandong Li, and Liqiong Xie. 2025. "A Truncated Endogenous U6 Promoter Enables High-Efficiency CRISPR Editing in Flax (Linum usitatissimum L.)" Plants 14, no. 7: 1142. https://doi.org/10.3390/plants14071142
APA StyleLi, F., Xue, M., Guo, D., Zhu, L., Li, Y., & Xie, L. (2025). A Truncated Endogenous U6 Promoter Enables High-Efficiency CRISPR Editing in Flax (Linum usitatissimum L.). Plants, 14(7), 1142. https://doi.org/10.3390/plants14071142






